Abstract
Pseudomonas aeruginosa isolates from cystic fibrosis (CF) patients undergo remarkable phenotypic divergence over time, including loss of pigmentation, hemolysis, motility, and quorum sensing and emergence of antibiotic hypersusceptibility and/or auxotrophism. With prolonged antibiotic treatment and steady decline in lung function in chronically infected patients, the divergent characteristics associated with CF isolates have traditionally been regarded as “adapted/unusual virulence,” despite the degenerative nature of these adaptations. We examined the phenotypic and genotypic diversity in clonally related isogenic strains of P. aeruginosa from individual CF patients. Our observations support a novel model of intra-airway pseudomonal syntrophy and accompanying loss of virulence. A 2007 calendar year collection of CF P. aeruginosa isolates (n = 525) from 103 CF patients yielded in vitro MICs of sulfamethoxazole-trimethoprim (SMX-TMP, which typically has no activity against P. aeruginosa) ranging from 0.02 to >32 μg/ml (median, 1.5). Coisolation of clonally related SMX-TMP-susceptible and -resistant P. aeruginosa strains from the same host was common (57%), as were isogenic coisolates with mutations in efflux gene determinants (mexR, mexAB-oprM, and mexZ) and genes governing DNA mismatch repair (mutL and mutS). In this cohort, complete in vitro growth complementation between auxotrophic and prototrophic P. aeruginosa isogenic strains was evident and concurrent with the coding sequence mosaicism in resistance determinants. These observations suggest that syntrophic clonal strains evolve in situ in an organized colonial structure. We propose that P. aeruginosa adopts a multicellular lifestyle in CF patients due to host selection of an energetically favorable, less-virulent microbe restricted within and symbiotic with the airway over the host's lifetime.
INTRODUCTION
Pseudomonas aeruginosa is found in many natural and domestic environments, including plants, soil, and surface water, and it is particularly prevalent in environments that are moist and contain organic material such as human or animal waste (17, 37). This opportunistic pathogen infects individuals deficient in host immunity or anatomic barriers, such as patients undergoing chemotherapy or those with skin damage due to burns (27). In patients with chronic lung diseases like cystic fibrosis (CF), P. aeruginosa causes airway infections that persist for months or years (7, 44). Strains from newly infected CF patients are polyclonal, while those from chronically infected individual CF patients are predominately monoclonal and unique to each patient, despite considerable intraspecific diversity (6, 10, 44, 49).
P. aeruginosa strains of environmental origin and those from acute human clinical infections are resistant to many classes of antimicrobial agents, including almost uniform resistance to sulfamethoxazole-trimethoprim (SMX-TMP) (14). In fact, SMX-TMP has been excluded from the Clinical and Laboratory Standards Institute (CLSI) P. aeruginosa antibiotic testing tables (see the M100 series) since 2007. However, hypersusceptibility to SMX-TMP is a notable feature of many P. aeruginosa isolates from CF patients, and SMX-TMP was once suggested as an oral treatment option (39, 42).
SMX-TMP is a combination of two antimicrobial agents that act synergistically against a wide variety of bacteria. SMX and TMP inhibit enzymes sequentially involved in the bacterial synthesis of tetrahydrofolic acid (THF). Reduced availability of THF inhibits de novo bacterial synthesis of methionine from l-homocysteine and thymidine from deoxyuridine and consequently impairs DNA synthesis and other cellular functions (13). Pseudomonal resistance to SMX-TMP in CF patients is due largely to drug efflux and to a lesser extent to the acquisition of integron cassettes carrying dfr and sul determinants (50). Köhler et al. have shown that an efflux complex encoded by mexAB-oprM is primarily responsible for SMX-TMP resistance in P. aeruginosa (22). The expression of mexAB-oprM is regulated by its own upstream mexR and other cross regulators, including mexZ, which is intimately involved in the regulation of another polyspecific efflux pump, MexXY-OprM (25, 33).
Microbial adaptation within a highly organized multicellular biofilm that lines the airways of patients with CF has been demonstrated by comparative genomic analysis, and this adaptation is reflected by marked differences in gene expression patterns within an organized colonial structure (8, 10, 21, 44, 45). Chronic infection of the CF airway by P. aeruginosa leads to evolution of adapted strains with extensive phenotypic changes that include the emergence of divergent mucoid colony morphology, loss of motility and green pigmentation, quorum-sensing mutations, and amino acid biosynthetic deficiencies (auxotrophism) (3, 10, 19, 20, 46). Auxotrophism, in particular, is often observed in strains isolated from chronically infected patients, and these auxotrophic strains are often found to coexist with their isogenic prototrophs (organisms that are able to grow in medium with glucose as the sole nutrient) (3).
Recent genomic studies have confirmed that P. aeruginosa isolates from CF individuals with advanced pulmonary infections show high degrees of genomic clonality that are unique to each patient but also show marked microdiversity at the level of genomic coding integrity (7, 10, 44). In the airways of CF patients, hypermutator P. aeruginosa strains with “relaxed” DNA mismatch repair systems associated with mutations in mutL and mutS have been found at very high frequencies (7, 10, 44). CF P. aeruginosa isolates in a hypermutable state appear to depart from their single-cell replicative mode of living and show a retarded growth rate and reduced virulence, as measured both by in vitro and in vivo characteristics (7, 21). Nonsynonymous and nonsense mutations arise in genes involved in DNA repair, metabolic activities, virulence, and efflux pumps, including mexAB (10, 44). One pattern in particular, intraspecific mutations (mutations in P. aeruginosa strains derived from a single founder clone) in both regulator genes of mexR and/or mexZ and the efflux structural genes mexAB, can be detected in simultaneous coisolates from the same CF patient that show antibiotic hypersusceptibility (10, 49, 51).
The emergence of degenerative characteristics, such as loss of pigmentation, hemolysis, motility, prototrophism, and quorum sensing, which are associated with CF P. aeruginosa has long been theorized to represent host airway adaptation and intraspecific gain of unusual adapted virulence (30, 45, 46). In contrast, the frequent development of antibiotic-susceptible and/or auxotrophic isogenic strains from a unique resistant prototrophic founder strain in chronically infected CF patients who undergo prolonged antibiotic treatment and exhibit steady decline in lung function seems paradoxical. In this study, coisolation of susceptible and resistant isogenic strains which show nutritional complementation suggests intra-airway evolution of a syntrophic lifestyle exhibiting reduced virulence. The resulting pseudomonal syntrophy is selected for by the enormous pressure imposed by host defense and antimicrobial treatment.
MATERIALS AND METHODS
Bacterial strains and culture standards.
Over 100 CF patients from birth to 21 years of age are cared for by the Seattle Children's Hospital CF pulmonary service annually. This study was approved by Seattle Children's Hospital Institutional Review Board as “nonhuman subject research.” A total of 525 P. aeruginosa strains isolated from CF clinical airway secretion cultures associated with 103 patients in 2007 were evaluated anonymously. Based on the Cystic Fibrosis Foundation Clinical Practice Guidelines, routine quarterly monitoring (4 times a year) includes laboratory evaluation for assessing pulmonary status, disease progression, and response to therapy. Microbiologic studies include quantitative culture of respiratory secretions and antimicrobial susceptibility testing of pathogenic isolates, such as Staphylococcus aureus, Streptococcus pneumoniae, Haemophilus influenzae, P. aeruginosa, Stenotrophomonas maltophilia, Burkholderia cepacia, and Achromobacter spp. (5). Isolates of P. aeruginosa are maximally separated with continued subculturing for visually homogeneous growth of a single colony type. Plated on various selective and nonselective agar media, mixtures of organisms are maximally separated and isolated based on colony morphology (metallic sheen, roughness, colony size, and texture consistency), hemolysis, soluble or nonsoluble pigment production, and key biochemical reactivity patterns (5, 35). Molecular methods using real-time PCR for species identification, as well as 16S rRNA partial sequencing, are used for effective and accurate resolution of bacterial isolates to the species level (5, 35; X. Qin, unpublished methods). All 525 P. aeruginosa isolates included in this study were able to grow on standard laboratory agar media, such as sheep blood agar, chocolate agar, and MacConkey agar (Remel, United States). P. aeruginosa ATCC 27853, PA-mucQC (an established laboratory mucoid CF P. aeruginosa isolate), and Escherichia coli ATCC 25922 were used as controls for MIC or disc diffusion reference ranges on Mueller-Hinton (MH) agar. UCBPP-PA14 (PA14) and its transposon insertion mutants PA14 (mexR), PA14 (mexA), PA14 (oprM), and PA14 (mexZ) were also included as P. aeruginosa control strains. PA14 is a reference clinical isolate that originated from a burn patient; its fully sequenced genome was completed in 2006, and its transposon library has also been made available (http://ausubellab.mgh.harvard.edu/cgi-bin/pa14/home.cgi) (24).
In vitro susceptibility testing.
