Abstract
GES-1 is a class A extended-spectrum β-lactamase conferring resistance to penicillins, narrow- and expanded-spectrum cephalosporins, and ceftazidime. However, GES-1 poorly hydrolyzes aztreonam and cephamycins and exhibits very low kcat values for carbapenems. Twenty-two GES variants have been discovered thus far, differing from each other by 1 to 3 amino acid substitutions that affect substrate specificity. GES-11 possesses a Gly243Ala substitution which seems to confer to this variant an increased activity against aztreonam and ceftazidime. GES-12 differs from GES-11 by a single Thr237Ala substitution, while GES-14 differs from GES-11 by the Gly170Ser mutation, which is known to confer increased carbapenemase activity. GES-11 and GES-12 were kinetically characterized and compared to GES-1 and GES-14. Purified GES-11 and GES-12 showed strong activities against most tested β-lactams, with the exception of temocillin, cefoxitin, and carbapenems. Both variants showed a significantly increased rate of hydrolysis of cefotaxime, ceftazidime, and aztreonam. On the other hand, GES-11 and GES-12 (and GES-14) variants all containing Ala243 exhibited increased susceptibility to classical inhibitors. The crystallographic structures of the GES-11 and GES-14 β-lactamases were solved. The overall structures of GES-11 and GES-14 are similar to that of GES-1. The Gly243Ala substitution caused only subtle local rearrangements, notably in the typical carbapenemase disulfide bond. The active sites of GES-14 and GES-11 are very similar, with the Gly170Ser substitution leading only to the formation of additional hydrogen bonds of the Ser residue with hydrolytic water and the Glu166 residue.
INTRODUCTION
Extended-spectrum β-lactamases (ESBLs) of the GES type are increasingly reported among a large variety of Gram-negative bacterial species, including Pseudomonas aeruginosa, Enterobacter cloacae, Klebsiella pneumoniae, and Acinetobacter baumannii (20). GES-type β-lactamases form a distinct branch of class A enzymes, but their origin is unknown. The first GES β-lactamase, GES-1, was described in 2000 (24). This enzyme confers resistance to penicillins, narrow- and expanded-spectrum cephalosporins, and ceftazidime. However, unlike most ESBLs, GES-1 poorly hydrolyzes monobactams. Moreover, although it contains the disulfide bond between Cys69 and Cys238 (using the standard numbering for class A β-lactamases [1]) characteristic of class A carbapenemases, GES-1 hydrolyzes carbapenems very slowly and is even inhibited by low concentrations of imipenem (25, 28). To date, 22 GES variants differing by one to three amino acid residues have been reported (http://lahey.org/studies/other.asp), and differences in the substrate spectra have been observed. The most alarming characteristic of the GES family of enzymes that distinguishes them from other ESBLs of the TEM and SHV superfamilies is their apparent ability to evolve into carbapenemases. GES-2, which differs from GES-1 by a single Gly170Asn substitution located inside the Ω-loop of the catalytic site, hydrolyzes carbapenems, but still at a low level (25). Variants with Ser at position 170 (GES-4, GES-5, GES-6, and GES-14) are of special clinical interest, since they exhibit increased carbapenemase activity (2–4, 30, 31). On the other hand, the Gly243Ser and Gly243Ala substitutions in GES-9 and GES-11, respectively, confer increased activity against aztreonam and ceftazidime (3, 18, 26).
Recently, three GES ESBLs (GES-11, GES-12, and GES-14) were observed in A. baumannii clinical isolates in Belgium. GES-12 and GES-14 differ from GES-11 by a single amino acid substitution, Thr237Ala and Gly170Ser, respectively (3) (Table 1). This paper details the kinetic characterization of the GES-11 and GES-12 variants, compares them to the GES-1 and GES-14 enzymes (4, 24), and describes the first crystallographic structures of the GES-11 and GES-14 variants.
Table 1.
Residues in positions 80, 170, 237, and 243 in GES β-lactamases
| β-Lactamase | Residue at position: |
|||
|---|---|---|---|---|
| 80 | 170 | 237 | 243 | |
| GES-1 | Val | Gly | Thr | Gly |
| GES-5 | —a | Ser | — | — |
| GES-11 | — | — | — | Ala |
| GES-12 | — | — | Ala | Ala |
| GES-14 | — | Ser | — | Ala |
| GES-18 | Ile | Ser | — | — |
—, the residue is the same as that in GES-1.
MATERIALS AND METHODS
Cloning, expression, and purification of GES-11, GES-12, and GES-14 β-lactamases.
To overproduce GES-11, GES-12, and GES-14 β-lactamases in Escherichia coli, the corresponding blaGES genes were amplified by using primers 5′-CCATGGCACGCTTCATTCACGCACTATTACTG-3′ and 5′-GAATTCCTATTTGTCCGTGCTCAGGATGAGTTG-3′ containing NcoI and EcoRI restriction sites (underlined). The PCR products were then cloned into pET-28a (Novagen, Madison, WI) in the corresponding insertion sites. The expression vectors, named pET28/GES11, pET28/GES12, and pET28/GES14, respectively, were introduced into E. coli BL21(DE3) (New England BioLabs, Ipswich, MA). Bacteria were grown overnight at 37°C with shaking in 50 ml LB medium supplemented with 50 μg/ml kanamycin. The bacterial suspension was diluted 100-fold into a total of 2 liters of fresh LB medium supplemented with kanamycin (50 μg/ml), and the expression of the corresponding GES β-lactamase was induced with isopropyl-β-d-thiogalactopyranoside (final concentration, 1 mM) when the culture reached an A600 of 0.6. The induced culture was incubated overnight at 28°C with shaking.
