Abstract
We previously demonstrated that aminoglycoside acetyltransferases (AACs) display expanded cosubstrate promiscuity. The enhanced intracellular survival (Eis) protein of Mycobacterium tuberculosis is responsible for the resistance of this pathogen to kanamycin A in a large fraction of clinical isolates. Recently, we discovered that Eis is a unique AAC capable of acetylating multiple amine groups on a large pool of aminoglycoside (AG) antibiotics, an unprecedented property among AAC enzymes. Here, we report a detailed study of the acyl-coenzyme A (CoA) cosubstrate profile of Eis. We show that, in contrast to other AACs, Eis efficiently uses only 3 out of 15 tested acyl-CoA derivatives to modify a variety of AGs. We establish that for almost all acyl-CoAs, the number of sites acylated by Eis is smaller than the number of sites acetylated. We demonstrate that the order of n-propionylation of the AG neamine by Eis is the same as the order of its acetylation. We also show that the 6′ position is the first to be n-propionylated on amikacin and netilmicin. By sequential acylation reactions, we show that AGs can be acetylated after the maximum possible n-propionylation of their scaffolds by Eis. The information reported herein will advance our understanding of the multiacetylation mechanism of inactivation of AGs by Eis, which is responsible for M. tuberculosis resistance to some AGs.
INTRODUCTION
The tuberculosis (TB) epidemic, caused primarily by Mycobacterium tuberculosis, kills millions of people worldwide each year. Taking into account the 2 billion currently infected and the current rate of infection, nearly 10 million people will become infected in the next year (15). The overuse of some drugs and the failure to comply with a proper therapeutic regimen, through either complete lack or intermittent availability of drugs and ineffective secondary care, have in part been responsible for the emergence of multidrug-resistant (MDR) M. tuberculosis strains, resistant to the first-line antibiotics isoniazid and rifampin, as well as extensively drug-resistant (XDR) M. tuberculosis strains that additionally render second-line anti-TB drugs ineffective, including the aminoglycosides (AGs) kanamycin A (KAN) and amikacin (AMK) (7, 17).
Resistance to the broad-spectrum AG antibiotics is a continuously increasing problem for the treatment of many serious bacterial infections (22). AG resistance results, in great part, from the evolution or acquisition of AG-modifying enzymes (AMEs) that acetylate (AG acetyltransferases [AACs]), phosphorylate (AG phosphotransferases [APHs]), or nucleotidylate (AG nucleotidyltransferases [ANTs]) various positions on the AG scaffolds, resulting in their deactivation as antibacterials (35). To broaden their AG resistance profile, bacteria have also evolved bifunctional AMEs, including AAC(6′)-30/AAC(6′)-Ib (46), AAC(6′)-Ie/APH(2″)-Ia (2, 6), AAC(3)-Ib/AAC(6′)-Ib′ (14, 19, 25), and ANT(3″)-Ii/AAC(6′)-IId (9, 24).
For a large fraction of M. tuberculosis clinical isolates, it was recently shown that upregulation of the enhanced intracellular survival (Eis) protein confers KAN resistance on the mycobacterium, a hallmark of XDR-TB (8, 45). This enzyme was initially found to be involved in intracellular survival of Mycobacterium smegmatis within the human macrophage-like cell line U-937 (42). Phase separation assays suggested that Eis appears primarily in the cytoplasm and in modest amounts in the cell envelope and in the culture supernatant (13). Further studies suggested that Eis inhibits T-cell proliferation in vitro, as well as subsequent production of tumor necrosis factor-α and interleukin-4 (28, 36). It was suggested that Eis is a mycobacterial effector that is released into the host cell to modulate macrophage autophagy, inflammatory responses, and cell death via a reactive oxygen species (ROS)-dependent pathway (37). This year, Kim et al. found that Eis is capable of acetylating Lys55 of dual-specificity protein phosphatase 16 (DUSP16)/mitogen-activated protein kinase phosphatase-7 (MKP-7), a Jun N-terminal protein kinase (JNK)-specific phosphatase (26). They proposed that Eis of M. tuberculosis suppresses JNK-dependent autophagy, phagosome maturation, and ROS generation through inhibition of lipopolysaccharide (LPS)-induced JNK phosphorylation via acetylation of DUSP16/MKP-7. We recently discovered that Eis is a unique AAC that can modify multiple amine functionalities on a variety of AG scaffolds (10). This novel enzyme and its homolog in M. smegmatis (11) are, to date, the only known monofunctional AMEs capable of catalyzing multiacetylation reactions. For this study, it is important to distinguish acetylation, which exclusively refers to the transfer of an acetyl group, from acylation, which refers to the general transfer of any acyl moiety (e.g., n-propionyl, malonyl, crotonyl, etc., including acetyl).
We previously demonstrated that AACs could display broad acyl-coenzyme A (CoA) cosubstrate promiscuity (20, 21, 34). Here, to gain insight into the mechanism of multiacetylation of Eis, we performed an in-depth study of the cosubstrate tolerance of Eis. Through sequential use of acyl-CoAs, we established the limits of Eis cosubstrate tolerance. We also investigated the effect that the nature of different acyl-CoA derivatives has on the multiplicity of acylation.
MATERIALS AND METHODS
Materials and instrumentation.
Eis and the AAC(6′)-Ie/APH(2″)-Ia, AAC(3)-IV (30), and AAC(2′)-Ic (39) enzymes were expressed and purified as previously described (10, 20). AAC(6′)-Ie/APH(2″)-Ia was used solely for its AAC(6′) activity and is referred to as AAC(6′) herein. The acyl-CoAs (acetyl-CoA [AcCoA], acetoacetyl-CoA, benzoyl-CoA, n-butyryl-CoA, crotonyl-CoA [CroCoA], glutaryl-CoA, d,l-β-hydroxybutyryl-CoA, isovaleryl-CoA, malonyl-CoA [MalCoA], palmitoyl-CoA, n-propionyl-CoA [ProCoA], hexanoyl-CoA, lauroyl-CoA, octanoyl-CoA, succinyl-CoA, and decanoyl-CoA) (see Fig. S1 in the supplemental material), 4,4′-dithiodipyridine (DTDP), ammonium molybdate, ammonium cerium nitrate, and the AGs (AMK, hygromycin [HYG], KAN, neomycin B [NEO], paromomycin [PAR], ribostamycin [RIB], sisomicin [SIS], and tobramycin [TOB]) (see Fig. S1 in the supplemental material) were purchased from Sigma-Aldrich (Milwaukee, WI). The AGs neamine (NEA) and netilmicin (NET) were purchased from AK Scientific (Mountain View, CA) (see Fig. S1). Ninety-six-well plates were purchased from Thermo Fisher Scientific (Waltham, MA). UV-Vis assays were monitored on a SpectraMax M5 plate reader. Liquid chromatography-mass spectrometry (LC-MS) was performed on a Shimadzu LCMS-2019EV equipped with a SPD-20AV UV-Vis detector and an LC-20AD liquid chromatograph.