All CF P. aeruginosa isolates were tested initially with a battery of antipseudomonal agents that included aztreonam, ceftazidime, meropenem, imipenem, ticarcillin-clavulanate, amikacin, tobramycin, ciprofloxacin, and SMX-TMP. All agents were tested by Etest MIC methodology (bioMérieux, France), except for tobramycin, which was tested by disc diffusion due to the lack of approved commercial MIC methodology for measuring this agent in high concentration ranges prior to April 2011. The MICs of sulfisoxazole (SX) (investigational use only; bioMérieux) and tobramycin (TM) were tested by the Etest strips against a subset of isolates for this study (see Table 3). MH agar plates (Remel, United States) were prepared by inoculating 0.5 McFarland saline suspensions of each isolate before the application of Etest strips or antibiotic-containing filter discs (Remel). The test plates were incubated at 35°C in ambient air for 18 to 24 h for the determination of MIC or zone-of-inhibition results. According to the Clinical and Laboratory Standards Institute (CLSI) document M100-S16 (7a), isolates with SMX-TMP MICs of ≤2 μg/ml were considered susceptible and those with MICs of ≥4 μg/ml were considered resistant.
Table 3.
Antimicrobial susceptibilities of the P. aeruginosa control isolates and 27 patient isolates measured by Etest
| Isolate | MIC (μg/ml)a |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SMX-TMP | SX | TLc | AT | TZ | MP | IP | AK | TM | CI | |
| PA-ATCC 27853 | >32 | >1,024 | 16 | 16 | 1.5 | 0.25 | 2 | 3 | 0.5 | 0.19 |
| PAmucQC | >32 | >1,024 | ||||||||
| PA14 | 16 | 128 | 16 | 3 | 1 | 0.125 | 1 | 2 | 0.38 | 0.094 |
| PA14 (mexR) | >32 | >1,024 | 48 | 16 | 3 | 1.5 | 1.5 | 3 | 0.5 | 0.38 |
| PA14 (mexA) | 0.190 | 6 | 0.38 | 0.19 | 0.5 | 0.047 | 0.75 | 1.5 | 0.38 | 0.032 |
| PA14 (oprM) | 0.250 | 6 | 0.5 | 0.25 | 0.75 | 0.047 | 1 | 1.5 | 0.38 | 0.016 |
| PA14 (mexZ) | >32 | >1,024 | ||||||||
| PA-A1 | 0.75 | 16 | >256 | 16 | >256 | 0.5 | 1.5 | >256 | 64 | 0.5 |
| PA-A2 | 0.25 | 3 | 1.5 | 0.25 | 0.75 | 0.094 | 1.5 | 16 | 2 | 0.38 |
| PA-A3 | 1 | 48 | 0.38 | 0.19 | 0.75 | 0.032 | 0.5 | 12 | 1.5 | 1 |
| PA-A4 | 0.5 | 12 | 1.5 | 0.5 | 1 | 0.064 | 1.5 | 48 | 4 | 0.094 |
| PA-A5 | 0.38 | 12 | 0.75 | 0.75 | 0.5 | 0.064 | 0.38 | 48 | 3 | 0.38 |
| PA-A6 | >32 | >1,024 | >256 | 12 | 3 | >32 | 12 | 32 | 3 | 0.75 |
| PA-A7 | >32 | >1,024 | >256 | 12 | 3 | 0.5 | 1.5 | 32 | 4 | 0.38 |
| PA-A8 | 0.5 | 3 | 96 | 12 | 2 | 3 | >32 | >256 | 24 | 4 |
| PA-A9 | >32 | >1,024 | >256 | 12 | 2 | >32 | >32 | 32 | 3 | 0.19 |
| PA-A10 | 0.5 | 12 | 1.5 | 0.5 | 1.5 | 8 | 1.5 | >256 | 8 | 0.38 |
| PA-A11 | 0.5 | 16 | 0.5 | 0.19 | 0.5 | 0.064 | 0.38 | >256 | 4 | 0.25 |
| PA-B1 | >32 | >1,024 | 16 | 4 | 2 | 0.75 | 3 | 2 | 0.38 | 0.125 |
| PA-B2 | 8 | 64 | 8 | 3 | 4 | 0.19 | 0.75 | 32 | 3 | 0.5 |
| PA-B3 | 3 | 24 | 3 | 0.25 | 1.5 | 0.094 | 2 | 32 | 4 | 0.75 |
| PA-B4 | 4 | 512 | 16 | 4 | 2 | 0.25 | 2 | 32 | 8 | 0.38 |
| PA-B5 | 1 | 12 | 0.5 | 0.25 | 2 | 0.064 | 0.38 | >256 | 32 | 0.5 |
| PA-B6 | 3 | 64 | 12 | 2 | 2 | 0.38 | 2 | 8 | 1.5 | 0.5 |
| PA-B7 | 6 | 48 | 3 | 1.5 | 0.75 | 0.064 | 0.5 | 12 | 2 | 0.19 |
| PA-B8 | 0.75 | 32 | 0.5 | 0.25 | 1.5 | 0.094 | 0.38 | >256 | 32 | 0.5 |
| PA-B9 | >32 | >1,024 | 8 | 2 | 3 | 0.19 | 0.75 | 48 | 4 | 1 |
| PA-C1 | 0.750 | 16 | 1 | 0.19 | 0.75 | 0.19 | 1.5 | 12 | 0.75 | 0.75 |
| PA-C2 | >32 | >1,024 | >256 | 24 | >256 | 6 | >32 | >256 | 192 | 16 |
| PA-C3 | 2.000 | 32 | 0.5 | 0.5 | 1 | 1.5 | >32 | 24 | 1.5 | 0.5 |
| PA-C4 | >32 | >1,024 | >256 | 6 | >256 | 4 | >32 | >256 | 64 | 6 |
| PA-C5 | 1.500 | 48 | 1.5 | 1 | 1 | 1.5 | >32 | 24 | 1.5 | 0.75 |
| PA-C6 | >32 | >1,024 | 48 F | 6 | 6 | 16 f | >32 | >256 | >1,024 | 4 |
| PA-C7 | 0.380 | 32 | >256 | 32 | >256 | >32 | >32 | >256 | 192 | >32 |
SMX-TMP, sulfamethoxazole-trimethoprim; SX, sulfisoxazole; TLc, ticarcillin-clavulanate; AT, aztreonam; TZ, ceftazidime; MP, meropenem; IP, imipenem; AK, amikacin; TM, tobramycin; CI, ciprofloxacin.
Studies of bacterial growth and auxotrophism.
Nutrient minimal agar medium M9 (Teknova, Hollister, CA) containing glucose as the only carbon source was used for all nutritional studies of auxotrophism in the P. aeruginosa isolates. Each P. aeruginosa isolate, at 0.5 McFarland concentration (∼1 × 107), was confluently swabbed onto an M9 agar plate. Sterile blank filter paper discs (6-mm diameter; Becton, Dickinson and Company, United States) containing 5 μl of a specific nutritional compound were used for evaluation of growth requirements. Nutritionally dependent growth of P. aeruginosa isolates was checked daily for a maximum of 14 days, with the first 48 h of incubation at 35°C and the remaining 12 days at room temperature (∼25°C). The agar plates were wrapped in plastic bags to minimize the desiccation of agar. Common biochemically active cofactors, such as heme (Remel), menadione, and thymidine, were used for growth studies as previously described (2). Preparation of amino acid solutions was as follows. Amino acid powders (Fluka BioChemika, Steinheim, Switzerland), including l-alanine, l-arginine hydrochloride, l-asparagine, l-aspartic acid, l-cysteine, l-cystine, l-glutamic acid, l-glutamine, glycine, l-histidine hydrochloride, l-4-hydroxyproline, l-isoleucine, l-leucine, l-lysine hydrochloride, l-methionine, l-phenylalanine, l-proline, l-serine, l-threonine, l-tryptophan, l-tyrosine, and l-valine, were diluted to a final concentration of 0.1 M. Amino acid solutions were prepared using molecular-grade water (Fisher Scientific, Fairlawn NJ), with the following exceptions for the purpose of increased solubility: l-aspartic acid was prepared with 0.1 N HCl, l-tryptophan with 0.5 N HCl, l-cystine and l-tyrosine with 1 N HCl, and l-glutamic acid with 2 N HCl. All amino acid solutions were stored at −20°C. Methionine and arginine were used for the first round of auxotrophic screening. For other specific deficiencies or possible multiple amino acid deficiencies, a pooled solution of all 22 amino acids and a series of “pool minus one” solutions (where each solution contained all the amino acids except one) were tested.
Auxotrophic complementation of the nutritionally deficient strains by prototrophic “feeder” strains included either isogenic strains from the same patient or nonisogenic strains from unrelated patients and quality control (QC) wild-type P. aeruginosa strains (ATCC 27853, PA-mucQC, and PA14) based on their growth in M9 medium. Similar to amino acid and cofactor growth complementation tests, the feeder strains were prepared at a McFarland concentration of 0.5 and 5 μl of the preparation was used on each blank filter paper disc for growth rescue of the auxotrophic isolates.