The GES β-lactamases were purified by ion-exchange chromatography and gel filtration. Briefly, the bacterial suspension was pelleted, resuspended in 60 ml of buffer A (20 mM Tris-HCl, pH 7.5), disrupted by sonication, and cleared by ultracentrifugation. The supernatant was then dialyzed against buffer A and loaded onto a Q-Sepharose FF column equilibrated with the same buffer. The proteins were eluted by a linear NaCl gradient (0 to 0.5 M). Fractions were analyzed by SDS-PAGE and by monitoring the hydrolysis of nitrocefin. The fractions containing the β-lactamase activity were pooled and dialyzed overnight against buffer A at 4°C. This partially purified enzyme was loaded onto a MonoQ column (Pharmacia Biotech) equilibrated with the same buffer. The proteins were eluted by a linear gradient of NaCl (0 to 0.5 M). The fractions containing the β-lactamase activity were pooled and concentrated to 2 ml by ultrafiltration using an Amicon Ultra 10 K. Size-exclusion chromatography was performed on a Hiload 16/60 Superdex 75 Prep-grade column (Pharmacia Biotech) equilibrated with buffer A supplemented with NaCl (250 mM). Finally, fractions containing the highest β-lactamase activities were pooled and subsequently dialyzed overnight at 4°C against 100 mM sodium phosphate buffer (pH 7.0). The enzymes were more than 95% pure as judged by SDS-PAGE and mass spectrometry.
Determination of kinetic parameters.
All experiments were carried out at 30°C in 100 mM sodium phosphate buffer (pH 7.0). Imipenem was obtained from Merck Sharp and Dohme Research Laboratories (Rahway, NJ), nitrocefin was from ProGenosis (Liège, Belgium), and temocillin (Negaban) was from Eumedica Pharmaceuticals (Brussels, Belgium); all other antibiotics and inhibitors were from Sigma-Aldrich (St. Louis, MO). The hydrolysis of the antibiotics was monitored by following the change of absorbance resulting from the opening of the β-lactam ring using a Uvikon 943 spectrophotometer equipped with thermostatically controlled cells. The kcat and Km parameters were determined from initial rates of reactions, using both Hanes' linearization of the Henri-Michaelis-Menten equation and a direct nonlinear regression with the hyperbolic equation. Low and very high Km values were determined as Ki values using nitrocefin as the reporter substrate. For low Km values, the kcat values were derived from the initial hydrolysis rates measured at saturating substrate concentrations, whereas for high Km values, kcat was derived directly from the kcat/Km ratio.
Specific β-lactamase activities (nmol min−1 mg−1) were obtained using 100 μM each aztreonam, ceftazidime, cefotaxime, cefoxitin, ertapenem, meropenem, and imipenem as the substrates.
Fifty percent inhibitory concentrations (IC50s) were determined using clavulanic acid and tazobactam as inhibitors as previously described (24, 25, 31).
GES-11 and GES-14 crystallization.
Initial crystallization trials with the GES-11 (10 mg/ml) and GES-14 (8 mg/ml) enzymes were set up using Crystal Screens I and II and a PEG/Ion screen (Hampton Research) (resulting in 146 different initial conditions). Both β-lactamases were crystallized at 21°C from 0.05 M NaI and 20 to 25% (wt/vol) polyethylene glycol 3350, using the sitting-drop method with crystallization plates designed by Taorad GmbH (Aachen, Germany). A volume of 0.5 μl of 20% (vol/vol) glycerol was added to the mixture of reservoir solution (1 μl) and protein solution (1 μl) before being left to equilibrate against the solution reservoir. Typically, prismatic crystals of GES-11 and GES-14 grew within a few days to dimensions of 200 by 80 by 80 μm and belong to space group P22121 (for GES-11, a = 76.680 Å, b = 85.240 Å, and c = 85.120 Å; for GES-11, a = 70.745 Å, b = 84.098 Å, and c = 85.026 Å).
Data collection and processing.
For data collection, crystals were transferred to a cryoprotectant solution containing reservoir solution supplemented with 30% (vol/vol) glycerol. The mounted crystals were flash-frozen in a liquid nitrogen stream. Near-complete X-ray data sets were collected using a Bruker FR591 rotating anode X-ray generator and a Mar345dtb detector. Diffraction data were processed with XDS (11). The scaling was done by XDS (11) for GES-11 and by SCALA from the CCP4 suite for GES-14 (5).
Structure determination and refinement.
The structures of GES-11 and GES-14 were solved using a molecular replacement approach with Phaser (17) and the structure of GES-1 (Protein Data Bank [PDB] code 2QPN) as the starting model. The resulting models were rebuilt with ARP/wARP (23). The structures were refined in a cyclic process including manual inspection of the electron density with Coot (6) and refinement with Refmac (19). The refinement to convergence was carried out with isotropic B values and with the TLS parameter (21). The positions of the iodide ions (coming from the crystallization condition) were verified by calculating anomalous maps, and the occupancies of the ions were determined by the disappearance of differences in electron densities. Alternative conformations were modeled for a number of side chains, and occupancies were adjusted to yield similar B values for the disordered conformations.
PDB accession codes.
Coordinates and structure factors were deposited in the Protein Data Bank (PDB) under accession codes 3V3R (GES-11) and 3TSG (GES-14).
RESULTS
Production and purification.
After production of the three enzymes in E. coli BL21(DE3) and their purification to homogeneity, a yield of 20 to 30 mg of β-lactamase per liter of culture was typically obtained. The mass of the different proteins was verified by electrospray ionization-mass spectrometry (ESI-MS). Within experimental errors, the variants were found to exhibit the expected masses (29,230 versus 29,231 Da for GES-11, 29,200 versus 29,201 Da for GES-12, and 29,260 versus 29,261 Da for GES-14).
