Abstract
A steady decline in macrolide resistance among Streptococcus pyogenes (group A streptococci [GAS]) in Portugal was reported during 1999 to 2006. This was accompanied by alterations in the prevalence of macrolide resistance phenotypes and in the clonal composition of the population. In order to test whether changes in the macrolide-resistant population reflected the same changing patterns of the overall population, we characterized both macrolide-susceptible and -resistant GAS associated with a diagnosis of tonsillo-pharyngitis recovered in the period from 2000 to 2005 in Portugal. Pulsed-field gel electrophoresis (PFGE) profiling was the best predictor of emm type and the only typing method that could discriminate clones associated with macrolide resistance and susceptibility within each emm type. Six PFGE clusters were significantly associated with macrolide susceptibility: T3-emm3-ST406, T4-emm4-ST39, T1-emm1-ST28, T6-emm6-ST382, B3264-emm89-ST101/ST408, and T2-emm2-ST55. Four PFGE clusters were associated with macrolide resistance: T4-emm4-ST39, T28-emm28-ST52, T12-emm22-ST46, and T1-emm1-ST28. We found no evidence for frequent ongoing horizontal transfer of macrolide resistance determinants. The diversity of the macrolide-resistant population was lower than that of susceptible isolates. The differences found between the two populations suggest that the macrolide-resistant population of GAS has its own dynamics, independent of the behavior of the susceptible population.
INTRODUCTION
Streptococcus pyogenes (Lancefield group A streptococci [GAS]), is the most common bacterial agent implicated in acute pharyngitis and can also cause a variety of skin and soft tissue infections, as well as severe invasive disease. Although penicillin remains the antibiotic of choice in the treatment of GAS infections, macrolides and lincosamides are recommended as suitable alternatives for patients who are allergic to penicillin (4).
High macrolide resistance in GAS was reported in most southern European countries, such as Spain, Greece, Italy, and Portugal (14, 15, 19, 22, 25, 27), but this was not a characteristic of all European countries (22). Although differences in the prevalence of particular macrolide resistance genotypes and emm types were documented, the majority of these emm types shared the same resistance determinants, suggesting a broad geographical dissemination of a few clones. Isolates fully susceptible to macrolides that share the same pulsed-field gel electrophoresis (PFGE) profiles or the same sequence types (STs) as these major clones have been described, and several resistance determinants have been associated with each resistant lineage. Taken together, these data imply that independent acquisition of resistance genes by the same prevalent clones followed by local dissemination could have also played a role in conditioning the successful macrolide resistance phenotypes and clones in particular geographic locations.
An association between certain emm types and macrolide resistance was documented (1, 7, 16, 17, 23, 29), but in most of these studies the characterization of the S. pyogenes isolates was limited to emm typing. However, it was recently shown that this typing technique is not sufficient to unambiguously identify GAS clones. For the precise identification of genetic lineages, emm typing should be complemented with other typing methods, such as PFGE or multilocus sequence typing (MLST) (5).
In Portugal, we noted a steady decline in macrolide resistance among S. pyogenes isolates in 1999 to 2006, decreasing from 20% in 1999 to 12% in 2006 (25). This was accompanied by large fluctuations of the macrolide resistance phenotypes, as well as changes in the clonal composition of the population (25–27). Given these results, we hypothesized that the changing patterns of macrolide resistance could also be reflecting fluctuations in the overall population, implying that important causes for this fluctuation lay outside antibiotic usage and reflected other selective forces acting on the entire GAS population.
The aims of this study were to characterize the macrolide-susceptible population of GAS and to compare it with the macrolide-resistant population to determine how much the dynamics of macrolide-resistant isolates could be mirroring the behavior of the overall GAS population.
MATERIALS AND METHODS
Bacterial isolates and identification.
A total of 1,606 S. pyogenes isolates recovered from throat swabs and associated with a diagnosis of tonsillo-pharyngitis were collected from 32 microbiology laboratories located throughout Portugal from January 2000 to December 2005. In this period, erythromycin resistance declined from 28% in 2000 to 11% in 2005 (25). Pharyngitis is frequently managed in Portugal without a microbiological investigation, with these being performed mostly for epidemiological purposes. On the other hand, the availability of rapid antigen tests means that an isolate will not always be recovered, even when an etiological diagnosis is sought. The participating laboratories were asked to submit all nonduplicate S. pyogenes isolates obtained from outpatients during the study period, but no audit was performed to ensure compliance, and this is known to be variable in this kind of study. The combination of these factors may have contributed to a lower number of isolates than anticipated, but we have no reason to suspect that there was a bias in the isolates that were submitted. The isolates were distributed in the study period as follows: 214 in 2000, 216 in 2001, 270 in 2002, 230 in 2003, 284 in 2004, and 392 in 2005. Isolates were identified to the species level by colony morphology, beta-hemolysis on sheep blood agar, and a commercial latex agglutination technique (Slidex Strepto A; bioMérieux, Marcy l'Etoile, France). A total of 803 isolates, randomly chosen and representing 50% of the total collection, were characterized. Among this group were 155 macrolide-resistant isolates (19.3%). The distribution of the macrolide-resistant isolates included in this study was as follows: 23 in 2000, 19 in 2001, 23 in 2002, 38 in 2003, 30 in 2004, and 22 in 2005. These isolates represent a subset of the total number of macrolide-resistant isolates found during 2000 to 2005 (n = 318).