Molecular amplification and sequencing of mexR, mexAB-oprM, mexZ, mutL, and mutS.
Primers for amplification and sequencing of the pertinent genetic determinants were optimally selected by using Primer3 (http://frodo.wi.mit.edu/primer3/). The gene coding regions of mexR, mexAB-oprM, mutL, and mutS were amplified, and some were fully sequenced. DNA sequencing was carried out using a BigDye terminator cycle sequencing ready reaction DNA sequencing kit and an ABI Prism 310 genetic analyzer (Applied Biosystems). The sequencing thermocycling steps used were 96°C for 30 s, 25 cycles of 96°C for 10 s, and 60°C for 10 s. Sequencing chromatographs generated from both forward and reverse strands of the PCR template were edited using Seqman, DNASTAR (DNASTAR, Inc.).
PCR product cloning and transformation.
Cloning of heterogeneous mexAB PCR product was carried out by gel purification of an ∼1,500-bp amplification product containing 131 bp of the 3′ end of mexA and 1,341 bp of the 5′ half of the mexB from all isolates, followed by plasmid ligation and transformation into E. coli Qiagen EZ competent cells (Qiagen, Germany). Heterogeneous mexAB products were successfully cloned from two isolates, PA-A5 (four clones) and PA-B8 (two clones) (see Fig. S1 in the supplemental material). The E. coli transformants were selected on Luria-Bertani (LB) agar containing ampicillin (100 μg/ml) (Remel, United States). Plasmid DNA was extracted from the E. coli transformants by using a QIAprep spin miniprep kit (Qiagen, Germany) and sequenced individually with dense primer coverage for each cloned product.
Bacterial genome typing for clonality.
Genetic DNA fingerprinting of P. aeruginosa strains was generated by pulsed-field gel electrophoresis (PFGE) using SpeI restriction fragmentation (service provided by IEH Laboratories & Consulting Group, Lake Forest Park, WA, United States).
RESULTS
P. aeruginosa clinical isolates and their patterns of susceptibility to SMX-TMP.
We examined the antimicrobial susceptibilities in 525 archived CF P. aeruginosa isolates collected from 103 patients in 2007. A subset of 498 (95%) isolates was obtained from 76 patients who had an average of 3 cultures per patient over the year with a median of 2 (range, 2 to 24) isolates per patient. The remaining 27 P. aeruginosa isolates were obtained from single-isolate cultures from 27 patients.
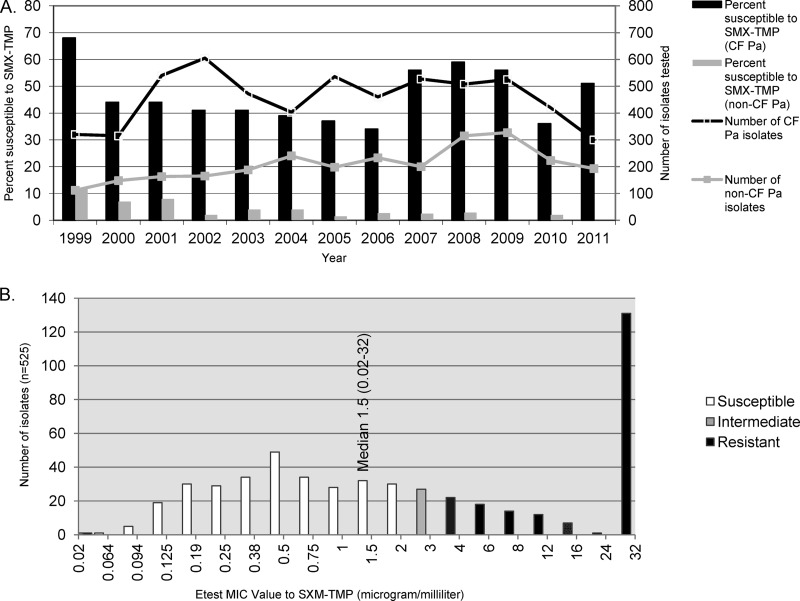
Antimicrobial susceptibility results generated from isolates of CF patients have been analyzed separately from those of non-CF patients in our hospital over the last 13 years using CLSI M100-S16 interpretive criteria (7a). The P. aeruginosa pseudomonal susceptibility results demonstrated overall higher rates of resistance to beta-lactams, aminoglycosides, and quinolones than did the results for P. aeruginosa isolates of non-CF origin (data not shown). However, an unusual proportion of CF P. aeruginosa isolates (34 to 68% from 1999 to 2011) was susceptible to SMX-TMP (Fig. 1A).
Fig 1.
(A) P. aeruginosa susceptibility to sulfamethoxazole-trimethoprim (Seattle Children's Hospital data). (B) Sulfamethoxazole-trimethoprim MIC distribution in CF P. aeruginosa isolates from 2007.
We observed a wide range of in vitro MICs of many antimicrobial agents among the 525 CF P. aeruginosa isolates, including SMX-TMP (Fig. 1B and unpublished data). SMX-TMP MICs ranged from 0.02 to ≥32 μg/ml (median, 1.5 μg/ml) using Etest methodology (Fig. 1B). Of these 525 isolates, 293 (56%) showed SMX-TMP MICs in the susceptibility range of ≤2 μg/ml, with a median of 0.38 μg/ml. The distribution of SMX-TMP MICs generated by the 525 P. aeruginosa isolates is nearly continuous in the concentration range measurable by Etest (Fig. 1B). Coexisting P. aeruginosa isolates exhibiting a wide range of SMX-TMP MICs (0.02 to ≥32 μg/ml) were present in CF airway secretions from 43 patients (57%) (Table 1). Vector-borne and integron-associated SMX-TMP resistance was excluded by negative int1 and sul1 amplification results obtained from these 525 P. aeruginosa isolates after they were divided into smaller sample pools (5 isolates in each PCR mix).
Table 1.
Groups of patient based on the number(s) of P. aeruginosa isolates with divergent SMX-TMP MICs
| Patient groupa | No. (%) of patients withb: |
||
|---|---|---|---|
| Coexisting isolates exhibiting S/I/R SMX-TMP MICs (0.002–32 μg/ml) | Isolate(s) exhibiting S/I SMX-TMP MICs (≤3 μg/ml) | Isolate(s) exhibiting R SMX-TMP MICs only (≥4 μg/ml) | |
| Two or more isolates (n = 76) | 43 (57) | 21 (28) | 12 (16) |
| Single isolate (n = 27) | NA | 15 (56) | 12 (44) |
| All patients (n = 103) | 43 (42) | 36 (35) | 24 (23) |
Patients are grouped based on cultures of their respiratory secretions and the number of P. aeruginosa isolates obtained in 2007 with divergent SMX-TMP MICs using CLSI M100-S6 breakpoints.
S, susceptible, I, intermediate; R, resistant; NA, not applicable .
Coisolation of patient-specific clonal P. aeruginosa strains with divergent susceptibilities to SMX-TMP.
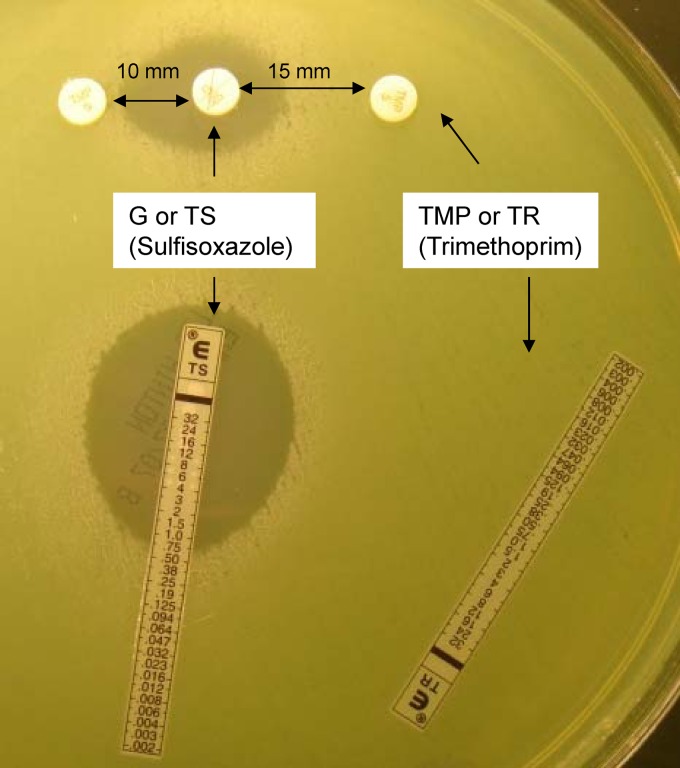
In order to understand the resistance and functional divergence associated with potentially chronically colonized patients, three sets of P. aeruginosa isolates (n = 27) with widely divergent SMX-TMP MICs from three patients (patients A, B, and C) were chosen for further molecular analysis and in vitro nutrient requirement studies (Table 2). In each set, organisms had been isolated from three to five sputum cultures collected over 2007. Two P. aeruginosa strains, P. aeruginosa ATCC 27853 and PA-mucQC (a laboratory-validated mucoid CF P. aeruginosa type strain), were used as susceptibility testing controls, and both consistently showed SMX-TMP MICs of ≥32 μg/ml and correlated sulfisoxazole (SX) MICs of ≥1,024 μg/ml. Strain PA14 and its corresponding mex gene-inactivated derivatives were used as additional controls for SMX-TMP MIC determinations (24). The MICs for PA14 mutational derivatives correlated with the functional predictions of their mutated genes; mexR mutation resulted in an elevated SMX-TMP MIC of ≥32 μg/ml (the PA14 wild-type MIC is 16 μg/ml), while mexA and oprM mutations resulted in significantly reduced of SMX-TMP MICs (0.19 to 0.25 μg/ml) (Table 2). For both control and patient isolates, TMP alone showed no activity against any of the P. aeruginosa isolates regardless of whether the isolate was susceptible to SX, although TMP appeared to have a synergistic effect in vitro, as was evident from MIC and spatial antibiotic disc approximation in the 10- to 15-mm range, suggesting that the folic acid biosynthetic pathway was intact (Table 2 and Fig. 2).