Comparison of hydrolytic activities.
We compared the activities of GES-1, GES-11, GES-12, GES-14, and GES-5 against compounds representing the four major families of β-lactam antibiotics, namely, penicillins, cephalosporins, carbapenems, and monobactams. The kinetic data and the specific activities are reported in Tables 2 and 3, respectively.
Table 2.
Kinetic parameters for GES-14, GES-12, GES-11, GES-1, and GES-5 β-lactamasesa
| Antibiotic | Kinetic parameters for: |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GES-14 |
GES-12 |
GES-11 |
GES-1 |
GES-5 |
|||||||||||
| kcat (s−1) | Km (μM) | kcat/Km (mM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (mM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (mM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (mM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (mM−1 s−1) | |
| Benzylpenicillin | 23 | 300 | 77 | 16 | 60 | 270 | 4 | 37 | 110 | 30 | 120 | 250 | 110 | 430 | 255 |
| Amoxicillin | 7 | 130 | 54 | 16 | 73 | 220 | 7 | 37 | 190 | 13 | 180 | 72 | 13 | 50 | 260 |
| Ticarcillin | 0.2 | 32 | 6 | 4.6 | 214 | 21 | 2.6 | 37 | 70 | 7 | 1,000 | 7 | 5 | 1,600 | 3 |
| Piperacillin | 60 | 750 | 80 | 96 | 300 | 320 | 27 | 120 | 225 | 8 | 900 | 9 | 160 | 1,000 | 160 |
| Temocillin | 0.02 | 400 | 0.05 | 0.16 | 1,000 | 0. 16 | 0.015 | 550 | 0.03 | 0.002 | 4,000 | 0.0005 | 0.003 | 4,500 | 0.0007 |
| Cephalothin | 1,100 | 6,000 | 180 | 80 | 5,300 | 15 | 740 | 4,000 | 185 | 900 | 5,300 | 170 | 900 | 9,000 | 100 |
| Cephaloridine | 150 | 4,400 | 34 | 75 | 640 | 120 | 455 | 3,600 | 130 | 540 | 4,600 | 120 | 725 | 5,000 | 145 |
| Nitrocefin | 56 | 135 | 415 | 72 | 215 | 330 | 320 | 530 | 600 | 380 | 340 | 1,100 | 260 | 550 | 470 |
| Cefoxitin | 0.35 | 220 | 1.6 | 0.03 | 10 | 3 | 0.02 | 13 | 1.5 | 0.08 | 60 | 1.3 | 4.5 | 860 | 5 |
| Cefotaxime | 14 | 7,900 | 1.8 | 586 | 9,800 | 60 | 1,600 | 5,500 | 290 | 370 | 9,000 | 40 | 36 | 25,000 | 1.4 |
| Ceftazidime | 2.5 | 20,000 | 0. 125 | 264 | 5,350 | 50 | 990 | 38,000 | 26 | 110 | 40,000 | 27.5 | 7 | 50,000 | 0. 14 |
| Cefepime | 0.4 | 600 | 0. 7 | 8 | 1,300 | 6 | 9 | 930 | 10 | 25 | 30,000 | 0. 8 | 8 | 90,000 | 0.09 |
| Aztreonam | 3.5 | 5,000 | 0. 7 | 25 | 740 | 34 | 53 | 3,200 | 17 | 4.8 | 4,000 | 1.2 | 0.06 | 1,800 | 0.03 |
| Imipenem | 0.09 | 2.9 | 31 | 0.02 | 1 | 20 | 0.015 | 1 | 15 | 0.007 | 1.5 | 5 | 0.7 | 4.6 | 150 |
| Ertapenem | 0.01 | 0.3 | 33 | 0.007 | 0.07 | 100 | 0.002 | 0.2 | 10 | 0.003 | 0.4 | 7.5 | 0.09 | 1.5 | 60 |
| Meropenem | 0.01 | 0.2 | 50 | 0.008 | 0.06 | 130 | 0.0035 | 0.1 | 35 | 0.0007 | 0.1 | 7 | 0.06 | 1 | 60 |
Data are the means from three independent experiments. Standard deviations were within 10% of the means. Values for GES-1 and GES-5 are from Bebrone et al. (unpublished).
Table 3.
Specific activity with 100 μM substratea
| Antibiotic | Sp act (nmol min−1 mg−1) of β-lactamase: |
||||
|---|---|---|---|---|---|
| GES-14 | GES-12 | GES-11 | GES-1 | GES-5 | |
| Imipenem | 193 | 42 | 31 | 15 | 1,330 |
| Meropenem | 22 | 17 | 7 | 1.5 | 120 |
| Ertapenem | 23 | 14 | 5 | 7 | 180 |
| Cefoxitin | 228 | 62 | 45 | 103 | 1,400 |
| Cefotaxime | 468 | 12,200 | 58,900 | 7,660 | 290 |
| Ceftazidime | 24 | 9,968 | 5,345 | 562 | 27 |
| Aztreonam | 141 | 6,220 | 3,248 | 234 | 7 |
Standard deviations were within 10% of the means. Values for GES-1 and GES-5 are from Bebrone et al. (unpublished).
