Abstract
Tildipirosin is a 16-membered-ring macrolide developed to treat bacterial pathogens, including Mannheimia haemolytica and Pasteurella multocida, that cause respiratory tract infections in cattle and swine. Here we evaluated the efficacy of tildipirosin at inhibiting protein synthesis on the ribosome (50% inhibitory concentration [IC50], 0.23 ± 0.01 μM) and compared it with the established veterinary macrolides tylosin, tilmicosin, and tulathromycin. Mutation and methylation at key rRNA nucleotides revealed differences in the interactions of these macrolides within their common ribosomal binding site.
TEXT
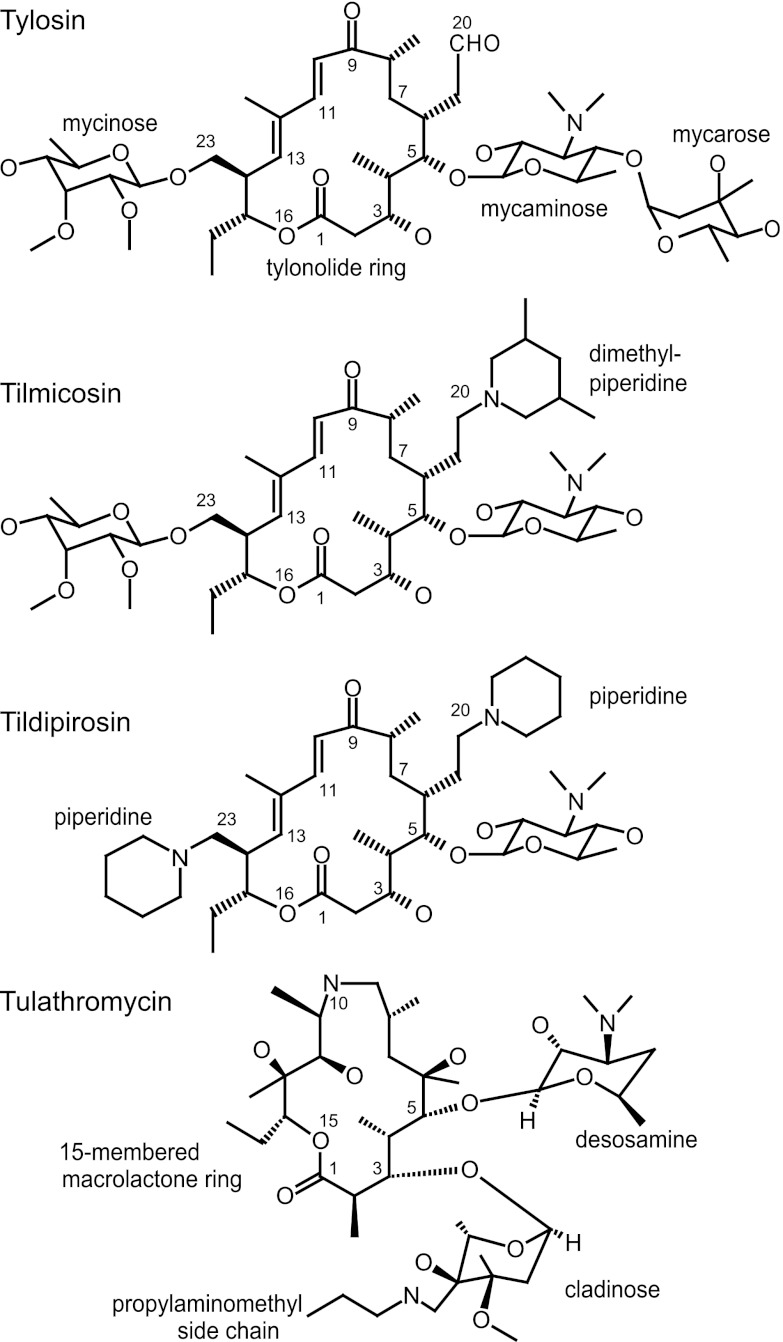
Recent approval has been given in Europe and the United States for the use of tildipirosin (20,23-dipiperidinyl-mycaminosyl-tylonolide; Zuprevo) in combating bovine and swine respiratory tract infections. Tildipirosin is a derivative of the natural compound tylosin with two piperidine rings and no mycarose sugar (Fig. 1). Despite its wide use as a veterinary macrolide, tylosin is not particularly effective at penetrating the outer membrane of Gram-negative pathogens (Table 1), and the substitutions in tildipirosin were made to improve efficacy against Mannheimia haemolytica and Pasteurella multocida, which are the two main etiological agents of bovine respiratory disease (8, 22). Tildipirosin has additionally proven effective against Histophilus somni, Bordetella bronchiseptica, Actinobacillus pleuropneumoniae, and Haemophilus parasuis (6), which can also be associated with animal respiratory diseases. Different arrangements of hydrophobic and basic substituents are present in an earlier tylosin derivative, tilmicosin (20-dimethylpiperidinyl-mycaminosyl-tylonolide; Micotil) and the 15-membered triamilide tulathromycin (Draxxin) (Fig. 1), which are used for similar indications.