Table 2.
Characteristics of the 27 clinical P. aeruginosa strains isolated from cultures in 2007a
| Isolate | Muc/pigb | MIC (μg/ml) |
Amino acid auxotrophy | Growth complementation by other P. aeruginosa strains | Sequencing resultc |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SMX-TMP | SX | mexR | mexA | mexB | mexZ | mutL | mutS | ||||
| PA-ATCC | N/g | >32 | >1,024 | Prototroph | NA | ||||||
| PAmucQC | M/n | >32 | >1,024 | Prototroph | NA | ||||||
| PA14 | N/g | 16 | 128 | Prototroph | NA | ||||||
| PA14 (mexR) | N/g | >32 | >1,024 | Prototroph | NA | mexR | |||||
| PA14 (mexA) | N/g | 0.190 | 6 | Prototroph | NA | mexA | |||||
| PA14 (oprM) | N/g | 0.250 | 6 | Prototroph | NA | ||||||
| PA14 (mexZ) | N/g | >32 | >1,024 | Prototroph | NA | mexZ | |||||
| PA-A1 | N/n | 0.75 | 16 | Lys, met | A3, B1, B3, B4, C6 | WT | Δ18 bp near 3′ end | m | NSS | m | Δ12 bp near 3′ end |
| PA-A2 | N/g | 0.25 | 3 | Arg | A3, B4, C6 | ΔG near 5′ end | WT | m | m | m | m |
| PA-A3 | N/n | 1 | 48 | Prototroph | NA | WT | WT | m | m | m | m |
| PA-A4d | N/g | 0.5 | 12 | Arg | A3, B4, C6 | ΔG near 5′ end | WT | m | m | m | m |
| PA-A5d | M/n | 0.38 | 12 | Lys, met | C6, PAmucQC | ΔG near 5′ end | Hetero: SS & NSS | Hetero: SS & NSS | m | m | m |
| PA-A6d | N/g | >32 | >1,024 | Arg | All prototrophs | ΔG & Δ10 bp | WT | m | m | m | m |
| PA-A7d | N/n | >32 | >1,024 | Arg | All prototrophs | ΔG near 5′ end | WT | m | m | m | m |
| PA-A8 | N/n | 0.5 | 3 | Unknown | A3, B1, B3, B4, C6 | WT | WT | m | m | m | m |
| PA-A9d | N/g | >32 | >1,024 | Arg | All prototrophs | ΔG near 5′ end | WT | m | m | m | m |
| PA-A10 | N/n | 0.5 | 12 | Arg | All prototrophs | ΔG near 5′ end | WT | m | m | m | m |
| PA-A11 | M/n | 0.5 | 16 | Arg | All prototrophs | ΔG near 5′ end | WT | m | m | m | m |
| PA-B1 | N/n | >32 | >1,024 | Prototroph | NA | WT | WT | m | Missing | m | m |
| PA-B2 | M/n | 8 | 64 | Prototroph | NA | WT | WT | m | m | m | m |
| PA-B3 | N/g | 3 | 24 | Prototroph | NA | WT | WT | m | m | m | m |
| PA-B4 | N/n | 4 | 512 | Prototroph | NA | WT | Δ7 bp near 5′ end | m | m | m | m |
| PA-B5 | N/n | 1 | 12 | Ile, val | A3, B4 | WT | Δ7 bp near 5′ end | m | m | m | Δ12 bp near 5′ end |
| PA-B6 | N/n | 3 | 64 | Ala, trp | A3, B4 | WT | WT | m | m | m | m |
| PA-B7 | M/g | 6 | 48 | Prototroph | NA | WT | WT | m | m | m | m |
| PA-B8 | N/n | 0.75 | 32 | Ile, leu, val | A3, B4 | WT | Δ7 bp near 5′ end | Hetero: SS & NSS, bp+ | m | m | Δ12 bp near 5′ end |
| PA-B9 | M/n | >32 | >1,024 | Prototroph | NA | WT | WT | m | m | m | m |
| PA-C1 | M/n | 0.750 | 16 | Met | All prototrophs | WT | WT | m | 6 bp+ & NSS | WT | m |
| PA-C2 | N/n | >32 | >1,024 | Arg | C6, A3, B1, B4 | WT | WT | m | m | ΔT near 5′ end | m |
| PA-C3 | M/n | 2.000 | 32 | Met | All prototrophs | WT | WT | m | m | WT | m |
| PA-C4 | N/n | >32 | >1,024 | Arg | C6, A3, B1, B4 | WT | WT | m | m | ΔT near 5′ end | m |
| PA-C5 | M/n | 1.500 | 48 | Met | All prototrophs | WT | WT | m | m | WT | m |
| PA-C6d | N/n | >32 | >1,024 | Prototrophd | NA | WT | WT | m | m | ΔT near 5′ end | m |
| PA-C7 | N/n | 0.380 | 32 | Arg, leu | C6, B4 | WT | WT | m | m | ΔT near 5′ end | m |
Summary of the SMX-TMP and SX MICs, growth properties, nutritional requirements, and sequencing results of efflux and DNA mismatch repair genes of the strains.
Muc/pig, bacterial growth on agar media exhibiting mucoid (Muc) or pigmented (pig) colonies; M, mucoid; N, nonmucoid; g, green pigment; n, no pigment.
WT, wild type, discounting any synonymous point mutation that resulted in no amino acid change; NA, not applicable; SS, a synonymous mutation due to a nucleotide mutation that resulted in no amino acid substitution; NSS, a nonsynonymous mutation due to a nucleotide mutation that resulted in amino acid substitution; Δ bp, deletion of indicated number of base pair(s); ΔT or ΔG, deletion of indicated nucleotide; bp+, base pair insertion; m, PCR-amplified materials that were mixed or heterogeneous; Hetero, cloned sequences show heterogeneity.
Isolate shows show nonhomogeneous colonies under ×60 stereoscope.
Fig 2.
An example of a CF P. aeruginosa isolate tested for its susceptibility to SMX-TMP and to TMP or sulfisoxazole alone. A green-pigmented CF P. aeruginosa isolate is showing susceptibility to SMX-TMP (TS) at a MIC of 0.75 μg/ml by Etest and to TMP (TR) alone at a MIC of >32 μg/ml. Disc diffusion with sulfisoxazole (G, 500 μg) in the top center approximated by TMP (5 μg) discs measured at 10 mm to the left and 15 mm to the right.
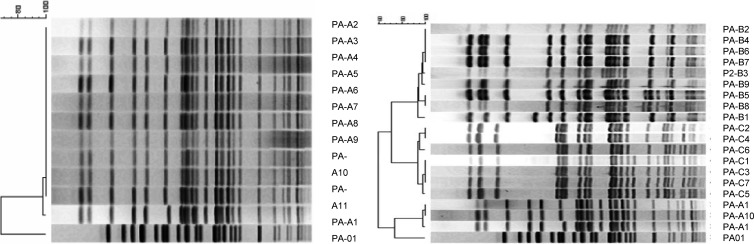
Divergent antimicrobial susceptibilities to our standard panel of antimicrobial agents were observed in these 27 CF P. aeruginosa isolates, as shown by wide MIC distributions (Table 3). The SMX-TMP susceptibilities exhibited by CF P. aeruginosa isolates correlated closely with their susceptibilities to SX alone (see the results for PA-A1 to PA-C7 in Table 2). In addition, antimicrobial pansusceptible strains were found in all three patients (see the results for PA-A2, -A3, -B6, and -C1 in Table 3). Despite divergence in resistance patterns, isolates of P. aeruginosa from a chronically infected individual are often closely related based on DNA fingerprinting patterns, which was confirmed by representative isolates from these three patients (Fig. 3) (6–8).