Antimicrobial susceptibility testing.
Susceptibilities to erythromycin and clindamycin were tested by disk diffusion according to the recommendations of CLSI (6) and were reported previously (25–27). The macrolide resistance phenotype was determined according to previously published procedures (27).
DNA extraction.
Total bacterial DNA was isolated according to the methodology described by the Centers for Disease Control and Prevention (CDC) (http://www.cdc.gov/ncidod/biotech/strep/protocols.htm).
T and emm typing.
emm typing was performed as described by the CDC (http://www.cdc.gov/ncidod/biotech/strep/protocols.htm). Amplification products were purified using the High Pure PCR purification kit (Roche, Mannheim, Germany) according to the manufacturer's instructions and sequenced using primer emmseq2 (2), and the DNA sequences were searched against the emm sequences deposited in the CDC database (http://www.cdc.gov/ncidod/biotech/strep/strepindex.htm).
PFGE and MLST.
Pulsed-field gel electrophoresis (PFGE) was performed as previously described (26). All the macrolide-susceptible isolates were digested with SmaI, and isoschizomer Cfr9I was used only for isolates presenting the M phenotype (26). The BioNumerics software (Applied-Maths, Sint-Martens-Latem, Belgium) was used to create unweighted-pair group method with arithmetic mean (UPGMA) dendrograms of the SmaI or Cfr9I fragment patterns. The Dice similarity coefficient was used, with optimization and position tolerance settings of 1.0 and 1.5, respectively. PFGE clusters were defined as groups of isolates with ≥80% similarity in the dendrogram (5). A PFGE-based cluster was considered to be a major lineage if it included ≥10 isolates. Clusters that included between 5 and 10 isolates were considered minor PFGE clusters. Major lineages were identified by uppercase letters and minor clones by lowercase letters. The subscript number in the PFGE cluster designation identifies the number of isolates included in the cluster.
Multilocus sequence typing (MLST) analysis was performed for at least 30% of randomly selected isolates of each emm type that was represented by more than 10 isolates (n = 282) as previously described (8), and allele and sequence type (ST) were attributed using the S. pyogenes MLST database (spyogenes.mlst.net). The relationships between the MLST STs were determined using the goeBURST algorithm (9) implemented in PHYLOViZ (10) with the complete S. pyogenes database available at spyogenes.mlst.net.
Statistical analysis.
In order to compare the probability of a given emm type or PFGE clone being more associated with macrolide resistance or susceptibility, the odds ratios (OR) and the corresponding P values, corrected for multiple testing through the false-discovery rate (FDR) linear procedure (3), were calculated by reference to all other emm types and PFGE clones.
Adjusted Wallace (AW) coefficients were used to compare partitions, and Simpson's index of diversity (SID) and the corresponding 95% confidence intervals (CIs) were used to evaluate the clonal diversity of the macrolide-susceptible and -resistant isolates (5, 24). Trends were evaluated using the Cochran-Armitage test of trend, and yearly variability was tested using the Fisher exact test. P values of <0.05 were considered significant.
RESULTS
Clonal characterization.
The characteristics of the 803 isolates included in this study, as well as their distribution during the study period, are summarized in Table 1. Among the 34 different emm types found, the most frequent were emm4 (15.2%, n = 122), emm1 and emm3 (9.6%, n = 77 each), emm12 (9.3%, n = 75), emm6 (7.2%, n = 58), and emm89 (7%, n = 56). The remaining 28 emm types accounted for 42.1% of the isolates (n = 338). Among the 16 T types found in the collection, the most prevalent were T4 (13.8%, n = 111), T12 (12.5%, n = 100), T1 (9.7%, n = 78), T28 (8.5%, n = 68), and B3264 (8.3%, n = 67). Slightly under 8% of the isolates (n = 64) were T nontypeable.
Table 1.