Fig 1.
Chemical structures of the macrolides used in the study. Tilmicosin and tildipirosin are derivatives of tylosin and have retained the 16-membered macrolactone (tylonolide) ring and the 5-mycaminose amino sugar. The 16-membered macrolides are distinguished by the mycarose sugar in tylosin, the 20-dimethylpiperidine in tilmicosin, and the 20- and 23-piperidines in tildipirosin. The 15-membered triamilide tulathromycin is chemically closely related to azithromycin (1).
Table 1.
MICs of macrolide antibiotics that prevent growth of Gram-negative strainsa
| Strain | MIC (μg/ml) of macrolide |
|||
|---|---|---|---|---|
| Tylosin | Tilmicosin | Tildipirosin | Tulathromycin | |
| M. haemolytica 11935 | 64 | 4 | 0.5 | 2 |
| P. multocida 4407 | 32 | 4 | 1 | 0.5 |
| E. coli ATCC 25922 | 512 | 128 | 16 | 16 |
| E. coli AS19rlmAI | 1 | 2 | 0.5 | 1 |
P. multocida 4407 and M. haemolytica 11935 are susceptible strains that contain none of the macrolide resistance determinants (4, 19). E. coli ATCC 25922 also lacks any resistance determinants and is a wild-type strain with regard to cell wall/membrane structures. The hyperpermeable strain AS19rlmAI is described in Table 2. Quality controls were included as previously described (19).
Here we quantified the inhibitory activity of tildipirosin (MSD Animal Health) on protein synthesis and compare it with tylosin, tilmicosin (Sigma), and tulathromycin (extracted from Draxxin [Pfizer]). The concentration of each macrolide that inhibits 50% of protein synthesis (IC50) was determined in an in vitro transcription/translation assay, and the effects of methylations and mutations at rRNA nucleotides within the macrolide binding site were evaluated in cell cultures. The data have been visualized using computationally calculated models of the binding site and reveal subtle differences in macrolide contacts, indicating how changes in the rRNA target have distinct effects on drug efficacy.
An in vitro transcription/translation system based on cell extracts containing susceptible, wild-type Escherichia coli ribosomes (Promega) was adapted to translate the 27-kDa green fluorescent protein (GFP). GFP mRNA was transcribed from 3 μg of plasmid pIVEX (Roche) in 50 μl of 100 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES)-KOH (pH 7.9), 5 mM MgCl2, 0.5 mM CaCl2, 100 mM KCl, 5 mM NH4Cl, 1 mM dithiothreitol, and 1 mM spermidine and translated to produce [35S]methionine-labeled protein. Synthesis was followed over 155 min with macrolide antibiotics at up to 2 μM, and full-length GFP was quantified by phosphorimager scanning (Typhoon; GE Healthcare) of polyacrylamide sodium dodecyl sulfate gels. Rates of protein synthesis were calculated from the initial linear portions of the slopes (GFP band intensity plotted against time) and normalized against reactions in the absence of drug. All the macrolides were effective inhibitors of GFP synthesis, with IC50 values of 0.36 ± 0.02 μM for tilmicosin, 0.26 ± 0.05 μM for tulathromycin, 0.31 ± 0.05 μM for tylosin, and 0.23 ± 0.01 μM for tildipirosin.
The IC50 values largely match previous estimates for tilmicosin (0.39 ± 0.04 μM) (7), tulathromycin (0.37 ± 0.02 μM) (7), and tylosin (0.25 μM) (23); tildipirosin had not been studied previously. Measurement of radioisotope incorporation, as opposed to GFP fluorescence (23), enabled quantification of aborted and extended products formed by translational drop-off and stop codon read-through, respectively. None of the drugs tested here aborted peptide chains longer than 10 residues, consistent with observations for other macrolides blocking synthesis after addition of 2 to 8 residues (13, 24). None of the drugs detectably enhanced UAA stop codon read-through.