GES-11 and GES-12 showed better affinity than GES-1 for all tested penicillins, as reflected by lower Km values. However, the Km values of GES-12 were somewhat higher than those of GES-11 (from 1.6- to 4.8-fold). GES-11 exhibited lower kcat values than GES-1, with the exceptions of those for piperacillin and temocillin (6-α-methoxy-ticarcillin). For these two penicillins, GES-12 also exhibited largely higher kcat values than GES-1. GES-14 exhibited higher Km values than GES-11, with the exceptions of ticarcillin and temocillin. Its kcat values were higher than or similar to those of GES-11, except for ticarcillin. Temocillin was a very poor substrate of the five enzymes, with the lowest kcat/Km ratios among all of the β-lactams tested.
As already observed for GES-1 (24), GES-11 and GES-12 showed poor imipenem hydrolysis. The three enzymes exhibited strong affinity for imipenem (Km values were ∼1.5, 1, and 1 μM for GES-1, GES-11, and GES-12, respectively) without efficient hydrolysis (kcat values were only 0.007, 0.015, and 0.02 s−1, respectively). On the other hand, for GES-14, which differs from GES-11 by the Gly170Ser substitution, the Km value was in the μM range (2.9 μM) but the kcat value (0.09 s−1) was approximately 13-fold higher than that for GES-1. GES-14, which differs from GES-5 by the presence of the Gly243Ala mutation, exhibited a kcat value for imipenem approximately 8-fold lower than that of the latter.
Although GES-1, GES-11, and GES-12 showed low kcat values for carbapenems, their kcat/Km ratios were rather large owing to the very high affinity of these enzymes for this family of β-lactams. Furthermore, for all carbapenems tested, the kcat/Km ratios of GES-12 were similar to or even higher than those of GES-14 (Table 2). However, as shown in Table 3, the carbapenem concentration (100 μM) was much higher than the Km values; thus, each variant hydrolyzed these substrates at Vmax. GES-14 exhibited higher kcat values and thus higher specific activities than GES-1 and GES-11 for the three carbapenems. On the other hand, the kcat values and specific activities of GES-12 were almost similar to those of GES-14 for meropenem and ertapenem. GES-5 exhibited the largest kcat values and thus the highest specific activities for the three carbapenems (Tables 2 and 3).
As observed for GES-1 (24), GES-11 and GES-12 showed poor activities against cefoxitin. The Km values determined as Ki values, with nitrocefin as the reporter substrate, were ∼60, 13, and 10 μM for GES-1, GES-11, and GES-12, respectively. The kcat values were only 0.08, 0.02, and 0.03 s−1, respectively. GES-14 showed higher Km and kcat values than GES-1 and the GES-11 and GES-12 variants. On the other hand, GES-14 showed lower Km and kcat values than GES-5.
GES-11 showed the highest kcat value for aztreonam (11-fold higher than GES-1) but a Km value similar to that of GES-1. GES-12 also showed a significantly increased level of hydrolysis of aztreonam compared to that of GES-1. The better efficiency of GES-12 was due to a 5.4-fold decreased Km value combined with a 5-fold increased kcat value compared to GES-1 (Table 2). At 100 μM aztreonam, the specific activities of GES-11 and GES-12 were approximately 14- and 27-fold higher than that of GES-1, respectively. GES-14 hydrolyzed aztreonam more efficiently than GES-5 (Tables 2 and 3). However, GES-14 showed a large decrease in aztreonam hydrolysis compared to GES-11 (Tables 2 and 3).
GES-11 also showed the highest kcat value for ceftazidime (9-fold increase compared to GES-1), with a Km value similar to that of GES-1. GES-12 showed a significantly increased level of hydrolysis of ceftazidime. The better efficiency of GES-12 was due to a 7.5-fold decreased Km value combined with a 2.4-fold increased kcat value compared to GES-1 (Table 2). At 100 μM ceftazidime, the specific activities of GES-11 and GES-12 were approximately 9.5- and 18-fold higher than that of GES-1, respectively (Table 3). A similar observation was made for cefotaxime (Tables 2 and 3). As observed for aztreonam, the presence of the Gly170Ser substitution in GES-14 was detrimental to the hydrolysis of ceftazidime and cefotaxime. At 100 μM substrate, the specific activities of GES-14 were 223- and 125-fold lower than those of GES-11 for ceftazidime and cefotaxime, respectively (Table 3). All three GES-11, GES-12, and GES-14 variants showed largely better affinities but lower kcat values for cefepime than GES-1 (Table 2).
Comparison of susceptibilities to inhibitors.
We tested two commercial β-lactamase inhibitors, and we found that tazobactam was the most active against all five GES variants. Lower IC50s for both inhibitors were found with GES-11 and GES-12, which contain the Gly243Ala substitution, with GES-12 being the most susceptible. In GES-14, the higher level of resistance to inhibitors conferred by the Gly170Ser mutation (8) already observed in GES-5 (13, 31) disappeared because of the presence of the single Gly243Ala mutation (Table 4).
Table 4.
IC50s of the commercial inhibitors clavulanic acid and tazobactam for GES-14, GES-12, GES-11, GES-1, and GES-5a
| Antibiotic | IC50 (μM) |
||||
|---|---|---|---|---|---|
| GES-14 | GES-12 | GES-11 | GES-1 | GES-5 | |
| Clavulanic acid | 7 | 0.25 | 1 | 5 | 20 |
| Tazobactam | 4 | 0.35 | 0.5 | 3 | 10 |
Data are the means from three independent experiments. Standard deviations were within 10% of the means. Values for GES-1 and GES-5 are from Bebrone et al. (unpublished).
Crystallographic structures.
The crystal structures of the GES-11 and GES-14 variants were solved by molecular replacement using the GES-1 structure (PDB accession number 2QPN) as a model. After refinement, almost all residues lay in the allowed or favored region in the Ramachandran plot. Stereochemical parameters were calculated by PROCHECK (15) and were in the range expected for structures with similar resolutions. The crystallographic and model statistics for GES-11 and GES-14 structures are summarized in Table 5. Attempts to generate good-quality GES-12 crystals have been unsuccessful thus far.