Fig 3.
Genetic DNA fingerprinting patterns of P. aeruginosa strains from patients A, B, and C were generated by pulsed-field gel electrophoresis (PFGE) using SpeI restriction fragmentation.
Characterization of critical mutations in efflux determinants mexR, mexAB-oprM, and mexZ, as well as the bacterial hypermutable state associated with mutL and mutS inactivation.
The 27 P. aeruginosa isolates with various MICs to SMX-TMP from the three selected patients were subjected to amplification and sequence examination, focusing primarily on mexR, mexAB-oprM, and mexZ, as well as on mutL and mutS coding regions. Among the isolates, we identified critical mutations in efflux determinants, such as mexR, mexAB, and mexZ, as well as bacterial hypermutable states associated with mutL and mutS inactivation. These results are summarized in Table 2. Despite the patient-specific clonal nature of these isolates, it is evident that their genetic integrity has undergone a process of erosion, as shown by deletions and insertions, as well as single-nucleotide polymorphisms, in mexR, mexAB, mexZ, mutL, and mutS, but not in oprM (affected only by a few silent mutations; data not shown). The patterns of micromutation consisted largely of small deletions/insertions resulting in frameshift and premature stop codons, as well as both synonymous (SS, or silent mutations) and nonsynonymous substitutions (NSS, or missense mutations) (Table 2; also see Fig. S1 in the supplemental material).
Molecular amplification produced seemingly homogeneous sequences for mexR, oprM, and for the most part, mexA from these 27 patient isolates (Table 2). In contrast, despite many in vitro passages for colony purification, the molecular amplification products associated with the coding sequences of mexB, mexZ, mutL, and mutS in all 27 isolates appeared to be composed of mixed or heterogeneous sequences which required cloning for resolution (Table 2; also see Fig. S1 in the supplemental material). Single or compound mutations, either in efflux regulatory genes such as mexR (or mexZ) or structural gene(s) such as mexAB or in both, were common in these isolates, which serves to explain their intraspecific divergent and continuous SMX-TMP MIC distribution (Table 2). For example, the three SMX-TMP-resistant strains (isolates PA-A2, PA-A4 to -A7, and PA-A9 to -A11) from patient A all contained a critical single base G deletion (Table 2, ΔG near 5′) that caused a frameshift resulting in an immediate premature termination codon in mexR, a well-studied negative regulator that controls mexAB-oprM expression (33, 49). In patient B, while mexR remained intact in all nine isolates (PA-B1 to -B9), a 7-bp deletion near the 5′ end of mexA (Table 2, Δ7 bp near the 5′) which was identical in each instance was found in three isolates (PA-B4, -B5, and -B8) that showed significantly reduced MICs to SMX-TMP. Compound mutations involving mexR, mexZ, and/or mexAB were common to all isolates (Table 2; also see Fig. S1). Moreover, mutations associated with the mex gene determinants clearly showed specific patterns indicative of isogenic origin associated with each patient (Table 2, ΔG near the 5′ in mexR and Δ7 bp near the 5′ in mexA, associated with isolates from patient A and B, respectively). Such massive mex gene sequence degeneration has led us to hypothesize that pseudomonal isogenic divergence is not limited to antimicrobial susceptibility alone. Indeed, heterogeneous mutations found simultaneously in genes that govern bacterial hypermutable states, namely, mutL and mutS, further support the concept that organized gene decay is associated with the process of pseudomonal symbiotic adaptation selected for by the persistent CF airway environment (Table 2).
Amino acid auxotrophism in CF P. aeruginosa.
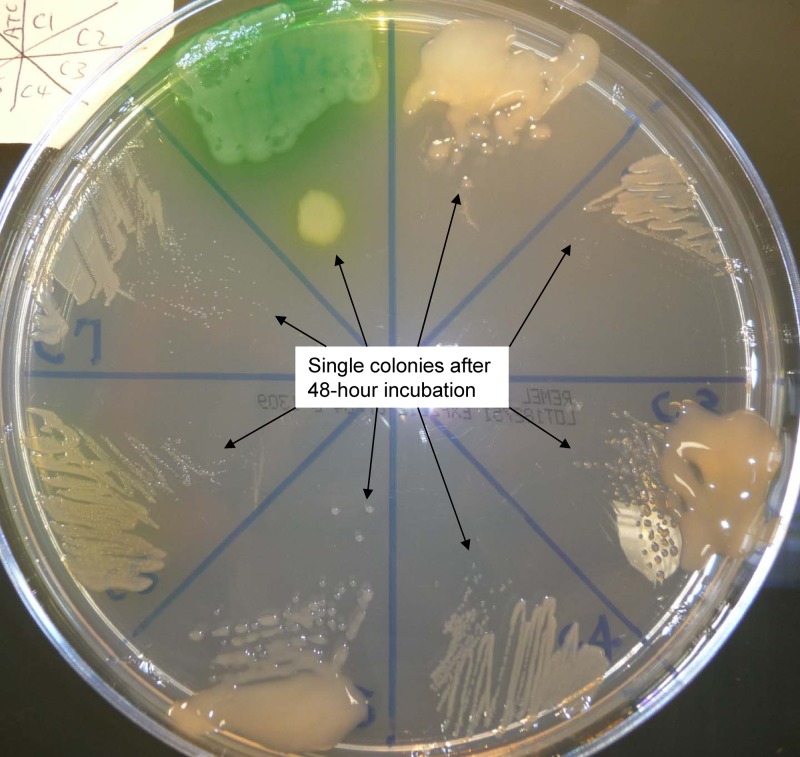
Compensatory effects of a multicellular mode of living are likely to allow nucleotide mismatch during DNA polymerization, which in turn may promote rapid intraspecific diversification within the colony (7, 10). Isolates of CF P. aeruginosa are well known to exhibit extraordinary in vitro growth and morphological divergence over the course of chronic infection (Tables 2 and 3 and Fig. 4) (21, 44). Typical P. aeruginosa isolates from chronically infected CF patient are mostly less or nonpigmented colonies with variable mucoid presentation and generally show a small colony phenotype resulting from a retarded growth rate on any nutrient medium, including Mueller-Hinton agar (see all seven isolates from patient C in Fig. 4). Nutritional deficiency, specifically amino acid auxotrophism, is common in CF P. aeruginosa isolates (3). Among the 27 isolates from the three patients, 19 were auxotrophic, as shown by their inability to grow on M9 medium containing only a simple sugar, glucose (Table 2). The remaining eight isolates (at least one from each patient) and all control organisms, including PA14 and its altered mex gene derivatives, were able to grow on M9 minimal medium (prototrophic). In isolates from patient A, a growth requirement for arginine alone was found in seven isolates (PA-A2, -A4, -A6, -A7, and -A9 to -A11) and for both lysine and methionine in two isolates (PA-A1 and -A5). In isolates from patient B, a growth requirement for both alanine and tryptophan was found in one (PA-B6), for both isoleucine and valine in one (PA-B5), and for isoleucine, leucine, and valine in one (PA-B8). For isolates from patient C, a growth requirement for methionine alone was found in three isolates (PA-C1, -C3, and -C5), for arginine alone in two (PA-C2 and -C4), and for both arginine and leucine in one (PA-C7). To rule out oxidative respiration deficiencies and/or folic acid biosynthetic deficiencies which may result in the bacterial small colony variant (SCV) phenotype and antimicrobial resistance to SMX-TMP, all 19 auxotrophic P. aeruginosa isolates were screened and determined not to require thymidine, heme, or menadione for growth. Moreover, no association was found between SMX-TMP susceptibility and amino acid auxotrophism, nor was there correlation with any other in vitro growth morphological phenotype (Table 2).
Fig 4.
Comparison of growth properties of P. aeruginosa isolates on Mueller-Hinton agar after a full 48-h incubation in 35°C ambient air. In clockwise sequence from the green wild-type ATCC 27853 at top left, the isolates are PA-C1 (mucoid), PA-C2, PA-C3 (mucoid), PA-C4, PA-C5 (mucoid), PA-C6, and PA-C7.
Complementation of auxotrophism by isogenic and nonisogenic P. aeruginosa prototrophic isolates.