Determination of cosubstrate profile for Eis by UV-Vis assays.
To determine which CoA derivatives were cosubstrates for Eis, 15 CoA derivatives (acetoacetyl-CoA, benzoyl-CoA, n-butyryl-CoA, CroCoA, glutaryl-CoA, d,l-β-hydroxybutyryl-CoA, isovaleryl-CoA, MalCoA, palmitoyl-CoA, ProCoA, hexanoyl-CoA, lauroyl-CoA, octanoyl-CoA, succinyl-CoA, and decanoyl-CoA) were tested against several AGs (KAN, NEO, NET, SIS, and TOB). Reaction mixtures with AcCoA were used as positive controls. The acylation activity of Eis was monitored at 324 nm (ε324 = 19,800 M−1 cm−1) by a UV-Vis assay using DTDP, as previously reported (20). Reaction mixtures (200 μl) containing a CoA derivative (0.5 mM, 5 eq), AG (0.1 mM, 1 eq), DTDP (2 mM), and Tris-HCl (50 mM, pH 8.0, adjusted at room temperature [RT]), were initiated by the addition of Eis (0.5 μM) at 25°C. Reactions were monitored by taking readings every 30 s for 60 min in 96-well plates. In addition to AcCoA, only CroCoA, MalCoA, and ProCoA were found to be cosubstrates of Eis and were further studied with all AGs (AMK, HYG, KAN, NEA, NEO, NET, PAR, RIB, SIS, and TOB) as described below.
Determination of cosubstrate profile for Eis by LC-MS.
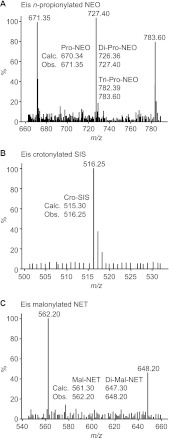
By using LC-MS, the results obtained by UV-Vis assays were confirmed and the degree of acylation was determined for all 10 AG substrates. Reaction mixtures (30 μl) containing AG (0.67 mM, 1 eq), CoA derivatives (3.35 mM, 5 eq), Tris-HCl (50 mM, pH 8.0, adjusted at RT), and Eis (10 μM) were incubated at RT for 48 h. The Eis protein was then precipitated by the addition of ice-cold methanol (MeOH) (30 μl) to the reaction mixture, which was then kept at −20°C for at least 20 min. The precipitated protein was removed by centrifugation (13,000 rpm, RT, 10 min). The supernatant (10 μl) was diluted with H2O (20 μl) and loaded onto the LC-MS. The masses of the acylated AGs present in each sample were determined in positive mode using H2O (0.1% formic acid). Mass spectra of all AGs modified by Eis are shown in Fig. 1; also see Fig. S2, S3, and S4 and Table S2 in the supplemental material.
Fig 1.
Representative mass spectra of AGs multiacylated by Eis. (A) Mono-, di-, and tri-n-propionyl-NEO (m/z [M+H]+ 671.35, 727.40, and 783.60, respectively) generated by reaction of NEO, Eis, and ProCoA. (B) Mono-crotonyl-SIS (m/z [M+H]+ 516.25) generated by reaction of SIS, Eis, and CroCoA. (C) Mono- and di-malonyl-NET (m/z [M+H]+ 562.20 and 648.20, respectively) generated by reaction of NET, Eis, and MalCoA.
Steady-state kinetic measurements for CoA derivatives.
The kinetic parameters for AcCoA and ProCoA were determined against 10 AGs (AMK, KAN, HYG, NEA, NEO, NET, PAR, RIB, SIS, and TOB) in reaction mixtures (100 μl) containing a fixed concentration of AG (0.5 mM), various concentrations of CoA derivatives (0, 20, 50, 100, 250, and 500 μM), and fixed concentrations of DTDP (2 mM), Tris-HCl (50 mM, pH 8.0, adjusted at RT), and Eis (0.25 μM). Using similar reaction conditions, the kinetic parameters for MalCoA and CroCoA were determined in reaction mixtures (100 μl) containing a fixed concentration of NEO (1 mM). Reactions were initiated by the addition of CoA derivatives and were carried out at least in duplicate at 25°C. The reactions were monitored as described above, taking measurements every 15 s for 15 min. The kinetic parameters Km and kcat were determined from Lineweaver-Burk plots (Table 1).
Table 1.
Kinetic parameters determined for Eis and acyl-CoA derivatives with various AGs
| CoA analog | AG | Value (mean ± SD) of: |
||
|---|---|---|---|---|
| Km (μM) | kcat (min−1) | kcat/Km (M−1 s−1) | ||
| Acetyl-CoA | AMK | 7.88 ± 1.35 | 1.14 ± 0.30 | 2,411 ± 757 |
| HYG | 176.62 ± 6.42 | 16.26 ± 0.48 | 1,534 ± 72 | |
| KAN | 9.44 ± 1.20 | 5.56 ± 0.36 | 9,816 ± 1,400 | |
| NEA | 47.30 ± 1.46 | 4.50 ± 0.36 | 1,586 ± 136 | |
| NEO | 24.44 ± 0.68 | 6.54 ± 0.18 | 4,460 ± 175 | |
| NET | 62.95 ± 2.44 | 14.82 ± 1.02 | 3,923 ± 310 | |
| PAR | 45.50 ± 5.52 | 3.24 ± 0.78 | 1,187 ± 320 | |
| RIB | 22.18 ± 1.72 | 3.54 ± 0.06 | 2,660 ± 211 | |
| SIS | 69.41 ± 0.80 | 4.20 ± 0.12 | 1,009 ± 31 | |
| TOB | 47.04 ± 1.31 | 16.92 ± 4.92 | 5,995 ± 1,751 | |
| n-Propionyl-CoA | AMK | 53.14 ± 6.18 | 1.14 ± 0.30 | 358 ± 103 |
| HYG | 30.84 ± 0.62 | 1.80 ± 0.01 | 973 ± 20 | |
| KAN | 55.66 ± 13.28 | 0.48 ± 0.06 | 144 ± 39 | |
| NEA | 55.53 ± 6.08 | 0.60 ± 0.06 | 180 ± 28 | |
| NEO | 53.22 ± 0.14 | 1.98 ± 0.12 | 620 ± 38 | |
| NET | 31.24 ± 3.76 | 3.23 ± 0.02 | 1,723 ± 208 | |
| PAR | 25.12 ± 2.62 | 0.48 ± 0.06 | 318 ± 52 | |
| RIB | 66.76 ± 3.61 | 1.32 ± 0.30 | 330 ± 77 | |
| SIS | 105.79 ± 4.79 | 6.42 ± 1.50 | 1,011 ± 241 | |
| TOB | 74.49 ± 7.78 | 6.36 ± 0.24 | 1,423 ± 158 | |
| Crotonyl-CoA | NEO | 88.86 ± 7.40 | 0.33 ± 0.04 | 62 ± 9 |
| Malonyl-CoA | NEO | 81.49 ± 5.07 | 1.51 ± 0.02 | 309 ± 20 |
TLC assays.