Properties of the 803 GAS isolates collected from tonsillo-pharyngitis in Portugal
| emm type (no. of isolates) | PFGE cluster (R/S)a | T type(s)b (no. of isolates) | ST(s) (no. of isolates)c | No. of isolates recovered in: |
|||||
|---|---|---|---|---|---|---|---|---|---|
| 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | ||||
| 4 (122) | B66 (1/64) | 4 (63), 3 (1), NT (1) | [39 (12), 566 (1), 635 (1)] | 2 | 2 | 10 | 12 | 10 | 29 |
| G49 (45/2) | 4 (39), B3264 (2), 13 (1), 2/4 (1), 5/27/44 (1), 6 (1), 12 (1), 1 (1) | [39 (16), 561 (1), 637 (1), 638 (1)] | 4 | 0 | 6 | 16 | 13 | 8 | |
| Other (2/8) | 4 (8), 6 (1), 2 (1) | 39 (6), 15 (1), 55 (1) | 0 | 0 | 1 | 2 | 0 | 7 | |
| 1 (77) | C62 (0/62) | 1 (61), NT (1) | [28 (16), 567 (1)] | 7 | 6 | 8 | 7 | 20 | 14 |
| R12 (12/0) | 1 (10), 13 (1), NT (1) | 28 (7) | 1 | 2 | 4 | 4 | 0 | 1 | |
| Other (1/2) | 1 (3) | 28 (1) | 0 | 1 | 2 | 0 | 0 | 0 | |
| 3 (77) | A76 (0/76) | 3 (40), NT (18), 3/13 (17), 5/27/44 (1) | [406 (14), 315 (5), 15 (4), 560 (1)] | 1 | 8 | 30 | 4 | 3 | 30 |
| Other (0/1) | NT (1) | 15 (1) | 0 | 0 | 0 | 0 | 0 | 1 | |
| 12 (75) | D59 (8/42) | 12 (40), NT (9), 2 (1) | [36 (14), 551 (1)] | 9 | 10 | 6 | 11 | 5 | 9 |
| S10 (5/5) | 12 (9), NT (1) | 36 (5) | 1 | 2 | 1 | 2 | 3 | 1 | |
| A9 (0/8) | 12 (6), NT (2) | 36 (3) | 1 | 0 | 2 | 2 | 1 | 2 | |
| Other (4/3) | 12 (6), 25 (1) | 36 (3), 150 (1) | 3 | 0 | 4 | 0 | 0 | 0 | |
| 6 (58) | E58 (1/56) | 6 (51), NT (4), 5/27/44 (1), 2 (1) | [382 (18), 411 (1)] | 13 | 10 | 8 | 4 | 2 | 20 |
| Other (0/1) | 6 (1) | 382 (1) | 1 | 0 | 0 | 0 | 0 | 0 | |
| 89 (56) | F53 (1/51) | B3264 (50), 3/13 (1), 28 (1) | [101 (5), 408 (5), 553 (1), 568 (3)] | 1 | 8 | 5 | 11 | 15 | 12 |
| Other (0/4) | B3264 (3), 5/27/44 (1) | [101 (2), 568 (1)] 555 (1) | 0 | 1 | 0 | 0 | 1 | 2 | |
| 75 (49) | L25 (8/16) | 25 (24) | [150 (7), 563 (1)] | 3 | 6 | 8 | 6 | 1 | 0 |
| Q13 (0/13) | 25 (13) | [150 (3), 564 (1), 639 (1)] | 1 | 1 | 2 | 2 | 5 | 2 | |
| Other (4/8) | 25 (11), NT (1) | 150 (5), 481 (1) | 1 | 1 | 0 | 3 | 5 | 2 | |
| 28 (45) | H38 (26/7) | 28 (32), NT (1) | 52 (9) | 4 | 8 | 5 | 3 | 5 | 8 |
| P13 (1/9) | 28 (10) | 52 (5) | 0 | 2 | 3 | 1 | 3 | 1 | |
| Other (0/2) | 28 (1), NT (1) | [52 (1), 456 (1)] | 0 | 0 | 2 | 0 | 0 | 0 | |
| 2 (38) | I32 (0/30) | 2 (29), 4 (1) | [55 (7), 634 (1)] | 4 | 6 | 3 | 3 | 9 | 5 |
| Other (2/6) | 2 (7), 12 (1) | [55 (4), 636 (1)] | 1 | 1 | 1 | 1 | 1 | 3 | |
| 22 (35) | K27 (21/4) | 12 (25) | [46 (6), 389 (1)] | 7 | 7 | 1 | 4 | 5 | 1 |
| Other (6/4) | 12 (10) | [46 (2), 389 (1)], 52 (2) | 0 | 2 | 1 | 5 | 2 | 0 | |
| 44/61 (33) | J32 (1/30) | 5/27/44 (28), NT (2), 12 (1) | [25 (10), 554 (1)] | 0 | 0 | 0 | 2 | 14 | 15 |
| Other (0/2) | 5/27/44 (2) | 555 (1) | 0 | 0 | 1 | 0 | 0 | 1 | |
| 87 (25) | N20 (0/18) | 28 (16), NT (2) | 62 (6) | 4 | 3 | 2 | 3 | 3 | 3 |
| i5 (0/5) | 28 (3), 1 (1), NT (1) | 62 (2) | 0 | 0 | 0 | 1 | 0 | 4 | |
| Other (0/2) | 28 (1), 6 (1) | 0 | 0 | 0 | 0 | 0 | 2 | ||
| 9 (23) | M24 (0/19) | 9 (19) | 75 (4) | 8 | 3 | 3 | 1 | 2 | 2 |
| Other (3/1) | 9 (4) | 75 (4) | 2 | 0 | 1 | 0 | 0 | 1 | |
| 77 (17) | b8 (0/8) | 13 (8) | 63 (3) | 2 | 1 | 1 | 2 | 2 | 0 |
| Other (0/9) | 13 (5), NT (2), 28 (1), 2 (1) | 63 (3) | 1 | 2 | 5 | 0 | 0 | 1 | |
| 78 (15) | O15 (0/15) | 11 (15) | [253 (5), 409 (1)] | 5 | 2 | 1 | 0 | 2 | 5 |
| 94 (11) | D59 (0/9) | B3264 (7), 1 (1), NT (1) | 89 (4) | 2 | 1 | 1 | 1 | 1 | 3 |
| Other (0/2) | B3264 (2) | 0 | 0 | 2 | 0 | 0 | 0 | ||
| Otherd | Multiplee | Multiplef | Multipleg | 18 | 12 | 5 | 2 | 9 | 1 |
PFGE clones defined as major lineages are designated by uppercase letters, and minor PFGE clones (10 > n ≥ 5) are designated by lowercase letters. Whenever a PFGE clone included fewer than 5 isolates of that particular emm type, it was not discriminated, and all such isolates were grouped under “other.” The numbers of resistant (R) and susceptible (S) isolates are indicated in parenthesis.