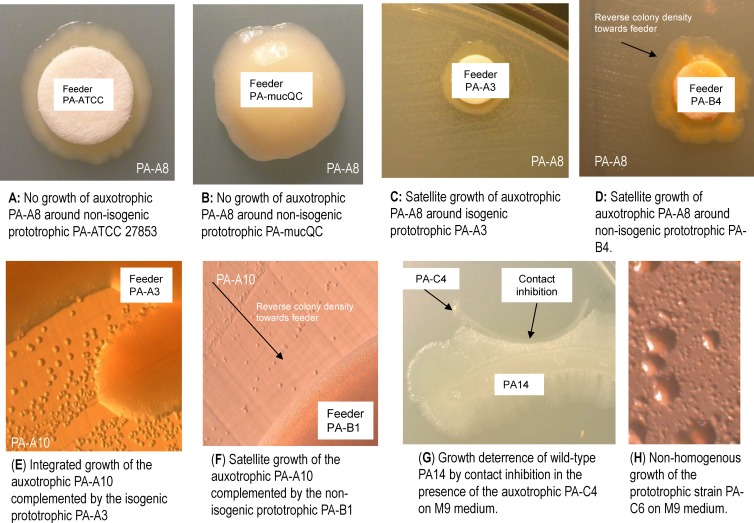
Intraspecific morphological, metabolic, and genetic heterogeneity in chronic CF isolates all suggest that growth complementation by isogenic and nonisogenic P. aeruginosa prototrophic isolates may reflect pseudomonal division of labor and a primitive multicellular lifestyle (36, 43). Prototrophic feeder strain(s) from each patient (Table 2) were tested on blank filter discs for growth complementation of the auxotrophic strains inoculated onto M9 minimal medium. Growth complementation was determined both by visual (Fig. 5A to D) and stereoscopic (Fig. 5E and F; ×30 magnification) examination. Auxotrophic isolates deficient in various specific amino acids from each patient were complemented by at least one of the prototrophic isogenic isolates as a feeder from the same patient, except for PA-A5. Some auxotrophs were complemented by the prototrophic nonisogenic feeder isolates from unrelated CF patients or even from control organisms with prolonged incubation (Table 2). The wild-type ATCC 27853 and PA14 appeared to be ineffective as feeder strains for growth complementation for many of the auxotrophs even with prolonged incubation (Fig. 5A and B). Most of the isogenic feeder effects were readily visible and diffusible into a radius ranging up to ∼15 mm within 7 days (Fig. 5C and D). The growth patterns with feeder complementation were highly diverse, as satellite colonies of the auxotrophs grown around the feeders showed integration with the feeder strain in pairs between isogenic strains (Fig. 5C and E) or showed reverse density radiating from the feeder strain in pairs between nonisogenic strains (Fig. 5D and F). The latter pattern was prominent during early examination (e.g., <7 days after plating). Moreover, the deterrence of growth of wild-type PA14 by contact inhibition in the presence of CF P. aeruginosa, such as by the auxotrophic PA-C4 on M9 medium, was apparent after 14 days of incubation (Fig. 5G). Nonhomogeneous growth on M9 medium was rather common among several patient strains (PA-C6, -A4, -A5, -A6, -A7, and -A9) and could be resolved only by stereoscopy at ×60 magnification (Table 2 and Fig. 5H). In addition, for isolates associated with patient C, none of the auxotrophic isolates deficient for arginine (PA-C2 and -C4) or methionine (PA-C1 and -C3) alone were able to grow on M9 medium, whereas either a mixture of PA-C1 and PA-C2 or a mixture of PA-C3 and PA-C4 was able to grow on M9 with prolonged incubation at room temperature for up to 14 days. In vitro nutrient and growth complementation between isogenic strains suggests pseudomonal development of a multicellular codependent lifestyle during the course of chronic infection in the CF airway.
Fig 5.
Growth complementation of auxotrophism by isogenic and nonisogenic P. aeruginosa prototrophic isolates (over a 6-mm sterile absorbent filter disc labeled “Feeder”) inoculated onto M9 agar medium. (A to F) Examples of negative (A and B) and positive (C, D, E, and F) growth complementation of an auxotrophic strain by various prototrophic P. aeruginosa strains. (E and F) Examples of close-up views (stereoscopy at ×30 magnification) of growth complementation patterns of an auxotrophic strain by prototrophic strains that were either isogenic (E) or nonisogenic (F). (G) An example of contact inhibition of wild-type PA14 growth by the auxotrophic PA-C4. (H) An example of nonhomogeneous growth of PA-C6 on M9 agar medium (×60 magnification).
DISCUSSION
The rates of antimicrobial resistance of CF P. aeruginosa strains reported in the past have been consistently higher than those of their non-CF counterparts (32, 47). Most reports used MIC aggregates generated from a cohort of CF clinical isolates grouped solely by interpretive MIC (or diameter of inhibition zone) criteria, which were established primarily for non-CF bacterial infections, and were expressed as “percent susceptible” rather than by MIC distribution. This clinical practice hinders appreciation of the concept that pseudomonal chronic infection in the CF airway results in a resistant syntrophy. This bacterial syntrophy is composed of a less free-living, less virulent, and intraclonally highly divergent bacterial population developed in each patient over his or her life time. Since environmental and non-CF P. aeruginosa strains are almost invariably SMX-TMP resistant, SMX-TMP-susceptible strains of P. aeruginosa must arise from a resistant parental clone via mutation. The wide range and continuous distribution of in vitro SMX-TMP MICs (similar MIC distribution patterns apply to all other agents tested; data not shown) in this P. aeruginosa collection confirmed that this resistance is not due to a single acquired determinant, such as integron-associated sul, which results in an “all-or-none” resistance pattern. Instead, our findings are consistent with independent, patient-specific clonal divergence involving both efflux regulation and structural determinants, primarily in mexR/mexAB-oprM and mexZ (Table 2 and Fig. 1B) (33, 49).
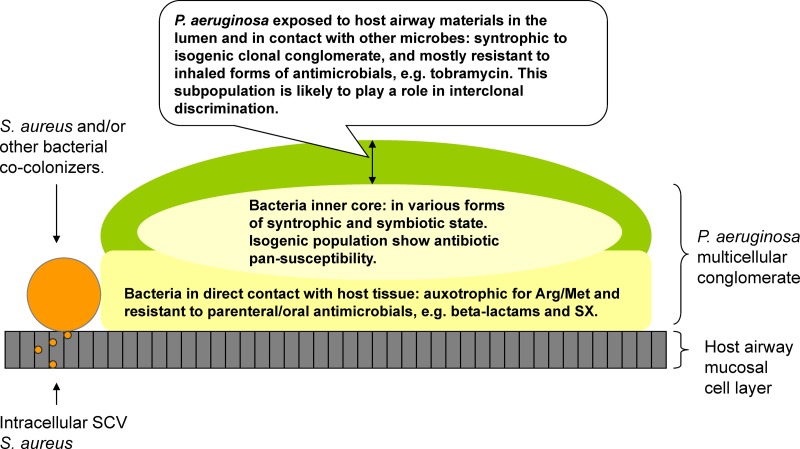
CF P. aeruginosa isogenic strains studied to date have long been regarded as “pure” cultures. However, poor reproducibility of in vitro susceptibility results in a single set of isolates suggests phenotypic variability (12). In addition to phenotypic variability, we have for the first time demonstrated that CF P. aeruginosa isogenic isolates are in fact not homogeneous despite the conventional practice of repeat single-colony passages in vitro. This conclusion is based on both the heterogeneous growth patterns (Fig. 5H) and heterogeneous mutations associated with mex gene products demonstrating sequence mosaicism generated from a “single colony” (see Fig. S1 in the supplemental material). Perhaps for these reasons, correlations between in vitro antimicrobial testing results and in vivo efficacy are inadequate to guide clinical practice in management of CF infection (11). Divergent intraclonal antimicrobial resistance patterns may in part reflect the complexity of treatment strategies that are uniquely employed in CF, as both parenteral and inhaled forms of antibiotics are used to control the bacterial burden in CF airways (40). These treatments, coupled with host defense mechanisms, lead to the formation of a pseudomonal biofilm or multicellular configuration in which susceptible and resistant isogenic organisms appear to coexist. Within this colonial lifestyle, intraspecific genetic variation may confer role differentiation between the bacteria closest to the host mucosal cells (in immediate contact with host defenses and oral/parenterally administered drugs) and those closest to the airway lumen (in contact with inhaled drugs) (Fig. 6). We hypothesize that divergently resistant P. aeruginosa isogenic strains evolve in situ to create a microenvironment that protects and fosters the existence of highly susceptible isogenic strains (specialized in other colonial functions) despite prolonged antimicrobial exposure.
Fig 6.
Diagram of CF airway-adapted P. aeruginosa: a multicellular syntrophy with specialized functional divergence within a colonial architecture.