The eluent system used for AMK reactions was 5:2 MeOH-NH4OH (∼25% in H2O). The eluent system utilized for NEA reactions was 3:0.8 MeOH-NH4OH (∼25% in H2O). The eluent system employed for NET reactions was 12:1 MeOH-NH4OH (∼25% in H2O). AGs were visualized on silica gel 60 F254 thin-layer chromatography (TLC) plates (Merck) by using a cerium-molybdate stain composed of (NH4)2Ce(NO3)6 (5 g), (NH4)6Mo7O24 · 4H2O (120 g) in 10% H2SO4 (1 liter). The observed Rf values are reported in Table S1 in the supplemental material. The exact reaction conditions are reported below.
(i) Control TLCs for mono-n-propionylated AGs at the 2′-, 3-, or 6′-position.
Reactions (10-μl mixtures) were performed at RT in morpholineethanesulfonic acid (MES) buffer [50 mM, pH 6.6, for AAC(3)-IV and AAC(6′)] or in potassium phosphate buffer [100 mM, pH 7.0, for AAC(2′)-Ic] with ProCoA (0.96 mM, 1.2 eq), AG (0.8 mM, 1 eq), and AAC enzyme (10 μM). After overnight incubation, aliquots (5 μl) of the reaction mixtures were loaded onto a TLC plate and eluted using the solvent system described above.
(ii) Control TLCs for di-n-propionylated NEA by sequential enzymatic reactions.
Reactions (10-μl mixtures) were performed at RT in MES buffer (50 mM, pH 6.6, adjusted at RT) with ProCoA (1.92 mM, 2.4 eq), NEA (0.8 mM, 1 eq), and AAC(6′) or AAC(3)-IV (10 μM). After overnight incubation, the second AAC enzyme [AAC(2′)-Ic or AAC(3)-IV] (10 μM) was added to the reaction mixture. After an additional 24 h of incubation, aliquots (5 μl) of each di-n-propionylation reaction mixture were loaded onto a TLC plate and eluted using the solvent system described above.
(iii) TLCs for n-propionylation of AMK and NET by Eis.
Reactions (20-μl mixture) were performed at RT in Tris-HCl buffer (50 mM, pH 8.0, adjusted at RT) with ProCoA (2.5 mM, 5 eq), AG (0.5 mM, 1 eq), and Eis (10 μM). After overnight incubation, aliquots (5 μl) of the reaction mixture were loaded onto a TLC plate and eluted using the solvent system described above (see Fig. S5 in the supplemental material).
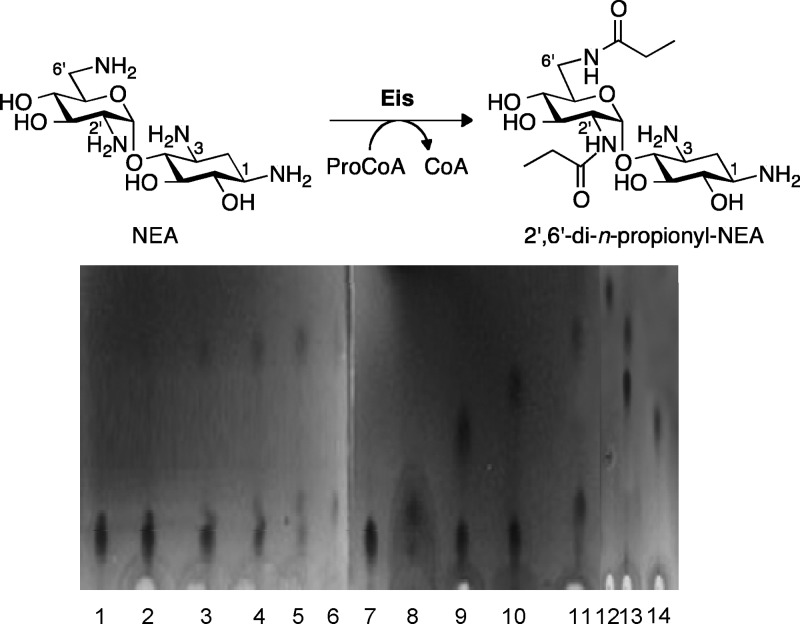
(iv) TLC time course for n-propionylation of NEA by Eis.
Reactions (30-μl mixtures) were performed at RT in Tris-HCl buffer (50 mM, pH 8.0, adjusted at RT) with ProCoA (2.5 mM, 5 eq), NEA (0.5 mM, 1 eq), and Eis (5 μM). Aliquots (4 μl) were loaded and run on a TLC plate after incubation times of 0, 1, 5, 10, 30, 120 min, and overnight using the solvent system described above (Fig. 2).
Fig 2.
TLC time course showing the 2′-mono- and 2′,6′-di-n-propionylated NEA products generated by Eis using 5 equivalents of ProCoA. Control reactions for mono- and di-n-propionylation were done using AAC(2′)-Ic, AAC(3)-IV, and AAC(6′) individually or sequentially. Lane 1, NEA (Rf 0.11); lane 2, NEA + ProCoA + Eis (1 min) (Rf 0.11, 0.13); lane 3, NEA + ProCoA + Eis (5 min) (Rf 0.11, 0.13, 0.41); lane 4, NEA + ProCoA + Eis (10 min) (Rf 0.11, 0.13, 0.41); lane 5, NEA + ProCoA + Eis (30 min) (Rf 0.11, 0.13. 0.41); lane 6, NEA + ProCoA + Eis (2 h) (Rf 0.11, 0.13. 0.41); lane 7, NEA (Rf 0.11); lane 8, 2′-n-propionyl-NEA (Rf 0.13); lane 9, 3-n-propionyl-NEA (Rf 0.24); lane 10, 6′-n-propionyl-NEA (Rf 0.33); lane 11, NEA + ProCoA + Eis (O/N) (Rf 0.41); lane 12, 6′,3-di-n-propionyl-NEA (Rf 0.53); lane 13, 6′,2′-di-n-propionyl-NEA (Rf 0.41); lane 14, 3,2′-di-n-propionyl-NEA (Rf 0.26).