NT, nontypeable.
Brackets indicate STs that belong to the same clonal complex defined by the goeBURST algorithm (http://goeburst.phyloviz.net/) with the complete S. pyogenes database available at spyogenes.mlst.net.
18 emm types (47 isolates): 11 (9), 102 (9), 58 (5), 48 (3), 74 (2), 18 (2), 29 (2), 68 (2), 53 (2), st7700 (2), 118 (2), 43 (1), 73 (1), 70 (1), 64 (1), 25 (1), 5 (1), and st38 (1).
Twenty-one PFGE clones.
12 T types: NT (15), 11 (8), 13 (5), 3/13 (5), B3264 (3), 28 (3), 9 (2), 3 (2), 25 (1), 12 (1), 18 (1), and 1 (1).
17 STs: 60 (3), 403 (2), 562 (2), 410 (2), 247 (1), 331 (1), 161 (1), 176 (1), 402 (1), 552 (1), 559 (1), 167 (1), 565 (1), 569 (1), 120 (1), 63 (1), and 12 (1).
The 803 isolates were classified into 62 PFGE clusters, including 19 major lineages representing 684 isolates (85%) (Fig. 1). Nine minor PFGE clusters were found, grouping 60 isolates (7.5%). In most cases, each PFGE cluster had a dominant macrolide resistance phenotype (susceptible, MLSB, or M), emm type, T type, and STs belonging to the same clonal complex (Table 1). The MLSB isolates were grouped mainly into PFGE cluster H38 (48%, n = 30) and cluster K27 (33%, n = 21), while 49% of isolates presenting the M phenotype (n = 45) were included into cluster G49.
Fig 1.
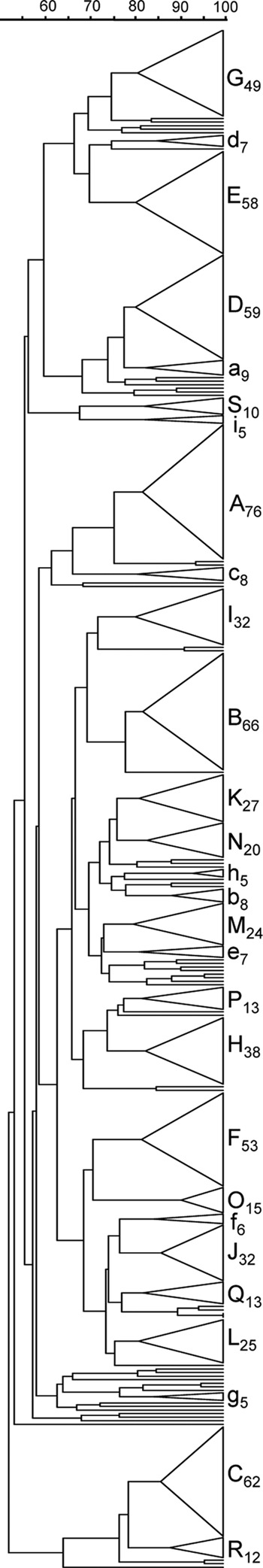
Dendrogram showing the PFGE profiles of the 803 GAS isolates. The dendrogram was constructed using the unweighted-pair group with arithmetic mean (UPGMA) method. Dice coefficients (percentages) are indicated in the scale above the dendrogram. For each PFGE cluster, a triangle proportional to the number of isolates is shown in the dendrogram. The clusters are designated by letters, with major clusters (n ≥ 10) identified by uppercase letters and minor PFGE clusters (10 > n ≥ 5) identified by lowercase letters. The subscript numbers indicate the number of isolates included in the cluster.
MLST analysis was performed for 282 isolates (35.1%). We identified 4 novel alleles among the genes used in MLST, two in gki (gki108 and gki109), one in mutS (muts63) and one in recP (recp101). New allele combinations producing novel STs were also noted and were numbered ST559 to ST569 and ST634 to ST639. The new alleles and the novel STs were submitted to the S. pyogenes MLST database (spyogenes.mlst.net). Isolates of the same emm type presented mostly the same ST or STs of the same clonal complex (Table 1).
