Abstract
Due to their abilities to form strong biofilms, Staphylococcus aureus and Staphylococcus epidermidis are the most frequently isolated pathogens in persistent and chronic implant-associated infections. As biofilm-embedded bacteria are more resistant to antibiotics and the immune system, they are extremely difficult to treat. Therefore, biofilm-active antibiotics are a major challenge. Here we investigated the effect of the lantibiotic gallidermin on two representative biofilm-forming staphylococcal species. Gallidermin inhibits not only the growth of staphylococci in a dose-dependent manner but also efficiently prevents biofilm formation by both species. The effect on biofilm might be due to repression of biofilm-related targets, such as ica (intercellular adhesin) and atl (major autolysin). However, gallidermin's killing activity on 24-h and 5-day-old biofilms was significantly decreased. A subpopulation of 0.1 to 1.0% of cells survived, comprising “persister” cells of an unknown genetic and physiological state. Like many other antibiotics, gallidermin showed only limited activity on cells within mature biofilms.
INTRODUCTION
Staphylococcus aureus and Staphylococcus epidermidis are widely involved in minor to severe infections. S. aureus has a wide range of virulence factors and can cause acute and chronic infections at many anatomical sites. In the last 2 decades, multidrug-resistant S. aureus and methicillin-resistant S. aureus (MRSA) have emerged as major causes of hospital-, community-, and livestock-acquired infections, which are increasingly difficult to manage (9, 20). In contrast to S. aureus, S. epidermidis strains produce fewer extracellular toxins than coagulase-positive strains and therefore produce fewer acute symptoms. However, despite their lower virulence, S. epidermidis is well adapted to forming biofilms (comprising adherence and intercellular aggregation) on surfaces of foreign bodies, such as vascular catheters, cardiac devices, ventricular catheters, and prosthetic joints (19), mainly due to its ability to form strong biofilms (18). It is therefore not surprising that S. epidermidis is normally associated with chronic infections. A major problem is the rising occurrence of highly virulent and multiply resistant strains. Especially when a biofilm is formed during infection, therapy is extremely difficult due to the general antibiotic resistance of a subpopulation in a biofilm community. Pathogenic staphylococci are now regarded in the scientific community as antibiotic-resistant “superbugs,” because they have an amazing capacity to acquire resistance traits. Possible targets for drug development are enzymes involved in the biosynthesis of cell envelope structures, such as peptidoglycan, teichoic acids, membrane lipids, or cell wall-associated adhesins (17).
Promising drugs could be the type A lanthionine-containing peptide antibiotics (lantibiotics), which are ribosomally synthesized antibiotic peptides that contain the nonprotein amino acids lanthionine and 3-methyllanthionine (51). Prominent representatives of lantibiotics are nisin (29), epidermin (1, 51), gallidermin (25), and mersacidin (3, 36). We have been working for some time to unravel the biosynthetic pathway of epidermin and gallidermin, which differ by only one amino acid (1, 14, 30, 31, 39, 41–43, 45, 49, 50). As gallidermin showed slightly higher antimicrobial activity than epidermin, we focused on gallidermin and developed a production system. The nuclear magnetic resonance structure of gallidermin is in excellent agreement with the amphiphilic and channel-forming properties of gallidermin on membranes and its tryptic cleavage activity at the exposed site between residues 13 and 14 (12).
The mode of action of the lantibiotics has been extensively studied by the Hans-Georg Sahl group in Bonn. Originally, it was assumed that type A lantibiotics primarily kill bacteria by permeabilization of the cytoplasmic membrane in a membrane potential-dependent way. However, more recently it has been shown that nisin and epidermin also interact with the membrane-bound peptidoglycan precursors lipid I and lipid II, suggesting that they also inhibit peptidoglycan synthesis (5). The activity of lantibiotics appears to be based on different killing mechanisms that are combined in one molecule. The prototype lantibiotic nisin inhibits peptidoglycan synthesis and forms pores through a specific interaction with the cell wall precursor lipid II. Gallidermin and epidermin possess the same putative lipid II binding motif as nisin; however, both peptides are considerably shorter (22 amino acids, compared to 34 in nisin). Indeed, it has been demonstrated that the pore formation by gallidermin depends on membrane thickness and that it is the interaction with lipids I and II rather than pore formation that contributes to bacterial killing. The superior activity of gallidermin over nisin in a cell wall biosynthesis assay may explain its high killing potency (4). Recently, it was shown that nisin and gallidermin not only bind to lipid II but also to the lipid intermediates lipid III (undecaprenol-pyrophosphate-N-acetylglucosamine) and lipid IV (undecaprenol-pyrophosphate-N-acetylglucosamine-N-acetyl-mannosamine) of the wall teichoic acid (WTA) biosynthesis pathway. It was shown that the specific interaction with WTA precursors promoted pore formation in artificial lipid bilayers (35).