Monitoring by LC-MS of sequential acylations by Eis using ProCoA followed by AcCoA.
Reaction mixtures (30 μl) containing AG (0.67 mM, 1 eq), ProCoA (3.35 mM, 5 eq), Tris-HCl (50 mM, pH 8.0, adjusted at RT), and Eis (5 μM) were incubated at RT for 48 h prior to the addition of AcCoA (3.35 mM, 5 eq) and an additional portion of Eis (5 μM), which brought the total volume of the reaction mixture to 50 μl. After another 48 h of incubation, the Eis protein was precipitated by the addition of ice-cold MeOH (50 μl) to the reaction mixture, which was then kept at −20°C for at least 20 min. The precipitated protein was removed by centrifugation (13,000 rpm, RT, 10 min). An aliquot of the supernatant (20 μl) was analyzed by LC-MS in the positive mode by using 0.1% (vol/vol) formic acid in H2O (see Fig. S6 and Table S3 in the supplemental material).
Monitoring by LC-MS of competition of modification reactions by Eis using ProCoA and AcCoA simultaneously.
Reaction mixtures (50 μl) containing AG (0.67 mM, 1 eq), AcCoA (3.35 mM, 5 eq), ProCoA (3.35 mM, 5 eq), Tris-HCl (50 mM, pH 8.0, adjusted at RT), and Eis (10 μM) were incubated at RT for 48 h, after which they were processed and analyzed by LC-MS as described above for the sequential acylations (see Fig. S7 and Table S3 in the supplemental material).
RESULTS
Acyl-CoA cosubstrate tolerance of Eis.
We previously reported that Eis is an AAC capable of multiacetylation of a variety of AG scaffolds by using AcCoA as a cosubstrate (10). Here, in order to determine the cosubstrate profile of Eis, we tested 15 additional acyl-CoA derivatives with the five Eis AG substrates with the best catalytic transfer efficiency (10). By UV-Vis assay, we established that only three of the acyl-CoA derivatives tested, CroCoA, MalCoA, and ProCoA, displayed any significant reactivity with an AG in the presence of Eis. To gain a better understanding of the catalytic efficiency of Eis toward its accepted cosubstrates, we first determined the steady-state kinetic parameters for AcCoA and ProCoA with all 10 AGs (AMK, HYG, KAN, NEA, NEO, NET, PAR, RIB, SIS, and TOB) (Table 1). Briefly, for AcCoA, the Km values ranged from 7.88 ± 1.35 μM (mean ± standard deviation) for AMK to 176.62 ± 6.42 μM for HYG, representing a 22-fold range. The catalytic turnover constants varied in a 15-fold range, with AMK displaying the lowest kcat value (1.14 ± 0.30 min−1) and TOB exhibiting the highest kcat value (16.92 ± 4.92 min−1). Overall, the catalytic efficiencies (kcat/Km) for AcCoA varied in a 10-fold range, with SIS displaying the lowest catalytic efficiency (1,009 ± 31 M−1 s−1) and KAN the highest (9,816 ± 1,400 M−1 s−1). For ProCoA, the Km values ranged from 25.12 ± 2.62 μM for PAR to 105.79 ± 4.79 μM for SIS, a 4.2-fold range. Interestingly, the higher variability of the Km values for AcCoA is due solely to the two AGs that are used against TB in clinical practice, KAN and AMK, which display significantly lower μM Km values than do the other AGs with AcCoA but not with ProCoA. The kcat values varied in a 13-fold range, with KAN exhibiting the slowest catalytic turnover (0.48 ± 0.06 min−1) and SIS the fastest (6.42 ± 1.50 min−1). Generally, the catalytic efficiencies for ProCoA spanned an 8.5-fold range, with KAN exhibiting the lowest kcat/Km value (144 ± 39 M−1 s−1) and NET the highest (1,723 ± 208 M−1 s−1). The order of AGs ranked by either their Km or kcat values was different for ProCoA than for AcCoA. We next determined the steady-state kinetic parameters for MalCoA and CroCoA using NEO as a substrate. MalCoA had a Km of 81.49 ± 5.07 μM and a kcat of 1.51 ± 0.02 min−1, resulting in a catalytic efficiency of 309 ± 20 M−1 s−1. CroCoA displayed a Km of 88.86 ± 7.40 μM and a kcat of 0.33 ± 0.04 min−1, yielding an efficiency of 62 ± 9 M−1 s−1.
Multiplicity of AG acylation by Eis.
We recently reported that Eis can acetylate between two to four amines on the same AG, depending on its scaffold (10). To investigate the maximum number of sites that can be n-propionylated, crotonylated, and malonylated by Eis, we monitored by mass spectrometry the reactions of 10 AGs (AMK, HYG, KAN, NEA, NEO, NET, PAR, RIB, SIS, and TOB) with ProCoA, CroCoA, and MalCoA, respectively (Table 2 and Fig. 1; also see Table S2 and Fig. S2 to S4 in the supplemental material). When using ProCoA as the cosubstrate, we observed that the 10 AGs tested were modified, but the number of acylations was generally different from the number of acetylations. HYG, NET, PAR, and SIS were mono-n-propionylated, AMK, KAN, NEA, RIB, and TOB became di-n-propionylated, and NEO turned out to be tri-n-propionylated. When utilizing the CroCoA cosubstrate, we found that Eis transferred a single propenyl group to eight AG scaffolds (AMK, KAN, NEA, NEO, NET, PAR, SIS, and TOB). We determined that MalCoA modified only four AGs, of which three were monomalonylated (NEO, PAR, and SIS) and one dimalonylated (NET).
Table 2.
Comparison of the numbers of acylations for reactions of Eis with various acyl-CoAs and AGs
| AG | No. of acylationsa for indicated reaction |
|||
|---|---|---|---|---|
| Acetylb (CH3) | n-Propionyl (CH2CH3) | Crotonyl (CH CHCH3) CHCH3) |
Malonyl (CH2CO2H) | |
| AMK | Tri | Di | Mono | — |
| HYG | Di | Mono | — | — |
| KAN | Di | Di | Mono | — |
| NEA | Tri | Di | Mono | — |
| NEO | Tri | Tri | Mono | Mono |
| NET | Di | Mono | Mono | Di |
| PAR | Di | Mono | Mono | Mono |
| RIB | Tri | Di | — | — |
| SIS | Tri | Mono | Mono | Mono |
| TOB | Tetra | Di | Mono | — |
Mono, Di, Tri, and Tetra indicate single, double, triple, and quadruple modifications, respectively; a dash indicates no modification.