Differences between PFGE clusters and emm types between macrolide-resistant and -susceptible isolates.
The SID values of all typing methods and the corresponding 95% CIs were determined for the macrolide-resistant and -susceptible subsets to ascertain whether there were differences in the diversities of the two populations (Table 2). For all the typing methods used to characterize the entire collection, the diversity of the macrolide-resistant population was lower than the diversity of the susceptible population. This not only is evident from the lower number of different types found among macrolide-resistant isolates but also is reflected in the significantly lower SID values (Table 2). For the subset of isolates analyzed by MLST, the diversity of the macrolide-resistant population was also lower.
Table 2.
Simpson's indices of diversity and 95% confidence intervals of the typing methods used in the analysis
| Typing method | SID (95% CI) [no. of partitions] |
|
|---|---|---|
| Macrolide-resistant isolates (n = 155) | Macrolide-susceptible isolates (n = 648) | |
| PFGE | 0.851 (0.817–0.884) [22] | 0.938 (0.932–0.944) [58] |
| emm typing | 0.823 (0.792–0.854) [12] | 0.927 (0.922–0.933) [34] |
| T typing | 0.819 (0.787–0.850) [14] | 0.924 (0.920–0.928) [16] |
| MLST | 0.889 (0.853–0.924) [17] | 0.959 (0.951–0.966) [52] |
Among the emm types that were represented by more than 10 isolates, none included solely resistant isolates. However, some were found only in macrolide-susceptible isolates: emm3, emm87, emm77, emm78, and emm94.
The calculation of OR supported the association of emm3 and emm87 with macrolide susceptibility (P < 0.001 and P = 0.007, respectively, both of which are significant for FDR) but not that of emm77, emm78, and emm94. emm types 6 and 89 were also associated with macrolide susceptibility, although both included one resistant isolate (both with a P value of <0.001, which is significant for FDR). On the other hand, emm types 4, 22, and 28 were associated with macrolide resistance (all with a P value of <0.001, which is significant for FDR).
Nine PFGE clusters that included only susceptible isolates were also identified (Table 1). In contrast, only one PFGE cluster, R12 (mostly T1-emm1-ST28), grouped exclusively resistant isolates. All the other PFGE clusters included both susceptible and resistant isolates.
Similarly to what was found for emm types, the calculation of OR for the PFGE major clusters supported only the association of some clusters with the macrolide resistance phenotype. The association with susceptibility of PFGE clusters A76 (mostly T3-emm3-ST406), C62 (mostly T1-emm1-ST28), and I32 (mostly T2-emm2-ST55) was statistically supported (P < 0.001, P < 0.001, and P = 0.002, respectively, all of which are significant for FDR). Additionally, PFGE clusters B66 (mostly T4-emm4-ST39), E58 (mostly T6-emm6-ST382), and F53 (mostly B3264-emm89-ST101/ST408), all including one resistant isolate, were also found to be associated with susceptibility (all with a P value of <0.001, which is significant for FDR). In contrast, PFGE clusters G49 (mostly T4-emm4-ST39), H38 (mostly T28-emm28-ST52), K27 (mostly T12-emm22-ST46), and R12 (mostly T1-emm1-ST28) were associated with macrolide resistance (all with a P value of <0.001, which is significant for FDR).
Correspondence between typing methods and differences between macrolide-susceptible and -resistant isolates.
The AW coefficient calculated for the relationship between PFGE and macrolide susceptibility was high (AWPFGE→Ery = 0.715; 95% CI, 0.642 to 0.788), while it was extremely low for emm type (AWemm→Ery = 0.095; 95% CI, 0.016 to 0.175). The AW coefficients also indicated that among the typing methods used in this work, PFGE was the best predictor of emm type. This was valid considering all the isolates included in the collection and the macrolide-resistant and -susceptible subsets (AWPFGE→emm, all = 0.914; 95% CI, 0.889 to 0.939). The inverse relationship was significantly weaker (AWemm→PFGE, all = 0.598; 95% CI, 0.565 to 0.630). When considering only the macrolide-resistant or -susceptible subsets, the relationship was much stronger (AWemm→PFGE, Eryr = 0.754 [95% CI, 0.658 to 0.850]; AWemm→PFGE, Erys = 0.787 [95% CI, 0.747 to 0.827]), reflecting the discrimination by PFGE of macrolide-resistant and macrolide-susceptible lineages of the same emm types. For the subset of isolates analyzed by MLST (n = 282), the ST was an excellent predictor of emm type (AWST→emm = 0.963; 95% CI, 0.937 to 0.988), but the inverse relationship was significantly weaker (AWemm→ST = 0.678; 95% CI, 0.602 to 0.753).
Changes of macrolide resistance within emm types with time.