The main inhibitory contacts of macrolide antibiotics are within the ribosomal tunnel at 23S rRNA nucleotide A2058 (5, 9, 20), and tylosin-like compounds make additional interactions around nucleotide G748 (9, 12). Mutations and methylations known to reduce macrolide binding were introduced at these and neighboring nucleotides in E. coli AS19rlmAI (11). Strain AS19rlmAI allows greater intracellular accumulation of drugs, resulting in macrolide MICs that are 16- to 500-fold lower than those for wild-type E. coli (Table 1).
MICs were measured by diluting overnight cultures of cells 105-fold and added to microtiter wells containing 2-fold dilution steps of the macrolides between 0.5 μg/ml and 2,048 μg/ml. Mutation or methylation of 23S rRNA nucleotide G745, G748, or A752 caused modest increases in the MICs in a manner that is typical of macrolide drugs (2, 15, 26). More severe increases in the MICs were brought about by substitution or methylation at nucleotide A2058 on the other side of the macrolide site (Table 2). We note that such rRNA mutations were introduced primarily to study drug interaction and are unlikely to arise in the field in bacteria with multiple rrn operons (25). However, methylation at nucleotide A2058 has been seen to confer macrolide resistance in P. multocida and M. haemolytica isolates and is added by the Pasteurellaceae-specific monomethyltransferase Erm(42) (3, 10, 14, 19). A2058 monomethylation causes a similar resistance profile in E. coli, while dimethylation at A2058 confers high resistance to all macrolides, including tylosin (Table 2). No Erm dimethyltransferase has so far been observed in P. multocida or M. haemolytica.
Table 2.
MICs for recombinant E. coli AS19rlmAI with rRNA methylation or mutation at the macrolide sitea
| Macrolide | MIC (μg/ml) for E. coli AS19rlmAI recombinant strain with: |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No change in 23S rRNAb | Methylations in 23S rRNAc |
Mutations in 23S rRNAd |
|||||||||||
| m1G745 | m1G748 | N6-A2058 | m1G745 N6-A2058 | m1G748 N6-A2058 | N6, N6-A2058 | A752U | Δ752 | G745U | G748U | A2058G | A2058G Δ752 | ||
| Tylosin | 1 | 4 | 2 | 8 | 32 | 256 | 2,048 | 1 | 2 | 2 | 2 | 128 | 128 |
| Tilmicosin | 2 | 8 | 8 | 32 | 128 | 256 | 512 | 2 | 8 | 4 | 4 | 256 | 256 |
| Tildipirosin | 0.5 | 4 | 4 | 128 | 1,024 | 2,048 | 2,048 | 0.5 | 1 | 4 | 0.5 | 1,024 | 512 |
| Tulathromycin | 1 | 8 | 4 | 16 | 32 | 16 | 2,048 | 1 | 2 | 4 | 1 | 1,024 | 1,024 |
AS19rlmAI was derived from the hyperpermeable strain AS19 (21) and lacks RlmAI methylation at 23S rRNA nucleotide G745 (11). AS19rlmAI was transformed with plasmids encoding either methyltransferases specific for 23S rRNA (11) or the rrnB operon with mutant versions of 23S rRNA (15). Cells were incubated at 37°C in triplicate, and MICs were scored after 20 h as the lowest concentrations at which no growth was observed.
Cells containing an empty plasmid.
Monomethylation at A2058 was added by the Erm(N) enzyme and conferred drug phenotypes identical to those of the Pasteurellaceae monomethyltransferase Erm(42) (3).
The wild-type bases are shown before the nucleotide numbers and the substituted bases after; nucleotide 752 is deleted in the Δ752 strain; the A2058G/Δ752 strain contains both these mutations.
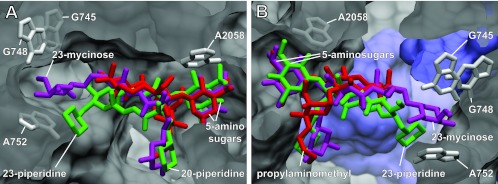
Although, on their own, monomethylation of G748 and that of A2058 have only a minor effect on tylosin binding, in combination they function synergistically to confer resistance (12). A similar synergistic effect is found here for tilmicosin and tildipirosin but not for tulathromycin. This difference is explained by the binding models (Fig. 2), where all the drugs are seen to make their primary ribosome interaction at nucleotide A2058, but in contrast to tilmicosin and tildipirosin, tulathromycin is too small to span the ribosomal tunnel to contact G748.