Data are from reference 10.
Order of AG n-propionylation by Eis.
Earlier, we demonstrated that Eis triacetylates NEA in a sequential manner by first modifying the 2′ position, followed by the 6′ and then 1 positions (10). Here, we showed that Eis can di-n-propionylate NEA. To determine whether the order of n-propionylation of NEA by Eis is consistent with that of the first two acetylations of this AG scaffold by this enzyme, we explored by TLC assay the regiospecificity and order of NEA n-propionylation by Eis (Fig. 2). By comparing the Rf values of n-propionylated Eis products formed over time to mono- and di-n-propionylated NEA standards obtained by using, individually or sequentially, AAC enzymes that catalyze a single acylation reaction [AAC(6′), AAC(3)-IV, and AAC(2′)-Ic], we established that the order of n-propionylation of NEA was similar to that of acetylation, with the 2′ position being modified first and the 6′ position second. Using a similar TLC approach, we also performed a preliminary investigation of the order of n-propionylation of two other AGs, AMK and NET (see Fig. S5 in the supplemental material). AMK appeared to be n-propionylated at the 6′ position and at a second site that could not be identified by TLC, as AMK could not be diacetylated using AAC(6′), AAC(3)-IV, and AAC(2′)-Ic sequentially or simultaneously (see Fig. S5A in the supplemental material). By comparing the retention factor of the product of the reaction of NET with ProCoA and Eis (Rf = 0.36) to that of a 6′-n-propionyl-NET standard (Rf = 0.36), we determined that 6′-n-propionyl-NET was the product of the enzymatic reaction (see Fig. S5B). These results indicate that the 6′ position is a common site of acylation of AGs by Eis.
Sequential modifications of AGs by Eis.
By close inspection of the data presented in Table 2, we observed that for almost all AGs studied, the number of sites of n-propionylation was lower than the number of sites of acetylation. To examine whether the n-propionylated AGs could be subsequently acetylated, we first incubated AGs with Eis and an excess of ProCoA, and after completion of the n-propionylation reactions, we added an excess of AcCoA. By mass spectrometry analysis, we established that the results of these sequential modification studies could be broken down into three scenarios (Table 3; also see Fig. S6 in the supplemental material) after complete n-propionylation, as follows. (i) Some AGs, intriguingly, were further acetylated to reach the number of modifications presented in the column for acetylation reactions in Table 3. This was the case for AMK, NEA, PAR, and TOB, whose final products were mono-Ac-di-Pro-AMK, mono-Ac-di-Pro-NEA, mono-Ac-mono-Pro-PAR, and di-Ac-di-Pro-TOB, respectively. (ii) Some AGs were not further derivatized, which was observed when the number of n-propionylations reached the reported number of acetylations. This was the scenario followed by KAN and NEO, where only di-Pro-KAN and tri-Pro-NEO, respectively, were produced. (iii) The other AGs were not further modified even though the amount of acetylation reported was larger than the amount of n-propionylation. This was the case for HYG, NET, RIB, and SIS, where mono-Pro-HYG, mono-Pro-NET, di-Pro-RIB, and mono-Pro-SIS were formed, respectively.
Table 3.
Comparison of the number of acylations for sequential or competition reactions of Eis with different acyl-CoAs and AGs
| AG | No. of acylations/propionylationsa for indicated reaction |
|||
|---|---|---|---|---|
| Acetylb | n-Propionyl | Pro → Acc | Ac + Prod | |
| AMK | Tri | Di | Mono-Ac-di-Pro | Di-Ac tri-Ac |
| HYG | Di | Mono | Mono-Pro | Mono-Ac mono-Ac-mono-Pro |
| KAN | Di | Di | Di-Pro | Di-Ac |
| NEA | Tri | Di | Mono-Ac-di-Pro | Tri-Ac di-Ac-mono-Pro |
| NEO | Tri | Tri | Tri-Pro | Tri-Ac tri-Pro |
| NET | Di | Mono | Mono-Pro | Mono-Ac di-Ac |
| PAR | Di | Mono | Mono-Ac-mono-Pro | Di-Ac |
| RIB | Tri | Di | Di-Pro | Di-Ac |
| SIS | Tri | Mono | Mono-Pro | Mono-Ac di-Ac mono-Ac-mono-Pro |
| TOB | Tetra | Di | Mono-Ac-di-Pro di-Ac-di-Pro | Di-Ac tri-Ac tetra-Ac |
Mono, Di, Tri, and Tetra indicate single, double, triple, and quadruple modifications, respectively.
Data are from reference 10.
ProCoA reactions were followed by incubation with AcCoA.
AcCoA and ProCoA were incubated with Eis and AG in a 1:1 ratio.
Competition assays using AcCoA and ProCoA simultaneously for AG modifications by Eis.
As both AcCoA and ProCoA are found in abundance inside bacterial cells, to corroborate our steady-state kinetic analysis studies, we performed cosubstrate competition assays where AGs were incubated in the presence of Eis and a 1:1 mixture of AcCoA and ProCoA (Table 3; also see Fig. S7 in the supplemental material). As expected, in all cases (AMK, KAN, NET, PAR, and RIB) where the catalytic efficiencies for transfer of an acetyl group were far superior to those for transfer of an n-propionyl moiety, multiacetylation occurred almost exclusively. Two exceptions to this pattern were observed with NEA and NEO. For NEA, with kcat/Km values of 1,586 ± 136 M−1 s−1 for AcCoA and 180 ± 32 M−1 s−1 for ProCoA, one would have expected multiacetylation to be almost exclusive. However, di-Ac-mono-Pro-NEA was detected by mass spectrometry when NEA was reacted with a 1:1 mixture of AcCoA and ProCoA. For NEO, with kcat/Km values of 4,460 ± 39 M−1 s−1 for AcCoA and 620 ± 38 M−1 s−1 for ProCoA, one would have expected multiacetylation to dominate. However, both tri-Ac-NEO and tri-Pro-NEO were detected by mass spectrometry. As predicted for HYG and SIS, which had similar catalytic efficiencies for AcCoA (1,534 ± 72 M−1 s−1 for HYG and 1,009 ± 31 M−1 s−1 for SIS) and ProCoA (973 ± 20 M−1 s−1 for HYG and 1,011 ± 241 M−1 s−1 for SIS), mono-Ac-mono-Pro-AG were detected by mass spectrometry.