Since particular genetic lineages were found to be associated with macrolide resistance, we tested whether the distribution of macrolide-resistant isolates changed with time within each of the emm types with ≥10 isolates in the studied period. Changes were noted in emm1 (P = 0.016) and emm4 (P < 0.001), but only the latter was significant for FDR. This indicates that while the proportion of macrolide-resistant isolates remained stable in time for most emm types, that for emm4 changed significantly. An increase in emm4 isolates was noted during the study period (Cochran-Armitage test for trend, P < 0.001). However, this increase reflects the underlying dynamics of its two major PFGE clusters. The increase in recent years of the macrolide-susceptible PFGE cluster B66, which peaked in the last year of the study, is in contrast to the dramatic increase of the macrolide-resistant PFGE cluster G49 in the early years, to a peak in 2003, and its current decline (Table 1).
DISCUSSION
The changes in the prevalence of macrolide resistance phenotypes noted in Portugal were accompanied by alterations in the clonal composition of the population (25). At this point, it remained unclear whether all these changes were a reflection of large fluctuations in the overall GAS population or whether macrolide-resistant S. pyogenes isolates have their own dynamics. In this work, we compared macrolide-resistant and -susceptible GAS isolates in the period from 2000 to 2005 in order to gain new insights into the reasons behind the fluctuations reported for the resistant population.
In this study, a statistically significant association was found between macrolide resistance and emm4, emm22, and emm28, of which emm4 and emm22 were also found in association with macrolide resistance elsewhere (1, 16, 23, 29). However, our findings are in contrast to a report from Korea that indicated a relationship between these emm types and macrolide susceptibility (12). It was also possible to establish significant associations between macrolide susceptibility and emm3, emm6, emm87, and emm89. While emm3 was also previously reported in association with macrolide susceptibility in Spain (1) and in Italy (29), emm89 was previously found in association with macrolide resistance (29), contrary to our findings. Associations between certain emm types and macrolide resistance and susceptibility were reported in other studies but were not detected in Portugal, such as for emm12 and emm2, which were often associated with macrolide resistance (7, 17, 29), and emm1 isolates, which were frequently found in association with macrolide susceptibility (1, 16).
PFGE was the only typing method capable of differentiating resistant isolates of the same emm type. For emm4 and emm1, PFGE grouped the isolates into two major PFGE clusters (B66 and G49 for emm4 isolates and C62 and R12 for emm1 isolates), associated with macrolide susceptibility and resistance, respectively. Associations between some PFGE clusters and macrolide susceptibility that could not be detected by emm type alone were identified. This was the case for clusters C62 and R12, including emm1 macrolide-susceptible and -resistant isolates, respectively, and for cluster I32, including emm2 isolates. None of these emm types was statistically associated with macrolide susceptibility. These observations reinforce the usefulness and the importance of using PFGE to characterize GAS isolates.
In order to compare the two populations, the SID values for all the typing methods used in this study were calculated (Table 2). There were differences in the number of types and SID values between the macrolide-resistant and -susceptible subsets for PFGE, emm typing, and T typing. The diversity of the resistant population was lower than that of the susceptible subset by all three methods. The SID values for the fraction of the isolates characterized by MLST were also lower in the macrolide-resistant population.
The association of certain emm types and PFGE clones with macrolide resistance or susceptibility and the differences in the diversity of the two groups indicate that the macrolide-resistant isolates represent lineages distinct from the susceptible population. The discrepancies in associations between emm types and macrolide resistance found in different geographical regions could be due to the circulation of different lineages expressing these emm types in different areas.
If the diversity of the resistant population previously reported (25–27) was a reflection of the diversity of the overall GAS population, the emm types and the PFGE clusters would not show these associations, and the diversities of the two populations would be approximately the same. Some emm types are never or rarely found in resistant isolates, even though their prevalence in susceptible isolates is high. This is the case for emm3, emm6, and emm89. In 2002 and 2005, 22% and 15% of the isolates, respectively, were emm3, and no macrolide-resistant isolates were detected. Fifty-seven out of the 58 emm6 isolates were macrolide susceptible, and while in 2005, 10% of the isolates presented this emm type, no acquisition of macrolide resistance determinants was detected. The same situation occurred with emm89, which increased throughout the study period, but a single resistant isolate was found. Taken together, these data suggest that there is limited ongoing transfer of macrolide resistance determinants.
The narrow group of resistant lineages is quite stable and has disseminated widely (11, 13, 18, 20, 21, 28). Whether their success is dependent solely on their association with macrolide resistance or may also reflect other clonal properties remains to be shown. The characterization of contemporaneous macrolide-resistant and -susceptible isolates revealed distinct populations that changed independently in the study period.