Table 5.
X-ray data collection and structure refinement for GES-11 and GES-14a
| Parameter | Value for: |
|
|---|---|---|
| GES-11 | GES-14 | |
| Data collection statistics | ||
| Unit cell dimensions (Å) | ||
| a | 70.7 | 70.7 |
| b | 85.2 | 84.1 |
| c | 85.1 | 85.0 |
| Space group | P22121 | P22121 |
| No. of molecules/asymmetric unit | 2 | 2 |
| Resolution range (Å) | 1.88–20 | 1.87–20 |
| No. of unique reflections | 40,335 | 40,567 |
| Overall completeness (%) | 94.1/87.2 | 99.9/97.2 |
| I/σ(I) | 8.98/2.35 | 11.6/3.6 |
| Multiplicity | 4.0/3.3 | 7.1/7.0 |
| Rsymb | 0.114/0.523 | 0.124/0.608 |
| Twin fraction | 0.07 | |
| Refinement statistics | ||
| Resolution range (Å) | 1.9–20 | 1.9–20 |
| No. of reflections | 37,055 | 38,560 |
| No. of atoms (non-H) | 4,453 | 4,461 |
| Rworkc | 0.19 | 0.16 |
| Rfreed | 0.23 | 0.20 |
| RMSD from ideal | ||
| Bond length (Å) | 0.010 | 0.009 |
| Bond angle (°) | 1.393 | 1.220 |
I is the intensity of a reflection, and (I) is the average intensity. For values separated by a slash, the first number corresponds to the average value for shells of all resolutions while the second number corresponds to the value for the highest-resolution shell.
Rsym = Σ|Ij − <I>|/Σ<I>, where Ij is the intensity for reflection j and <I> is the mean intensity.
Rwork = Σ‖Fo| − |Fc‖/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
Rfree was similarly calculated, with approximately 10% of the data excluded from the calculation of Rwork.
GES-11.
The crystal structure of GES-11 was refined in a resolution range of 1.9 to 20 Å, yielding an Rwork of 0.1939 and Rfree of 0.2317 (Table 5). The crystals adopted a P22121 space group, with two molecules of GES-11 in the asymmetric unit. The final crystal structure contains 265 residues for molecule A (Ser24-Ala291) and 264 residues for molecule B (Ser24-Leu290). Furthermore, the structure contains 264 water molecules, 23 iodide ions, and 1 sodium ion from the crystallization buffer. Both molecules in the asymmetric unit are related by a 2-fold noncrystallographic symmetry. The interface (935 Å2) is dominated by hydrogen bonds and bridging water molecules. Superimposition of the individually refined models showed that they are essentially identical in structure, with a root mean square deviation (RMSD) value for the Cα position of 0.283 Å.
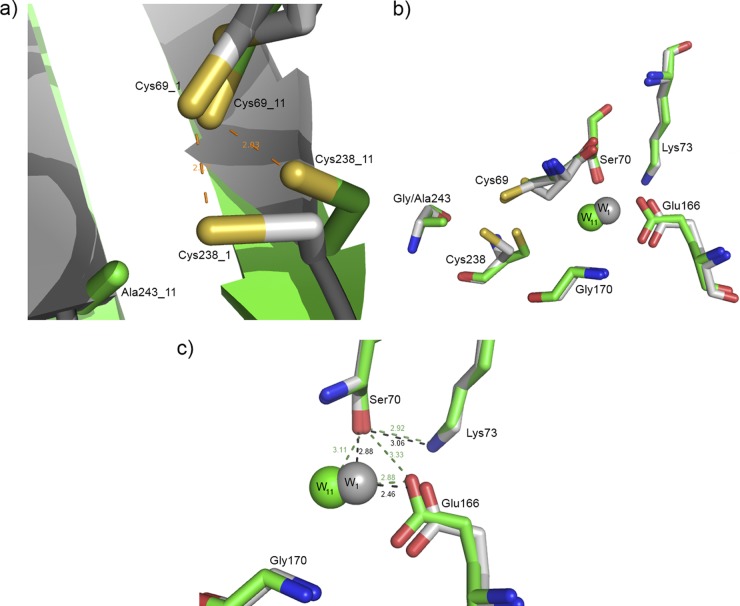
The Gly243Ala substitution has no effect on the overall structure of the GES β-lactamase. GES-11, like GES-1, shows the typical class A serine β-lactamase fold composed of two domains, one α/β and the other all α (Fig. 1). The methyl side chain (Cβ) of the Ala243 residue is oriented in the direction of the disulfide bond (Cys69-Cys238). This disulfide, which is characteristic of the carbapenemases, links the N terminus of helix H2 with the C terminus of β-strand S3. In GES-11, the Cys238 residue adopts a different conformation from that of GES-1 (SG is shifted from 1.7 Å) (Fig. 2a). The hydrolytic water molecule and the Glu166 general base are also slightly affected (Fig. 2b) (the water molecule is shifted 0.96 Å from its position in GES-1, while the carboxylate group of Glu166 is displaced by 0.6 Å). The distance between the hydroxyl group of the Ser70 side chain (OG) and the oxygen (OE2) of the carboxyl group of Glu166 is now 3.33 Å, while it is 3.96 Å in GES-1. Due to the displacement of the hydrolytic water molecule, the distances between the latter and Ser70 and the latter and Glu166 are slightly larger in GES-11 than in GES-1 (Fig. 2c).
Fig 1.

Superposition of the overall crystallographic structures of GES-1 (in gray), GES-11 (in green), and GES-14 (in orange).