DISCUSSION
Many enzymes display natural and/or engineered substrate and/or cosubstrate promiscuity/tolerance. A few examples of naturally catalytically promiscuous enzymes include hydrolases (5), the enzyme macrophomate synthase (18), the strictosidine synthase family of enzymes (1), ketoreductase enzymes involved in polyketide biosynthesis (33), the acyltransferase CouN7 (16), and AACs (20, 38). Cytochrome P450 monooxygenases have been engineered to expand their cosubstrate promiscuity (29). In addition, the broad substrate profile of antibiotic-inactivating enzymes has often resulted from mutations in the genes encoding the enzymes, as exemplified by β-lactamases (32). We have demonstrated that Eis is an AAC capable of multiacetylating a large number of AGs (10). Here, we performed a detailed study to establish the acyl-CoA cosubstrate tolerance of Eis in order to gain insight into its multiacetylation mechanism.
We first examined the cosubstrate profile of Eis using 16 acyl-CoAs with five AGs. In doing so, we observed that in contrast to other AACs that exhibit broad cosubstrate promiscuity, Eis catalyzed transfer only from a small set of the acyl-CoA derivatives. In addition to the natural cosubstrate AcCoA, only CroCoA, MalCoA, and ProCoA were found to act as cosubstrates of Eis. To rationalize the limited cosubstrate tolerance of Eis, we closely examined its structure and those of other AACs with bound cosubstrate, including AAC(3)s (23, 27, 43), AAC(2′)-Ic from M. tuberculosis (39), and AAC(6′)s (3, 4, 31, 40, 41, 44). Even though the AG-binding pocket of Eis is larger, the access to it is more limited than in other AACs due to the presence of the central GNAT region, absent in other AACs, whose surface contributes to the substrate-binding pocket (see Fig. S8A in the supplemental material) (10). This suggests that in AACs other than Eis, the substrate/cosubstrate promiscuity is due to the relatively accessible active sites of these enzymes. For example, in AAC(6′)-Ii from Enterococcus faecium (3), the AG-binding pocket is open to entry both in the direction orthogonal to the Ppant arm of AcCoA (from the side of the viewer in Fig. S8B) and in the direction along the Ppant arm toward the thiol moiety (from the left-hand side in Fig. S8B), whereas for Eis, access is possible only in the orthogonal direction. Therefore, it is likely that the larger acyl groups of cosubstrates block the access of the AG to the binding pocket in Eis. It is important to note that most AMEs were acquired to reduce the toxicity of AGs in AG-producing organisms (12), while Eis evolved through selective pressure to modify AGs (45). Interestingly, while Eis could not transfer the acyl moieties of CoA derivatives with bulkier (benzoyl, bromo-thiophene-2-carbonyl, and fluoro-picolinyl) or longer (butyryl and glutaryl) acyl groups that AAC(3)-IV and AAC(6′)/APH(2″) could transfer to a variety of AGs, Eis was found to readily transfer the crotonyl moiety to eight AGs that the other two AACs could not. With the exception of KAN and NEO, for which the numbers of sites n-propionylated and acetylated were identical, we observed that in every other case, the number of sites acylated was less than the number of sites acetylated (Table 2). Not surprisingly, while the smaller and less rigid n-propionyl group could be transferred to more than one position on 6 of the 10 AGs tested, the larger malonyl and the less-flexible crotonyl moieties were generally only transferred to one amine on some of the AGs tested.
We next determined the kinetic parameters (Km and kcat) for Eis and acyl-CoAs with a variety of AGs and established that for the majority of the AGs tested, the binding affinity of ProCoA to Eis was less than that of AcCoA (Table 1). We first corroborated our kinetic data by exploring the sequential modifications of AGs by Eis, using first an excess of ProCoA, followed by an excess of AcCoA (Table 3; also see Fig. S6 in the supplemental material). In the cases where the number of n-propionylated sites was smaller than the number of acetylated sites and where the catalytic efficiency (kcat/Km) of transfer of the acyl group was much lower for ProCoA than for AcCoA, we observed that after maximum n-propionylation, the AGs were further acetylated to reach the number of modified sites equal to the maximum number of acetylations observed previously. With RIB being the exception, as it did not get further acetylated after di-n-propionylation, this was the case for AMK, NEA, PAR, and TOB. AMK, with kcat/Km values of 2,411 ± 757 M−1 s−1 and 358 ± 103 M−1 s−1 for AcCoA and ProCoA, respectively, was transformed into mono-Ac-di-Pro-AMK. NEA, with kcat/Km values of 1,586 ± 136 M−1 s−1 and 180 ± 28 M−1 s−1 for AcCoA and ProCoA, respectively, was converted into mono-Ac-di-Pro-NEA. PAR, with kcat/Km values of 1,187 ± 320 M−1 s−1 and 318 ± 52 M−1 s−1 for AcCoA and ProCoA, respectively, was modified into mono-Ac-mono-Pro-PAR. TOB, with kcat/Km values of 5,995 ± 1,751 M−1 s−1 and 1,423 ± 158 M−1 s−1 for AcCoA and ProCoA, respectively, was transformed into di-Ac-di-Pro-TOB. In the cases of HYG, NET, and SIS, where the numbers of sites n-propionylated were lower than the numbers of positions acetylated and the catalytic efficiency (kcat/Km) of transfer of the acyl group was similar for ProCoA and AcCoA, we observed that the AGs were not further acetylated after complete n-propionylation. Finally, as expected, regardless of the catalytic efficiency of acyl transfers, we observed that KAN and NEO were not further acetylated by Eis after di- and tri-n-propionylation, as the level of number of sites acetylated and n-propionylated were equal. We conclude that modification with a large acyl group must, at least in some cases, prevent binding of the acylated AG to allow further acylation or acetylation for steric reasons.