ACKNOWLEDGMENTS
Members of the Portuguese Group for the Study of Streptococcal Infections are Teresa Vaz, Marília Gião, Rui Ferreira, and Iryna Klyeshtorna (Centro Hospitalar do Barlavento Algarvio), Ana Buschy Fonseca (Hospital de Cascais), Henrique Oliveira (Centro Hospitalar de Coimbra), Ana Cristina Silva, Hermínia Costa, Maria Fátima Silva, and Maria Amélia Afonso (Centro Hospitalar de Entre Douro e Vouga), Margarida Pinto, Odete Chantre, João Marques, Isabel Peres, Isabel Daniel, and Cristina Marcelo (Centro Hospitalar de Lisboa Central), Lurdes Monteiro and Luís Marques Lito (Centro Hospitalar Lisboa Norte), Teresa Marques, Maria Ana Pessanha, and Elsa Gonçalves (Centro Hospitalar Lisboa Ocidental), Paulo Lopes, Luísa Felício, and Angelina Lameirão (Centro Hospitalar de Vila Nova de Gaia/Espinho), Ana Paula Mota Vieira and Margarida Tomaz (Centro Hospitalar do Alto Ave), Rosa Bento (Centro Hospitalar do Baixo Alentejo), Maria Helena Ramos and Ana Apula Castro (Centro Hospitalar do Porto), Fernando Fonseca (Centro Hospitalar da Póvoa do Varzim/Vila do Conde), Ana Paula Castro (Centro Hospitalar Trás-os-Montes e Alto Douro), Graça Ribeiro, Luísa Boaventura, Catarina Chaves, and Teresa Reis (Hospitais da Universidade de Coimbra), Nuno Canhoto and Teresa Afonso (Hospital Central do Funchal), Teresa Pina and Helena Peres (Hospital Curry Cabral, Lisbon), Ilse Fontes and Paulo Martinho (Hospital de Santa Luzia, Elvas), Ana Domingos and Gina Marrão (Hospital de Santo André, Leiria), Manuela Ribeiro and Helena Gonçalves (Hospital de São João, Porto), Maria Alberta Faustino, Maria Carmen Iglesias, and Adelaide Alves (Hospital de Braga), Maria Paula Pinheiro and R. Semedo (Hospital Dr. José Maria Grande, Portalegre), Adriana Coutinho (Hospital do Espírito Santo, Évora), Luísa Cabral and Olga Neto (Hospital dos SAMS, Lisbon), Luísa Sancho (Hospital Dr. Fernando da Fonseca, Amadora/Sintra), José Diogo, Ana Rodrigues, and Isabel Nascimento (Hospital Garcia de Orta, Almada), Elmano Ramalheira and Raquel Diaz (Hospital Infante D. Pedro, Aveiro), José Miguel Ribeiro, Isabel Vale, and Ana Carvalho (Hospital de São Teotónio, Viseu), Maria Antónia Read, Margarida Monteiro, and Valquíria Alves (Hospital Pedro Hispano, Matosinhos), Engrácia Raposo, Maria Lurdes Magalhães, Helena Rochas, and Anabela Silva (Instituto Nacional de Saúde Ricardo Jorge, Porto), Margarida Rodrigues (Hospital Reynaldo dos Santos, Vila Franca de Xira), Eulália Carvalho and Karine Hyde (Hospital do Divino Espírito Santo, Ponta Delgada), and Clotilde Roldão (Hospital Distrital de Abrantes).
This work was partially supported by Fundação para a Ciência e Tecnologia, Portugal (PTDC/SAU-ESA/72321/2006).
We thank Filipa Vaz and Paulo Lopes for technical support.
Footnotes
Published ahead of print 20 August 2012
REFERENCES
- 1. Albertí S, et al. 2003. Survey of emm gene sequences from pharyngeal Streptococcus pyogenes isolates collected in Spain and their relationship with erythromycin susceptibility. J. Clin. Microbiol. 41:2385–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beall B, et al. 1998. Streptococcal emm types associated with T-agglutination types and the use of conserved emm gene restriction fragment patterns for subtyping group A streptococci. J. Med. Microbiol. 47:893–898 [DOI] [PubMed] [Google Scholar]
- 3. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate—a practical and powerful approch to multiple testing. J. R Stat. Soc. Ser. B Stat. Methodol. 57:289–300 [Google Scholar]
- 4. Bisno AL, Gerber MA, Gwaltney JM, Jr, Kaplan EL, Schwartz RH. 2002. Practice guidelines for the diagnosis and management of group A streptococcal pharyngitis. Clin. Infect. Dis. 35:113–125 [DOI] [PubMed] [Google Scholar]
- 5. Carriço JA, et al. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 44:2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clinical and Laboratory Standards Institute 2006. Performance standards for antimicrobial susceptibility testing, sixteenth informational supplement. CLSI document M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7. Creti R, et al. 2005. Association of group A streptococcal emm types with virulence traits and macrolide-resistance genes is independent of the source of isolation. J. Med. Microbiol. 54:913–917 [DOI] [PubMed] [Google Scholar]