Fig 2.
The macrolide site in the ribosome tunnel. (A and B) Two views of superimposed binding sites for tildipirosin (green), tilmicosin (magenta), and tulathromycin (red). Parameters for calculating tildipirosin and tilmicosin (and tylosin) binding using Schrödinger 2010(U1) Suite software were described recently (18). The model presented here for tulathromycin on the E. coli ribosome was calculated in a similar manner based on the Thermus thermophilus ribosome-azithromycin cocrystal structure PDB 3OI1 (1) aligned against the E. coli 50S structure PDB 3OAT (5). Clear overlap in the positions of the 5-amino sugars mycaminose (tildipirosin and tilmicosin) and desosamine (tulathromycin) is evident. The 20-piperidine (tildipirosin), the 20-dimethylpiperidine (tilmicosin), and the 3-cladinose propylaminomethyl side chain (tulathromycin) reach into the tunnel lumen with slightly different orientations. More distinct differences are seen in the locations of the 23-piperidine (tildipirosin) and the 23-mycinose (tilmicosin), while tulathromycin has no equivalent structure. Ribosomal nucleotides interacting with the drug substituents are indicated as gray sticks; ribosomal proteins are shown in blue.
These models, while explaining much of the genetic data, nevertheless represent static pictures of ribosome-drug interaction and leave a number of questions unanswered. For instance, despite the lack of any obvious contact, individual mutations and methylation around G748 have small but significant effects on tulathromycin binding (Table 2). This could reflect the manner in which the drug is accommodated into its binding site. The similarly structured azithromycin binds in a two-step process (17) that could thus involve transient interactions with nucleotides other than those contacted in the crystal structure (1). Tylosin also binds to its ribosomal site in a two-step process (16), and the methylations at G748 and A2058 probably interfere with this process prior to impeding the final orientation of the drug in its binding site (9). In the case of tildipirosin, a 23-piperidine replaces the 23-mycinose of tylosin and tilmicosin (Fig. 2), and this part of the drug has been calculated to be slightly further away from G748 (18). While the IC50 and growth studies indicate that tildipirosin binds tightly to an unmodified ribosomal target site, the data (Table 2) also suggest that modification of nucleotides within or adjacent to this site might interfere with accommodation of the drug into its optimal binding conformation.
We note that the nascent peptide in the ribosome tunnel was by necessity left out of the modeling calculations (18) and remains an important parameter that could contribute to the antimicrobial efficacy of tildipirosin that has been observed in the laboratory and in the field.
ACKNOWLEDGMENTS
S.D. gratefully acknowledges support from MSD Animal Health, the Danish Research Agency (FNU-rammebevillinger 09-064292/10-084554), and the Nucleic Acid Center of the Danish Grundforskningsfond.
We thank Simon Rose for advice on MICs and Michael Linder, Heinz-Jörg Wennesheimer, Karl-Heinz Grimm, and colleagues at MSD Animal Health for macrolide drug purification.