We also corroborated our steady-state kinetic analysis studies and gained valuable knowledge about Eis by investigating the competition between n-propionylation and acetylation of AGs by this enzyme (Table 3; also see Fig. S7 in the supplemental material). In agreement with their much higher kcat/Km values for AcCoA, we observed that AMK, KAN, NET, PAR, RIB, and TOB were solely acetylated when reacted with a 1:1 mixture of ProCoA and AcCoA. The aforementioned AGs have a higher catalytic efficiency, lower Km value, higher catalytic turnover rate, or a combination of these properties, which all can explain the dominating effect of AcCoA in the competition assays. Interestingly in the case of RIB, we observed only diacetylation of the AG in the presence of a 1:1 mixture of AcCoA and ProCoA, potentially suggesting that the presence of ProCoA inhibits the transfer of the third acetyl group to di-Ac-RIB and n-propionylation cannot occur at the third acylation site. Interestingly, in the case of NEO, where the kcat/Km value for AcCoA (4,460 ± 175 M−1 s−1) was much larger than that for ProCoA (620 ± 38 M−1 s−1), one would have expected multiacetylation to be dominant. However, both tri-Ac-NEO and tri-Pro-NEO were generated when NEO was reacted with a 1:1 mixture of AcCoA and ProCoA. This implies that after the first modification with a particular cosubstrate, only the second and third modifications by a different cosubstrate are strongly disfavored. In addition, the modifications do not follow the same rank order of catalytic efficiency. A plausible model for this effect is that after a NEO molecule is modified once by either AcCoA or ProCoA, the modified AG does not leave the enzyme active site but, rather, the second and third modifications occur before the singly modified AG can dissociate from the binding site, resulting in the homogeneity of the modifications of NEO. Finally, as expected, in the case of HYG and SIS, where the kcat/Km values for AcCoA (1,534 ± 72 M−1 s−1 for HYG and 1,009 ± 31 M−1 s−1 for SIS) and ProCoA (973 ± 20 M−1 s−1 for HYG and 1,011 ± 241 M−1 s−1 for SIS) were very similar, both acetylation and n-propionylation occurred. The reaction of a 1:1 AcCoA-ProCoA mixture with HYG in the presence of Eis resulted in mono-Ac-mono-Pro-HYG. From our sequential experiments, we established that acetylation of HYG must occur first in order for mono-Ac-mono-Pro-HYG to be formed. We also previously reported that HYG can be diacetylated (10). Therefore, it can be hypothesized that mono-Ac-HYG is a better substrate for n-propionylation than for a second acetylation. We can also draw a similar conclusion from the results obtained with SIS. The reaction of SIS with a 1:1 AcCoA-ProCoA mixture in the presence of Eis resulted in mono-Ac-SIS, di-Ac-SIS, and mono-Ac-mono-Pro-SIS. Since we know that mono-n-propionylation of SIS results in an unreactive species in our sequential experiments, we can conclude that acetylation must occur before n-propionylation in order to generate mono-Ac-mono-Pro-SIS. Based on the competition assay (Table 3) and the kinetic parameters determined (Table 1), we also conclude that mono-Ac-SIS has a similar propensity to become acetylated the second time or to become n-propionylated.
In summary, we have presented evidence that, albeit more limited than that of other AACs, Eis displays some cosubstrate tolerance, accepting AcCoA, ProCoA, CroCoA, and MalCoA for the multiacylation of AGs. We have established the number of sites acylated by these cosubstrates on a variety of AG scaffolds. We have demonstrated that the order of multiacylation for NEA is identical to the order of multiacetylation. We have shown that after complete n-propionylation, further acetylations can be allowed up to the level of multiacetylation allowed by Eis. Finally, by steady-state kinetic assays and acylation competition assays, we have gained insight into the mechanism of multiacylation of AGs by Eis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by start-up funds from the Life Sciences Institute and the College of Pharmacy at the University of Michigan (S.G.-T), by a grant from the Firland Foundation (S.G.-T.), and by National Institutes of Health (NIH) grant AI090048 (S.G.-T.).
We thank Oleg V. Tsodikov for critical reading of the manuscript and help with generating Figure S8 in the supplemental material.
Footnotes
Published ahead of print 4 September 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Bernhardt P, Usera AR, O'Connor SE. 2010. Biocatalytic asymmetric formation of tetrahydro-beta-carbolines. Tetrahedron Lett. 51:4400–4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boehr DD, Daigle DM, Wright GD. 2004. Domain-domain interactions in the aminoglycoside antibiotic resistance enzyme AAC(6′)-APH(2″). Biochemistry 43:9846–9855 [DOI] [PubMed] [Google Scholar]
- 3. Burk DL, Ghuman N, Wybenga-Groot LE, Berghuis AM. 2003. X-ray structure of the AAC(6′)-Ii antibiotic resistance enzyme at 1.8 A resolution; examination of oligomeric arrangements in GNAT superfamily members. Protein Sci. 12:426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burk DL, Xiong B, Breitbach C, Berghuis AM. 2005. Structures of aminoglycoside acetyltransferase AAC(6′)-Ii in a novel crystal form: structural and normal-mode analyses. Acta Crystallogr. D Biol. Crystallogr. 61:1273–1279 [DOI] [PubMed] [Google Scholar]
- 5. Busto E, Gotor-Fernandez V, Gotor V. 2010. Hydrolases: catalytically promiscuous enzymes for non-conventional reactions in organic synthesis. Chem. Soc. Rev. 39:4504–4523 [DOI] [PubMed] [Google Scholar]
- 6. Caldwell SJ, Berghuis AM. 2012. Small-angle X-ray scattering analysis of the bifunctional antibiotic resistance enzyme aminoglycoside (6′) acetyltransferase-ie/aminoglycoside (2″) phosphotransferase-ia reveals a rigid solution structure. Antimicrob. Agents Chemother. 56:1899–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caminero JA, Sotgiu G, Zumla A, Migliori GB. 2010. Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis. Lancet Infect. Dis. 10:621–629 [DOI] [PubMed] [Google Scholar]
- 8. Campbell PJ, et al. 2011. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 55:2032–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Centron D, Roy PH. 2002. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob. Agents Chemother. 46:1402–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen W, Biswas T, Porter VR, Tsodikov OV, Garneau-Tsodikova S. 2011. Unusual regioversatility of acetyltransferase Eis, a cause of drug resistance in XDR-TB. Proc. Natl. Acad. Sci. U. S. A. 108:9804–9808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen W, Green KD, Tsodikov OV, Garneau-Tsodikova S. 2012. The aminoglycoside multi-acetylating activity of the enhanced intracellular survival (Eis) protein from Mycobacterium smegmatis and its inhibition. Biochemistry 51:4959–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Courvalin P, Weisblum B, Davies J. 1977. Aminoglycoside-modifying enzyme of an antibiotic-producing bacterium acts as a determinant of antibiotic resistance in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 74:999–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dahl JL, Wei J, Moulder JW, Laal S, Friedman RL. 2001. Subcellular localization of the intracellular survival-enhancing Eis protein of Mycobacterium tuberculosis. Infect. Immun. 69:4295–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dubois V, et al. 2002. Molecular characterization of a novel class 1 integron containing blaGES-1 and a fused product of aac3-Ib/aac6′-Ib′ gene cassettes in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dye C, Williams BG. 2010. The population dynamics and control of tuberculosis. Science 328:856–861 [DOI] [PubMed] [Google Scholar]