Fig 2.
(a) Conformation of the Cys238 residue is slightly modified in GES-11 (Cys238_11) compared to GES-1 (Cys238_1). (b) Superposition of the active sites of GES-11 (in green) and GES-1 (in gray). The hydrolytic water molecule and the Glu166 general base are slightly displaced. (c) The distances between Ser70 and the hydrolytic water molecule and between Glu166 and the hydrolytic water molecule are slightly larger in GES-11 than in GES-1. In GES-11, Glu166 forms an additional hydrogen bond with Ser70.
GES-14.
The crystal structure of GES-14 was refined in a resolution range of 1.9 to 20 Å, yielding an Rwork of 0.163 and Rfree of 0.205 (Table 5). The crystals adopted a P22121 space group, with two molecules of GES-14 in the asymmetric unit. The final crystal structure contains 265 residues for molecule A and molecule B (Ser24-Ser291). Furthermore, the structure contains 281 water molecules. Additionally, we observed 55 iodide ions from the crystallization buffer and 11 glycerol molecules from the cryoprotecting solution. Both molecules in the asymmetric unit are related by a 2-fold noncrystallographic symmetry. The interface (943 Å2) is dominated by hydrogen bonds and by bridging water molecules. Superimposition of the individually refined models showed that they are essentially identical in structure, with an root mean square deviation (RMSD) value for the Cα position of 0.277 Å.
GES-14 and GES-11 structures show the same dimer formation and are mostly identical (the RMSD value is 0.337 Å for the superposition of the respective dimers). They are also identical to the dimer formation in the other GES structures, GES-1 (27), GES-2 (7), and GES-18 (PDB code 3V3S).
Compared to GES-1 and GES-11, the side chain of the Ser170 residue forms an additional hydrogen bond with the hydrolytic water. A new hydrogen bond is also formed by the side chain oxygen of Ser170 to the carboxyl group of Glu166 (3.02 Å) (Fig. 3a). This was already observed for GES-18 (PDB code 3V3S) (C. Bebrone, P. Bogaerts, H. Delbrück, S. Bennink, M. B. Kupper, R. Rezende de Castro, Y. Glupczynski, and K. M. Hoffmann, unpublished data). If one excepts these two new hydrogen bonds due to the Gly170Ser substitution, the active site of GES-14 does not differ from that of GES-11. The hydrolytic water molecule occupies similar positions in the two active sites (Fig. 3b). On the other hand, the hydrolytic water molecule of GES-14 (Gly170Ser-Gly243Ala) is shifted by 1.5 Å from its counterpart position in the GES-18 (Val80Ile-Gly170Ser) variant and is closer to that of GES-1. The Cys238 residue also adopts a different conformation from that of GES-18 (SG is shifted from 2.3 Å) (Fig. 3c).
Fig 3.
(a) Active site of GES-14. (b) Superimposition of the active sites of GES-14 (in orange) and GES-11 (in green). (c) Superimposition of the active sites of GES-14 (in orange) and GES-18 (in cyan).
DISCUSSION
The kinetic parameters measured for both GES-11 and GES-12 variants showed that they exhibited strong activity against most β-lactams tested, with the exception of temocillin, cefoxitin, and carbapenems. Furthermore, temocillin was the poorest substrate for all of the GES variants tested.
The Gly243 residue present in GES-1 is not conserved in other class A β-lactamases, where it is mainly a threonine (e.g., in NMCA, SME-1, KPC-2, or BLIC or in the β-lactamase from Proteus vulgaris K1) or a serine (e.g., in TEM-1). GES-9, with a Gly243Ser substitution, hydrolyzes aztreonam and ceftazidime despite high Km values (26). As already suggested by measurements of crude extracts (3), detailed kinetic measurements with the purified enzyme demonstrate that GES-11, bearing a Gly243Ala mutation (located on the β-strand B4), displays a higher hydrolytic activity than GES-1 against oxyimino-β-lactams such as cefotaxime, ceftazidime, and aztreonam. For these three substrates, the better efficiency of GES-11 is mainly due to higher turnover rates. The Km values remain very high. On the other hand, GES-11 presents better affinities for penicillins than GES-1. As observed for GES-9 (26), the substitution at position 243 does not allow GES-11 to efficiently hydrolyze cefoxitin or carbapenems. Also, the Gly243Ala substitution increases the sensitivity of the enzyme to β-lactam inhibitors such as clavulanic acid and tazobactam.
From a structural point of view, the Gly243Ala mutation has no effect on the global structure of the enzyme. This position is not directly in contact with the catalytic active residues but is situated behind the typical Cys69-Cys238 disulfide bond. The conformation of the Cys238 residue seems affected by the substitution. However, a superposition of all of the known structures of GES variants and molecules from the asymmetric unit showed a large variety of different conformations for this residue.