Isogenic P. aeruginosa strains with divergent SMX-TMP susceptibilities could not be distinguished from each other based on their colonial morphology, auxotrophism, or other in vitro growth properties. Similar to bacterial SCVs of Staphylococcus aureus and Escherichia coli, CF P. aeruginosa isolates often show slow growth that results in small colonies on standard agar media (Fig. 4) (18). The bacterial SCV phenotype is frequently associated with chronic infections, even in non-CF immunocompetent hosts. Bacterial SCVs are characterized by their retarded growth rate in vitro, due in part to deficiencies in electron transport chain components and/or blockage in the folic acid biosynthetic pathway, which require not only heme or menadione but both methionine and thymidine to restore rapid growth (2, 34). However, auxotrophism in our CF P. aeruginosa isolates differs from other bacterial SCV phenotypes in that none of the 19 auxotrophic isolates, including those that were methionine dependent (PA-A1, -A5, -C1, -C3, and -C5), showed a thymidine, heme, or menadione requirement for growth (18). Therefore, the folic acid biosynthetic pathway appears to be intact in CF P. aeruginosa. Instead, multicellular metabolic and resistance interdependency maturating from a single P. aeruginosa clone appears to be the result of evolution under host and antimicrobial pressures and may be vital to promote long-term chronic airway colonization via symbiosis.
Despite divergence in resistance patterns, isolates of P. aeruginosa from a single individual are often closely related based on DNA fingerprinting patterns (6–8). Nutritional complementation of in vitro growth in M9 minimal medium suggests that the survival of these auxotrophic strains is also possible in vivo because CF-adapted P. aeruginosa engages in a cooperative multicellular lifestyle. Bacterial intracolonial organization and cooperative metabolic interdependency is termed “syntrophy,” and this condition can only be supported by a multicellular mode of living (23, 43). Notably, the in situ development of a pseudomonal conglomerate resembles a multicellular organism that is highly tolerant of host defenses and antibiotic inhibitory actions through division of labor (36, 43). Auxotrophic strains may in fact be more common than has been observed because the severe auxotrophism associated with highly adapted organisms may prohibit their viability in vitro and culture-based microbiology may thus underrepresent these highly adapted auxotrophs. If bacterial syntrophy is responsible for the evolution of antibiotic-susceptible and/or auxotrophic bacteria in multicellular conglomerates found in airways of CF patients, these adaptive changes must offer some advantage to P. aeruginosa. For example, the net result of syntrophy may be bacterial energy conservation, better resource utilization, and/or, most importantly, antimicrobial and host tolerance.
The selective loss of certain amino acid biosynthesis capabilities in CF P. aeruginosa cannot be explained by the nutrient abundance in the diseased airway environment. Host immunodefense must play a role in selection of pseudomonal auxotrophism and in counterselection of bacterial virulence. Amino acid auxotrophism can be found in up to 86% of adult CF patients who were colonized by P. aeruginosa, with arginine and methionine being the most common auxotrophisms detected (1, 3, 30). Arginine auxotrophism is directly beneficial to the adapting organism for evasion of a harsh ancient mechanism of host defense, as l-arginine is the common substrate for nitric oxide (NO) synthases and arginase (15, 26, 31, 38). Pseudomonal development of arginine auxotrophism may thus explain a highly studied but underappreciated airway immunodeficiency in CF patients, who typically show decreased concentrations of exhaled nitric oxide (NO) (16). The elimination of methionine biosynthesis in CF P. aeruginosa is also unlikely to be the result of random events but may instead reflect host counterselection against bacterial virulence factors and their upstream regulators, such as the quorum-sensing molecule homoserine lactone and its precursor molecule methionine (19). The emergence and accumulation of quorum-sensing-negative mutants (lasR) in 30 to 63% of CF isolates further supports the concept of host counterselection against virulence during chronic infection (10, 20, 44). Clearly, gene retention or loss, as well as the processes by which gene decay proceeds in CF P. aeruginosa, cannot be random events but, rather, reflect a regulated pathway that gives rise to a spectrum of coding mosaicism and functional divergence during an unfinished pseudomonal journey to symbiosis.
Pseudomonal clonal success in the CF airway may protect the host from repeat acquisition of unrelated strains from the environment. Pseudomonal clonal discrimination and intraspecific coadaptation are supported by complete growth complementation in vitro between prototrophic and auxotrophic isogenic P. aeruginosa strains, as opposed to partial or no complementation or growth inhibition between clonally unrelated P. aeruginosa strains (Fig. 5D, F, and G). As they compete for the same pool of nutrient resources, free-living bacteria usually produce secondary metabolites and other weaponry, affecting both inter- and intrabacterial species (4, 48, 52). Such inhibitory properties are visible even in the presence of nutrient complementation between nonisogenic strains in vitro. Cellular interactions of this nature may preserve a strict P. aeruginosa clonal monopoly in the host airway. Thus, the absence of environmental nonisogenic strains of P. aeruginosa in the airway secretions of CF patients who are chronic CF P. aeruginosa carriers may not be explained simply by acquired host immunity to P. aeruginosa.
In many respects, the adaptations that occur in CF P. aeruginosa mimic aspects of evolving symbiosis, chiefly involving genomic degenerative patterns that are found in myriad forms of animal symbionts (29, 41). For example, the animal symbionts studied to date are in various stages of adaptation and show degrees of genome degradation typically involving mutL and mutS. Pseudomonal adaptation via multicellularity associated with CF airway chronic infection may provide a model for a common pathway by which evolution of pseudogenes and minimal genomes in ancient animal symbionts can be achieved (28). Many ancient bacterial symbionts provide their insect animal hosts with essential nutrients or ancillary benefits, such as resistance to environmental stress or natural enemies, and even affect host reproduction to favor their own matriline (9). However, during the early phases of symbiotic evolution that are possible over the life span of a CF patient, adaptation of CF P. aeruginosa does not prevent irreversible damage to the host airway and neither is it able to proceed further down the symbiotic pathway in order to yield benefit to the host. Moreover, the highly matured CF airway-specific P. aeruginosa conglomerate would have minimal chance of either horizontal or vertical transmission due to its inability to compete with wild-type P. aeruginosa outside the CF airway, the rarity of CF-affected hosts in the general population (1/2,500 in the Caucasian population), and a relatively short average life expectancy of these hosts, 38.3 years (http://www.cff.org/UploadedFiles/LivingWithCF/CareCenterNetwork/PatientRegistry/2010-Patient-Registry-Report.pdf).
In conclusion, pseudomonal persistence and antimicrobial resistance properties in the CF airway are consistent with a model based on the principle of bacterial syntrophy. The in vitro hypersusceptibility to SMX-TMP and to other antimicrobials associated with individual isogenic strains may not predict the therapeutic potential of SMX-TMP against the P. aeruginosa multicellular conglomerate in vivo. The net result is a biologically less free-living and less virulent P. aeruginosa strain which is selected by host defense, as this bacterial parasite possesses features symbiotic with the CF pulmonary environment. In some respects, a uniclonal and highly adapted P. aeruginosa syntrophy in the airway of chronically infected CF patients may benefit the host by preventing the acquisition of another environmental wild-type strain, which would result in a deleterious replay of the infectious process in the same host airway. These traits suggest that genetic manipulation of CF pulmonary P. aeruginosa into a probiotic symbiont, as opposed to eradication of the organism, may be an alternative therapeutic strategy in the clinical management of CF and related disorders.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daniel J. Wolter, Hemantha D. Kulasekara, Adam Griffith, and Lucas R. Hoffman for their kind assistance in providing PA14 reference strains and other technical assistance. We thank Jenny Stapp, Anne Marie Buccat, Scott Anderson, Lynn Stapp, Shannon Rich, Patrick Abe, Joan Guzzo, Patti Mau, Deena Motherwell, Catherine Jerome, Brian Poon, Treva Tsosie, Amy Samuelson, Aster Tesfaye, Jeremy Rowe, and Rosemary Martin in the Microbiology Laboratory, Seattle Children's Hospital, for their technical expertise and years of perseverance in working up CF respiratory cultures.
This study was supported in part by a Cystic Fibrosis Foundation Therapeutics Development Core Lab Clinical Toolkit award to J. L. Burns and by the Marie Meyer endowment to the Microbiology Laboratory, Seattle Children's Hospital.