- 16. Fridman M, et al. 2007. Chemoenzymatic formation of novel aminocoumarin antibiotics by the enzymes CouN1 and CouN7. Biochemistry 46:8462–8471 [DOI] [PubMed] [Google Scholar]
- 17. Georghiou SB, et al. 2012. Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: a systematic review. PLoS One 7:e33275 doi:10.1371/journal.pone.0033275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gillingham DG, Stallforth P, Adibekian A, Seeberger PH, Hilvert D. 2010. Chemoenzymatic synthesis of differentially protected 3-deoxysugars. Nat. Chem. 2:102–105 [DOI] [PubMed] [Google Scholar]
- 19. Green KD, Chen W, Garneau-Tsodikova S. 2011. Effects of altering aminoglycoside structures on bacterial resistance enzyme activities. Antimicrob. Agents Chemother. 55:3207–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Green KD, Chen W, Houghton JL, Fridman M, Garneau-Tsodikova S. 2010. Exploring the substrate promiscuity of drug-modifying enzymes for the chemoenzymatic generation of N-acylated aminoglycosides. Chembiochem 11:119–126 [DOI] [PubMed] [Google Scholar]
- 21. Green KD, Fridman M, Garneau-Tsodikova S. 2009. hChAT: a tool for the chemoenzymatic generation of potential acetyl/butyrylcholinesterase inhibitors. Chembiochem 10:2191–2194 [DOI] [PubMed] [Google Scholar]
- 22. Houghton JL, Green KD, Chen W, Garneau-Tsodikova S. 2010. The future of aminoglycosides: the end or renaissance? Chembiochem 11:880–902 [DOI] [PubMed] [Google Scholar]
- 23. Hu X, Norris AL, Baudry J, Serpersu EH. 2011. Coenzyme A binding to the aminoglycoside acetyltransferase (3)-IIIb increases conformational sampling of antibiotic binding site. Biochemistry 50:10559–10565 [DOI] [PubMed] [Google Scholar]
- 24. Kim C, Hesek D, Zajicek J, Vakulenko SB, Mobashery S. 2006. Characterization of the bifunctional aminoglycoside-modifying enzyme ANT(3″)-Ii/AAC(6′)-IId from Serratia marcescens. Biochemistry 45:8368–8377 [DOI] [PubMed] [Google Scholar]
- 25. Kim C, Villegas-Estrada A, Hesek D, Mobashery S. 2007. Mechanistic characterization of the bifunctional aminoglycoside-modifying enzyme AAC(3)-Ib/AAC(6′)-Ib′ from Pseudomonas aeruginosa. Biochemistry 46:5270–5282 [DOI] [PubMed] [Google Scholar]
- 26. Kim KH, et al. 2012. Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7. Proc. Natl. Acad. Sci. U. S. A. 109:7729–7734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klimecka MM, et al. 2011. Structural analysis of a putative aminoglycoside N-acetyltransferase from Bacillus anthracis. J. Mol. Biol. 410:411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lella RK, Sharma C. 2007. Eis (enhanced intracellular survival) protein of Mycobacterium tuberculosis disturbs the cross regulation of T-cells. J. Biol. Chem. 282:18671–18675 [DOI] [PubMed] [Google Scholar]
- 29. Li S, et al. 2009. Selective oxidation of carbolide C-H bonds by an engineered macrolide P450 mono-oxygenase. Proc. Natl. Acad. Sci. U. S. A. 106:18463–18468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Magalhaes ML, Blanchard JS. 2005. The kinetic mechanism of AAC3-IV aminoglycoside acetyltransferase from Escherichia coli. Biochemistry 44:16275–16283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Magalhaes ML, et al. 2008. Kinetic and structural analysis of bisubstrate inhibition of the Salmonella enterica aminoglycoside 6′-N-acetyltransferase. Biochemistry 47:579–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paterson DL, Bonomo RA. 2005. Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Piasecki SK, et al. 2011. Employing modular polyketide synthase ketoreductases as biocatalysts in the preparative chemoenzymatic syntheses of diketide chiral building blocks. Chem. Biol. 18:1331–1340 [DOI] [PubMed] [Google Scholar]
- 34. Porter VR, Green KD, Zolova OE, Houghton JL, Garneau-Tsodikova S. 2010. Dissecting the cosubstrate structure requirements of the Staphylococcus aureus aminoglycoside resistance enzyme ANT(4′). Biochem. Biophys. Res. Commun. 403:85–90 [DOI] [PubMed] [Google Scholar]
- 35. Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist. Updat. 13:151–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Samuel LP, et al. 2007. Expression, production and release of the Eis protein by Mycobacterium tuberculosis during infection of macrophages and its effect on cytokine secretion. Microbiology 153:529–540 [DOI] [PubMed] [Google Scholar]
- 37. Shin DM, et al. 2010. Mycobacterium tuberculosis eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLoS Pathog. 6:e1001230 doi:10.1371/journal.ppat.1001230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsitovich PB, Pushechnikov A, French JM, Disney MD. 2010. A chemoenzymatic route to diversify aminoglycosides enables a microarray-based method to probe acetyltransferase activity. Chembiochem 11:1656–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vetting MW, Hegde SS, Javid-Majd F, Blanchard JS, Roderick SL. 2002. Aminoglycoside 2′-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates. Nat. Struct. Biol. 9:653–658 [DOI] [PubMed] [Google Scholar]
- 40. Vetting MW, Magnet S, Nieves E, Roderick SL, Blanchard JS. 2004. A bacterial acetyltransferase capable of regioselective N-acetylation of antibiotics and histones. Chem. Biol. 11:565–573 [DOI] [PubMed] [Google Scholar]
- 41. Vetting MW, et al. 2008. Mechanistic and structural analysis of aminoglycoside N-acetyltransferase AAC(6′)-Ib and its bifunctional, fluoroquinolone-active AAC(6′)-Ib-cr variant. Biochemistry 47:9825–9835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wei J, et al. 2000. Identification of a Mycobacterium tuberculosis gene that enhances mycobacterial survival in macrophages. J. Bacteriol. 182:377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wolf E, et al. 1998. Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase. Cell 94:439–449 [DOI] [PubMed] [Google Scholar]
- 44. Wybenga-Groot LE, Draker K, Wright GD, Berghuis AM. 1999. Crystal structure of an aminoglycoside 6′-N-acetyltransferase: defining the GCN5-related N-acetyltransferase superfamily fold. Structure 7:497–507 [DOI] [PubMed] [Google Scholar]
- 45. Zaunbrecher MA, Sikes RD, Jr, Metchock B, Shinnick TM, Posey JE. 2009. Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 106:20004–20009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang W, Fisher JF, Mobashery S. 2009. The bifunctional enzymes of antibiotic resistance. Curr. Opin. Microbiol. 12:505–511 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.