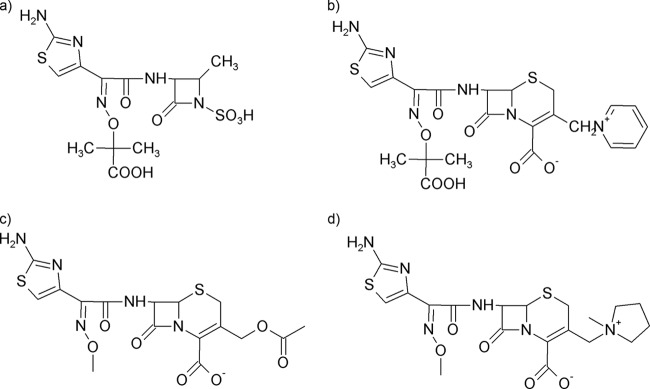
GES-12 differs from GES-11 by a single Thr237Ala substitution. The Thr237 in GES-type enzymes is atypical, with this residue commonly being alanine in class A β-lactamases. A serine residue at position 237 is conserved in the CTX-M group and also in other ESBLs (27). The side chain of residue 237 is on the outer, exposed side of the B3 β-strand that forms the right edge of the binding site. It is already known that residue 237 plays a role in substrate selectivity in class A β-lactamases (10, 12). In the TEM, KOXY-2, and SME-1 enzymes, the replacement of the typical Ala237 residue with a hydrogen bond acceptor such as threonine enhances the catalytic efficiency against cefotaxime (12, 16, 32). Accordingly, KPC-2, CTX-M-4, the β-lactamase from P. vulgaris K1, and Toho-1 show reduced catalytic efficiencies against oxyiminocephalosporins when Ser/Thr237 is replaced by Ala (9, 22, 27, 29). On the basis of the crystallographic observation of hydrogen bonding of cefotaxime to the side chain OH group of the Thr237 residue in the related d-Ala-d-Ala peptidase (14), it is likely that the ESBL variants exhibiting the Ala237Ser mutation utilize the side chain OH group of Ser237 rather than the backbone CO group for hydrogen bonding to rigid expanded-spectrum cephalosporins (12, 29). Here, the introduction of the Thr237Ala mutation in GES-11, yielding GES-12, led to a 2-fold better efficiency against aztreonam and ceftazidime, even if the kcat values were decreased, due to a strong positive effect on the affinities of the enzyme for both antibiotics. On the other hand, GES-11 was the most potent enzyme against cefotaxime and cefepime. This different effect could be explained by the fact that these two antibiotics contain a less complex imino side chain (OCH3) than ceftazidime and aztreonam (containing two methyl groups and a carboxylic acid group) (Fig. 4). Also, as already observed in Toho-1 (27) and CTX-M-4 (9), the Ser237Ala mutation resulted in increased Km and kcat values for penicillins. For meropenem and ertapenem, the kcat values of GES-12 were somewhat similar to those of GES-14.
Fig 4.
Some of the antibiotics used in this study. (a) Aztreonam; (b) ceftazidime; (c) cefotaxime; and (d) cefepime.
GES-14, bearing both Gly243Ala and Gly170Ser mutations, was compared to GES-11 (Gly243Ala) and to GES-5 (Gly170Ser) to evaluate the individual effect of each mutation on the activity. The presence of the Gly243Ala mutation in GES-14 decreased the catalytic efficiency against imipenem, other tested carbapenems, and cefoxitin compared to that of GES-5, while the efficiency against ceftazidime, aztreonam, or cefotaxime was similar to or increased compared to that of GES-5 (Tables 2 and 3). On the other hand, compared to GES-11, due to the presence of the Gly170Ser mutation, GES-14 showed an increased hydrolytic activity against imipenem, other tested carbapenems, and cefoxitin together with a decrease in activity toward ceftazidime, aztreonam, and cefotaxime. Thanks to the presence of these two mutations and their opposite effects, GES-14 displays a broad-spectrum activity against most β-lactams, including, in particular, oxyiminocephalosporins, monobactams, carbapenems, and cefoxitin. Cefoxitin usually is not hydrolyzed by class A ESBLs. As already observed for GES-18 (PDB code 3V3S; Bebrone et al., unpublished), the change from Gly to Ser in position 170 was connected to some subtle local rearrangements in the active site. Ser170 is now forming a hydrogen bridge with Glu166, and the hydrolytic water molecule presents an additional H-bond with Ser170.
It is interesting that both Gly170Ser and Gly243Ala mutations have effects (decreased or increased hydrolysis, respectively) that are identical for the aztreonam monobactam and the ceftazidime third-generation cephalosporin. This phenomenon could be attributed to the fact that these two antibiotics are similar, having the same (C3/C7) side chain, even if the latter is attached to a different nucleus.
All variants showed high Km values for cephalosporins and even very high Km values for cefepime and ceftazidime, maybe due to their similar C3 side chains (Fig. 4).
Unfortunately, the solved structures of the GES-11 and GES-14 apoenzymes do not explain the differences in substrate specificity or in sensitivity to inhibitors induced by the Gly243Ala substitution, and these challenges should be addressed by solving the crystal structures of enzyme complexes with oxyiminocephalosporin and carbapenem substrates and clavulanic acid or tazobactam inhibitors.
Conclusion.
In extended-spectrum GES β-lactamases, the replacement of the Gly243 residue by an alanine leads to an approximately 10-fold increase of enzyme efficiency toward aztreonam and ceftazidime. On the other hand, this substitution has a detrimental effect on the resistance to inhibitors. It has a negligible or modest effect on the rate of imipenem hydrolysis. This effect can become more significant for ertapenem and meropenem in the presence of an additional Thr237Ala (GES-12) or Gly170Ser (GES-14) substitution.
ACKNOWLEDGMENTS
Grateful thanks to Jean-Marie Frère (CIP, ULg, Belgium) for helpful discussions and for reviewing the manuscript.
The work in Aachen (Germany) was supported by the European Regional Development Fund (ERDF), the European Union (“Die Europäische Kommission investiert in Ihre Zukunft”), and the Alma in Silico project, financed by the Interreg IV Program. C. Bebrone was a fellow of the Alexander von Humboldt Foundation (Bonn, Germany). The work in Belgium was supported by EU grant FP7-HEALTH-2009-SINGLE-STAGE TEMPOtest-QC, project 241742, and the INAMI-RIZIV/WIV-ISP funded national reference center for antibiotic-resistant Pseudomonas and Acinetobacter.