Footnotes
Published ahead of print 10 September 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Agarwal G, Kapil A, Kabra SK, Das BK, Dwivedi SN. 2005. Characterization of Pseudomonas aeruginosa isolated from chronically infected children with cystic fibrosis in India. BMC Microbiol. 5:43 doi:10.1186/1471-2180-5-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson SW, Stapp JR, Burns JL, Qin X. 2007. Characterization of small-colony-variant Stenotrophomonas maltophilia isolated from the sputum specimens of five patients with cystic fibrosis. J. Clin. Microbiol. 45:529–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barth AL, Pitt TL. 1995. Auxotrophic variants of Pseudomonas aeruginosa are selected from prototrophic wild-type strains in respiratory infections in patients with cystic fibrosis. J. Clin. Microbiol. 33:37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burns JL, et al. 1998. Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clin. Infect. Dis. 27:158–163 [DOI] [PubMed] [Google Scholar]
- 6. Burns JL, et al. 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 183:444–452 [DOI] [PubMed] [Google Scholar]
- 7. Ciofu O, Mandsberg LF, Bjarnsholt T, Wassermann T, Hoiby N. 2010. Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants. Microbiology 156:1108–1119 [DOI] [PubMed] [Google Scholar]
- 7a. Clinical and Laboratory Standards Institute 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. CLSI document M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Cramer N, Wiehlmann L, Tummler B. 2010. Clonal epidemiology of Pseudomonas aeruginosa in cystic fibrosis. Int. J. Med. Microbiol. 300:526–533 [DOI] [PubMed] [Google Scholar]
- 9. Dale C, Moran NA. 2006. Molecular interactions between bacterial symbionts and their hosts. Cell 126:453–465 [DOI] [PubMed] [Google Scholar]
- 10. Feliziani S, et al. 2010. Mucoidy, quorum sensing, mismatch repair and antibiotic resistance in Pseudomonas aeruginosa from cystic fibrosis chronic airways infections. PLoS One 5:e12669 doi:10.1371/journal.pone.0012669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Foweraker J. 2009. Recent advances in the microbiology of respiratory tract infection in cystic fibrosis. Br. Med. Bull. 89:93–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Foweraker JE, Laughton CR, Brown DF, Bilton D. 2005. Phenotypic variability of Pseudomonas aeruginosa in sputa from patients with acute infective exacerbation of cystic fibrosis and its impact on the validity of antimicrobial susceptibility testing. J. Antimicrob. Chemother. 55:921–927 [DOI] [PubMed] [Google Scholar]
- 13. Gleckman R, Blagg N, Joubert DW. 1981. Trimethoprim: mechanisms of action, antimicrobial activity, bacterial resistance, pharmacokinetics, adverse reactions, and therapeutic indications. Pharmacotherapy 1:14–20 [DOI] [PubMed] [Google Scholar]
- 14. Gordon KA, Jones RN. 2003. Susceptibility patterns of orally administered antimicrobials among urinary tract infection pathogens from hospitalized patients in North America: comparison report to Europe and Latin America. Results from the SENTRY Antimicrobial Surveillance Program (2000). Diagn. Microbiol. Infect. Dis. 45:295–301 [DOI] [PubMed] [Google Scholar]
- 15. Grasemann H, Kurtz F, Ratjen F. 2006. Inhaled L-arginine improves exhaled nitric oxide and pulmonary function in patients with cystic fibrosis. Am. J. Respir. Crit. Care Med. 174:208–212 [DOI] [PubMed] [Google Scholar]
- 16. Grasemann H, Michler E, Wallot M, Ratjen F. 1997. Decreased concentration of exhaled nitric oxide (NO) in patients with cystic fibrosis. Pediatr. Pulmonol. 24:173–177 [DOI] [PubMed] [Google Scholar]
- 17. Green SK, Schroth MN, Cho JJ, Kominos SK, Vitanza-jack VB. 1974. Agricultural plants and soil as a reservoir for Pseudomonas aeruginosa. Appl. Microbiol. 28:987–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haussler S, Tummler B, Weissbrodt H, Rohde M, Steinmetz I. 1999. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin. Infect. Dis. 29:621–625 [DOI] [PubMed] [Google Scholar]
- 19. Heurlier K, et al. 2005. Quorum-sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J. Bacteriol. 187:4875–4883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoffman LR, et al. 2009. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J. Cyst. Fibros. 8:66–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hogardt M, Heesemann J. 2010. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int. J. Med. Microbiol. 300:557–562 [DOI] [PubMed] [Google Scholar]
- 22. Kohler T, et al. 1996. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40:2288–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lane N. 2010. Chance or necessity? Bioenergetics and the probability of life. J. Cosmol. 10:19 [Google Scholar]
- 24. Lee DG, et al. 2006. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 7:R90 doi:10.1186/gb-2006-7-10-r90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Llanes C, et al. 2004. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 48:1797–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luiking YC, Poeze M, Dejong CH, Ramsay G, Deutz NE. 2004. Sepsis: an arginine deficiency state? Crit. Care Med. 32:2135–2145 [DOI] [PubMed] [Google Scholar]
- 27. Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051–1060 [DOI] [PubMed] [Google Scholar]
- 28. McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10:13–26 [DOI] [PubMed] [Google Scholar]
- 29. Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42:165–190 [DOI] [PubMed] [Google Scholar]
- 30. Mowat E, et al. 2011. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am. J. Respir. Crit. Care Med. 183:1674–1679 [DOI] [PubMed] [Google Scholar]
- 31. Munder M. 2009. Arginase: an emerging key player in the mammalian immune system. Br. J. Pharmacol. 158:638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pitt TL, Sparrow M, Warner M, Stefanidou M. 2003. Survey of resistance of Pseudomonas aeruginosa from UK patients with cystic fibrosis to six commonly prescribed antimicrobial agents. Thorax 58:794–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Poole K, Srikumar R. 2001. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Top. Med. Chem. 1:59–71 [DOI] [PubMed] [Google Scholar]
- 34. Proctor RA, et al. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4:295–305 [DOI] [PubMed] [Google Scholar]
- 35. Qin X, et al. 2003. Use of real-time PCR with multiple targets to identify Pseudomonas aeruginosa and other nonfermenting gram-negative bacilli from patients with cystic fibrosis. J. Clin. Microbiol. 41:4312–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ratcliff WC, Denison RF, Borrello M, Travisano M. 2012. Experimental evolution of multicellularity. Proc. Natl. Acad. Sci. U. S. A. 109:1595–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Regnath T, Kreutzberger M, Illing S, Oehme R, Liesenfeld O. 2004. Prevalence of Pseudomonas aeruginosa in households of patients with cystic fibrosis. Int. J. Hyg. Environ. Health 207:585–588 [DOI] [PubMed] [Google Scholar]
- 38. Ricciardolo FL, Sterk PJ, Gaston B, Folkerts G. 2004. Nitric oxide in health and disease of the respiratory system. Physiol. Rev. 84:731–765 [DOI] [PubMed] [Google Scholar]
- 39. Richards RM, Hamilton VE, Thomas MR. 1998. In-vitro investigation of the antibacterial activity of agents which may be used for the oral treatment of lung infections in CF patients. J. Antimicrob. Chemother. 42:171–178 [DOI] [PubMed] [Google Scholar]
- 40. Rosenfeld M, Ramsey BW, Gibson RL. 2003. Pseudomonas acquisition in young patients with cystic fibrosis: pathophysiology, diagnosis, and management. Curr. Opin. Pulm. Med. 9:492–497 [DOI] [PubMed] [Google Scholar]
- 41. Ruby EG. 2008. Symbiotic conversations are revealed under genetic interrogation. Nat. Rev. Microbiol. 6:752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Seale TW, Thirkill H, Tarpay M, Flux M, Rennert OM. 1979. Serotypes and antibiotic susceptibilities of Pseudomonas aeruginosa isolates from single sputa of cystic fibrosis patients. J. Clin. Microbiol. 9:72–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Searcy DG. 2004. Nutritional syntrophies and consortia as models for the origin of mitochondria, p 163–183 In Seckbach J. (ed), Symbiosis: mechanisms and model systems, vol 4 Cellular origin, life in extreme habitats and astrobiology. Kluwer Academic Publishers, New York, NY [Google Scholar]
- 44. Smith EE, et al. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103:8487–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Son MS, Matthews WJ, Jr, Kang Y, Nguyen DT, Hoang TT. 2007. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect. Immun. 75:5313–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taylor RF, Hodson ME, Pitt TL. 1993. Adult cystic fibrosis: association of acute pulmonary exacerbations and increasing severity of lung disease with auxotrophic mutants of Pseudomonas aeruginosa. Thorax 48:1002–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Traczewski MM, Brown SD. 2006. In vitro activity of doripenem against Pseudomonas aeruginosa and Burkholderia cepacia isolates from both cystic fibrosis and non-cystic fibrosis patients. Antimicrob. Agents Chemother. 50:819–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vetsigian K, Jajoo R, Kishony R. 2011. Structure and evolution of Streptomyces interaction networks in soil and in silico. PLoS Biol. 9:e1001184 doi:10.1371/journal.pbio.1001184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vettoretti L, et al. 2009. Efflux unbalance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 53:1987–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weldhagen GF. 2004. Integrons and beta-lactamases—a novel perspective on resistance. Int. J. Antimicrob. Agents 23:556–562 [DOI] [PubMed] [Google Scholar]
- 51. Wolter DJ, Black JA, Lister PD, Hanson ND. 2009. Multiple genotypic changes in hypersusceptible strains of Pseudomonas aeruginosa isolated from cystic fibrosis patients do not always correlate with the phenotype. J. Antimicrob. Chemother. 64:294–300 [DOI] [PubMed] [Google Scholar]
- 52. Yim G, Wang HH, Davies JFRS. 2007. Antibiotics as signalling molecules. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362:1195–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.