- 8. Enright MC, Spratt BG, Kalia A, Cross JH, Bessen DE. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Francisco AP, Bugalho M, Ramirez M, Carriço JA. 2009. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10:152 doi:10.1186/1471-2105-10-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Francisco AP, et al. 2012. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 13:87 doi:10.1186/1471-2105-13-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grivea IN, Al-Lahham A, Katopodis GD, Syrogiannopoulos GA, Reinert RR. 2006. Resistance to erythromycin and telithromycin in Streptococcus pyogenes isolates obtained between 1999 and 2002 from Greek children with tonsillopharyngitis: phenotypic and genotypic analysis. Antimicrob. Agents Chemother. 50:256–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim S, Yong Lee N. 2004. Antibiotic resistance and genotypic characteristics of group a streptococci associated with acute pharyngitis in Korea. Microb. Drug Resist. 10:300–305 [DOI] [PubMed] [Google Scholar]
- 13. Malhotra-Kumar S, et al. 2005. Macrolide- and telithromycin-resistant Streptococcus pyogenes, Belgium, 1999-2003. Emerg. Infect. Dis. 11:939–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Malli E, et al. 2010. Macrolide-resistant Streptococcus pyogenes in Central Greece: prevalence; mechanism and molecular identification. Int. J. Antimicrob. Agents 35:614–615 [DOI] [PubMed] [Google Scholar]
- 15. Melo-Cristino J, et al. 2010. The Viriato study: update on antimicrobial resistance of microbial pathogens responsible for community-acquired respiratory tract infections in Portugal. Paediatr. Drugs 12(Suppl 1):11–17 [DOI] [PubMed] [Google Scholar]
- 16. Michos AG, et al. 2009. Macrolide resistance in Streptococcus pyogenes: prevalence, resistance determinants, and emm types. Diagn. Microbiol. Infect. Dis. 64:295–299 [DOI] [PubMed] [Google Scholar]
- 17. Nir-Paz R, et al. 2006. Streptococcus pyogenes emm and T types within a decade, 1996-2005: implications for epidemiology and future vaccines. Epidemiol. Infect. 138:53–60 [DOI] [PubMed] [Google Scholar]
- 18. Pérez-Trallero E, Garcia C, Orden B, Marimon JM, Montes M. 2004. Dissemination of emm28 erythromycin-, clindamycin- and bacitracin-resistant Streptococcus pyogenes in Spain. Eur. J. Clin. Microbiol. Infect. Dis. 23:123–126 [DOI] [PubMed] [Google Scholar]
- 19. Pérez-Trallero E, et al. 2010. Antimicrobial resistance among respiratory pathogens in Spain: latest data and its changes over 11 years (1996–1997 to 2006–2007). Antimicrob. Agents Chemother. 57:2953–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pérez-Trallero E, et al. 2007. Phenotypic and genotypic characterization of Streptococcus pyogenes isolates displaying the MLSB phenotype of macrolide resistance in Spain, 1999 to 2005. Antimicrob. Agents Chemother. 51:1228–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reinert RR, et al. 2004. Clonal relatedness of erythromycin-resistant Streptococcus pyogenes isolates in Germany. Antimicrob. Agents Chemother. 48:1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richter SS, et al. 2008. Increasing telithromycin resistance among Streptococcus pyogenes in Europe. J. Antimicrob. Chemother. 61:603–611 [DOI] [PubMed] [Google Scholar]
- 23. Rivera A, et al. 2006. Superantigen gene profile, emm type and antibiotic resistance genes among group A streptococcal isolates from Barcelona, Spain. J. Med. Microbiol. 55:1115–1123 [DOI] [PubMed] [Google Scholar]
- 24. Severiano A, Pinto FR, Ramirez M, Carriço JA. 2011. Adjusted Wallace coefficient as a measure of congruence between typing methods. J. Clin. Microbiol. 49:3997–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Silva-Costa C, Pinto FR, Ramirez M, Melo-Cristino J, Portuguese surveillance group for the study of respiratory pathogens 2008. Decrease in macrolide resistance and clonal instability among Streptococcus pyogenes in Portugal. Clin. Microbiol. Infect. 14:1152–1159 [DOI] [PubMed] [Google Scholar]
- 26. Silva-Costa C, Ramirez M, Melo-Cristino J. 2006. Identification of macrolide-resistant clones of Streptococcus pyogenes in Portugal. Clin. Microbiol. Infect. 12:513–518 [DOI] [PubMed] [Google Scholar]
- 27. Silva-Costa C, Ramirez M, Melo-Cristino J, Portuguese Surveillance Group for the Study of Respiratory Pathogens 2005. Rapid inversion of the prevalences of macrolide resistance phenotypes paralleled by a diversification of T and emm types among Streptococcus pyogenes in Portugal. Antimicrob. Agents Chemother. 49:2109–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Szczypa K, Sadowy E, Izdebski R, Hryniewicz W. 2004. A rapid increase in macrolide resistance in Streptococcus pyogenes isolated in Poland during 1996-2002. J. Antimicrob. Chemother. 54:828–831 [DOI] [PubMed] [Google Scholar]
- 29. Zampaloni C, Cappelletti P, Prenna M, Vitali LA, Ripa S. 2003. emm gene distribution among erythromycin-resistant and -susceptible Italian isolates of Streptococcus pyogenes. J. Clin. Microbiol. 41:1307–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]