Footnotes
Published ahead of print 20 August 2012
REFERENCES
- 1. Ambler RP, et al. 1991. A standard numbering scheme for the class A beta-lactamases. Biochem. J. 276:269–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bae IK, et al. 2007. Genetic and biochemical characterization of GES-5, an extended-spectrum class A beta-lactamase from Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 58:465–468 [DOI] [PubMed] [Google Scholar]
- 3. Bogaerts P, et al. 2010. GES extended-spectrum β-lactamases in Acinetobacter baumannii isolates in Belgium. Antimicrob. Agents Chemother. 54:4872–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bonnin RA, et al. 2011. Carbapenem-hydrolyzing GES-type extended spectrum β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55:349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collaborative Computational Project Number 4 1994. CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 6. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 7. Frase H, et al. 2011. Identification of products of inhibition of GES-2 beta-lactamase by tazobactam by X-ray crystallography and spectrometry. J. Biol. Chem. 286:14396–14409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frase H, Toth M, Champion MM, Antunes NT, Vakulenko SB. 2011. Importance of position 170 in the inhibition of GES-type β-lactamases by clavulanic acid. Antimicrob. Agents Chemother. 55:1556–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gazouli M, Tzelepi E, Sidorenko SV, Tzouvelekis LS. 1998. Sequence of the gene encoding a plasmid-mediated cefotaxime-hydrolyzing class A beta-lactamase (CTX-M-4): involvement of serine 237 in cephalosporin hydrolysis. Antimicrob. Agents Chemother. 42:1259–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Healey WJ, Labgold MR, Richards JH. 1989. Substrate specificities in class A beta-lactamases: preference for penams versus cephems. The role of residue 237. Proteins 6:275–283 [DOI] [PubMed] [Google Scholar]
- 11. Kabsch W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795–800 [Google Scholar]
- 12. Knox JR. 1995. Extended-spectrum and inhibitor-resistant TEM-type beta-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob. Agents Chemother. 39:2593–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kotsakis SD, Miriagou V, Tzelepi E, Tzouvelekis LS. 2010. Comparative biochemical and computational study of the role of naturally occurring mutations at Ambler positions 104 and 170 in GES β-lactamases. Antimicrob. Agents Chemother. 54:4864–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuzin AP, Liu H, Kelly JA, Knox JR. 1995. Binding of cephalothin and cefotaxime to d-ala-d-ala-peptidase reveals a functional basis of a natural mutation in a low-affinity penicillin-binding protein and in extended-spectrum beta-lactamases. Biochemistry 34:9532–9540 [DOI] [PubMed] [Google Scholar]
- 15. Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 16. Majiduddin FK, Palzkill T. 2005. Amino acid residues that contribute to substrate specificity of class A beta-lactamase SME-1. Antimicrob. Agents Chemother. 49:3421–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCoy AJ, et al. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moubareck C, Brémont S, Conroy MC, Courvalin P, Lambert T. 2009. GES-11, a novel integron-associated GES variant in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:3579–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 20. Naas T, Poirel L, Nordmann P. 2008. Minor extended-spectrum β-lactamases. Clin. Microbiol. Infect. 14:42–52 [DOI] [PubMed] [Google Scholar]
- 21. Painter J, Merritt EA. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62:439–450 [DOI] [PubMed] [Google Scholar]
- 22. Papp-Wallace KM, et al. 2010. Substrate selectivity and a novel role in inhibitor discrimination by residue 237 in the KPC-2 beta-lactamase. Antimicrob. Agents Chemother. 54:2867–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perrakis A, Morris R, Lamzin VS. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458–463 [DOI] [PubMed] [Google Scholar]
- 24. Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum beta-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Poirel L, et al. 2001. GES-2, a class A beta-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob. Agents Chemother. 45:2598–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poirel L, Brinas L, Fortineau N, Nordmann P. 2005. Integron-encoded GES-type extended-spectrum beta-lactamase with increased activity toward aztreonam in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:3593–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shimizu-Ibuka A, et al. 2011. Roles of residues Cys69, Asn104, Phe160, Gly232, Ser237, and Asp240 in extended-spectrum beta-lactamase Toho-1. Antimicrob. Agents Chemother. 55:284–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith CA, Caccamo M, Kantardjieff KA, Vakulenko S. 2007. Structure of GES-1 at atomic resolution: insights into the evolution of carbapenemase activity in the class A extended-spectrum β-lactamases. Acta Crystallogr. D Biol. Crystallogr. 63:982–992 [DOI] [PubMed] [Google Scholar]
- 29. Tamaki M, Nukaga M, Sawai T. 1994. Replacement of serine 237 in class A beta-lactamase of Proteus vulgaris modifies its unique substrate specificity. Biochemistry 33:10200–10206 [DOI] [PubMed] [Google Scholar]
- 30. Vourli S, et al. 2004. Novel GES/IBC extended-spectrum beta-lactamase variants with carbapenemase activity in clinical enterobacteria. FEMS Microbiol. Lett. 234:209–213 [DOI] [PubMed] [Google Scholar]
- 31. Wachino J, et al. 2004. Molecular characterization of a cephamycin-hydrolyzing and inhibitor-resistant class A beta-lactamase, GES-4, possessing a single G170S substitution in the omega-loop. Antimicrob. Agents Chemother. 48:2905–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Younes A, Hamouda A, Amyes SG. 2011. First report of a novel extended-spectrum beta-lactamase KOXY-2 producing Klebsiella oxytoca that hydrolyses cefotaxime and ceftazidime. J. Chemother. 23:127–130 [DOI] [PubMed] [Google Scholar]