Footnotes
Published ahead of print 27 August 2012
REFERENCES
- 1. Bulkley D, Innis CA, Blaha G, Steitz TA. 2010. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc. Natl. Acad. Sci. U. S. A. 107:17158–17163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Canu A, et al. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Desmolaize B, Rose S, Warrass R, Douthwaite S. 2011. A novel Erm monomethyltransferase in antibiotic resistance isolates of Mannheimia haemolytica and Pasteurella multocida. Mol. Microbiol. 80:184–194 [DOI] [PubMed] [Google Scholar]
- 4. Desmolaize B, Rose S, Wilhelm C, Warrass R, Douthwaite S. 2011. Combinations of macrolide resistance determinants in field isolates of Mannheimia haemolytica and Pasteurella multocida. Antimicrob. Agents Chemother. 55:4128–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dunkle JA, Xiong L, Mankin AS, Cate JH. 2010. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. U. S. A. 107:17152–17157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. European Medicines Agency 2009. CVMP assessment report. Zuprevo (EMEA/V/C/002009). European Medicines Agency, London, United Kingdom: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_product_information/veterinary/002009/WC500106578.pdf [Google Scholar]
- 7. Gootz TD. 2005. In vitro activity of triamilide CP-. 472,295(e) against ribosomes isolated from E. coli. In Draxxin injectable solution (tulathromycin). NADA 141-244 FDA, Silver Spring, MD [Google Scholar]
- 8. Griffin D. 2010. Bovine pasteurellosis and other bacterial infections of the respiratory tract. Vet. Clin. North Am. Food Anim. Pract. 26:57–71 [DOI] [PubMed] [Google Scholar]
- 9. Hansen JL, et al. 2002. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10:117–128 [DOI] [PubMed] [Google Scholar]
- 10. Kadlec K, et al. 2011. Molecular basis of macrolide, triamilide, and lincosamide resistance in Pasteurella multocida from bovine respiratory disease. Antimicrob. Agents Chemother. 55:2475–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu M, Douthwaite S. 2002. Activity of the ketolide antibiotic telithromycin is refractory to Erm monomethylation of bacterial rRNA. Antimicrob. Agents Chemother. 46:1629–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu M, Douthwaite S. 2002. Resistance to the macrolide antibiotic tylosin is conferred by single methylations at 23S rRNA nucleotides G748 and A2058 acting in synergy. Proc. Natl. Acad. Sci. U. S. A. 99:14658–14663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Menninger JR, Otto DP. 1982. Erythromycin, carbomycin, and spiramycin inhibit protein synthesis by stimulating the dissociation of peptidyl-tRNA from ribosomes. Antimicrob. Agents Chemother. 21:811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Michael GB, et al. 2012. Increased MICs of gamithromycin and tildipirosin in the presence of the genes erm(42) and msr(E)-mph(E) for bovine Pasteurella multocida and Mannheimia haemolytica. J. Antimicrob. Chemother. 67:1555–1557 [DOI] [PubMed] [Google Scholar]
- 15. Novotny GW, Jakobsen L, Andersen NM, Poehlsgaard J, Douthwaite S. 2004. Ketolide antimicrobial activity persists after disruption of interactions with domain II of 23S rRNA. Antimicrob. Agents Chemother. 48:3677–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Petropoulos AD, Kouvela EC, Dinos GP, Kalpaxis DL. 2008. Stepwise binding of tylosin and erythromycin to Escherichia coli ribosomes, characterized by kinetic and footprinting analysis. J. Biol. Chem. 283:4756–4765 [DOI] [PubMed] [Google Scholar]
- 17. Petropoulos AD, et al. 2009. Time-resolved binding of azithromycin to Escherichia coli ribosomes. J. Mol. Biol. 385:1179–1192 [DOI] [PubMed] [Google Scholar]
- 18. Poehlsgaard J, Andersen NM, Warrass R, Douthwaite S. 2012. Visualizing the 16-membered ring macrolides tildipirosin and tilmicosin bound to their ribosomal site. ACS Chem. Biol. 7:1351–1355 [DOI] [PubMed] [Google Scholar]
- 19. Rose S, Desmolaize B, Jaju P, Warrass R, Douthwaite S. 2012. Multiplex PCR to identify macrolide resistance determinants in Mannheimia haemolytica and Pasteurella multocida. Antimicrob. Agents Chemother. 56:3664–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schlünzen F, et al. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814–821 [DOI] [PubMed] [Google Scholar]
- 21. Sekiguchi M, Iida S. 1967. Mutants of Escherichia coli permeable to actinomycin. Proc. Natl. Acad. Sci. U. S. A. 58:2315–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Snowder GD, et al. 2007. Bovine respiratory disease in feedlot cattle: phenotypic, environmental, and genetic correlations with growth, carcass, and longissimus muscle palatability traits. J. Anim. Sci. 85:1885–1892 [DOI] [PubMed] [Google Scholar]
- 23. Starosta AL, et al. 2010. Interplay between the ribosomal tunnel, nascent chain, and macrolides influences drug inhibition. Chem. Biol. 17:504–514 [DOI] [PubMed] [Google Scholar]
- 24. Tenson T, Lovmar M, Ehrenberg M. 2003. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 330:1005–1014 [DOI] [PubMed] [Google Scholar]
- 25. Vester B, Douthwaite S. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xiong L, Shah S, Mauvais P, Mankin AS. 1999. A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase centre. Mol. Microbiol. 31:633–639 [DOI] [PubMed] [Google Scholar]