In light of the unique mode of action of gallidermin, which targets not only peptidoglycan but also wall teichoic acid biosynthesis, we aimed to study its activity on biofilm cells. Here we show that gallidermin is able to very efficiently inhibit the formation of staphylococcal biofilms; however, its killing activity on preformed biofilms was markedly decreased, as a small percentage of a “persister” subpopulation survived.
MATERIALS AND METHODS
Determination of the MIC and MBC of gallidermin.
The MICs of gallidermin against S. aureus and S. epidermidis were determined by the microdilution method, using Mueller-Hinton broth (MHB) and following the method described by the Clinical and Laboratory Standards Institute (6). Gallidermin was diluted in a 96-well microtiter plate to final concentrations ranging from 128 to 0.06 μg/ml. A 100-μl aliquot of the bacterial suspension (106 CFU/ml) was inoculated and incubated at 37°C for 18 h. The MIC was determined as the lowest concentration that completely inhibited bacterial growth. The minimal bactericidal concentration (MBC) was assessed as the extract concentration that gave significant MIC values after streaking the culture on Trypticase soy agar (TSA). Experiments were carried out in triplicate.
Growth inhibition by gallidermin added to mid-logarithmic-phase cultures.
Twenty-milliliter aliquots of S. aureus SA113 and S. epidermidis O47 at an optical density at 578 nm (OD578) of 0.1 were cultured in basic medium (BM; 1% soy peptone, 0.5% yeast extract, 0.5% NaCl, 0.1% K2HPO4, 0.1% glucose) at 37°C with shaking at 150 rpm until mid-logarithmic phase was reached (approximately 4 h). At that time, gallidermin was added at a final concentration of 1×, 2×, 4×, and 8× MIC. The OD of the cultures was measured at the indicated time intervals. The experiment was carried out in triplicate, and the results are presented as the mean OD ± the standard deviation.
Effect of gallidermin on biofilm formation.
Overnight cultures grown in Trypticase soy broth (TSB) were diluted to 106 CFU/ml. A 100-μl aliquot was transferred to a 96-well microtiter plate, and 100 μl of a subinhibitory concentration of gallidermin, dissolved in TSB, was added. After incubation at 37°C for 24 h without agitation, the bacterial growth (based on the OD600) was determined by using a microplate reader. A biofilm assay was carried out essentially as described earlier (7). Briefly, the culture supernatant was discarded, and the wells were washed twice with phosphate-buffered saline (PBS) to remove nonadherent cells. The plates were air dried, and the surface-attached cells were stained with 200 μl of 0.1% crystal violet for 30 min. Subsequently, the crystal violet was removed and the plate was washed with water. After air drying, photos were taken from the surface-attached biofilm. For quantification of the biofilm cells, 200 μl of dimethyl sulfoxide (DMSO) was added, and crystal violet-stained biofilm cells were determined at 570 nm with the microplate reader.
Viability assays of preformed biofilms cells treated with gallidermin.
Microtiter plates were filled with 200 μl of 106 CFU/ml in TSB and incubated at 37°C for 24 h or 5 days without shaking. For establishment of 5-day biofilms, planktonic cells were discarded daily and replaced with fresh TSB. After incubation, the medium was removed and the wells were rinsed twice with PBS. Gallidermin (200 μl of various MICs) dissolved in TSB was added. After cultivation at 37°C for 24 h, the supernatant was discarded and replaced with 200 μl of PBS supplemented with 50 μg methylthiazoltetrazolium (MTT; Sigma). This kind of indirect viability assay is based on the formation of insoluble purple formazan due to the reduction of MTT by (respiratory) reductases of living staphylococcal cells. The formazan crystals were dissolved in DMSO, and the absorbance was determined at 570 nm with a microplate reader. Viability of staphylococcal cells was also tested by the viable count method (CFU). The biofilms were prepared as described above, with the only difference that larger flat-bottom microtiter plates were used. The formed biofilms were scratched off with a sterile spatula, sonicated, and then vortexed. The suspension was diluted and plated onto a TSA plate to detect the viable count after incubation.
Transcriptional analysis of atl and ica genes.
Staphylococcal cells were cultured in TSB until mid-exponential phase (after approximately 4 h). At this time, 4× MIC gallidermin dissolved in TSB was added; the control received the same amount of TSB only (see Fig. 1, below). Samples for RNA isolation were taken at 4, 6, and 8 h. Total RNA was isolated by using the acid-phenol method (16, 34) with some modifications, as described earlier (13). Briefly, 35-ml aliquots of collected samples were mixed with 15 ml of ice-cold killing buffer (20 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 20 mM NaN3) and immediately centrifuged at 5,000 × g at 4°C for 10 min. The cells were resuspended in 1 ml of the same buffer and centrifuged at 8,000 × g at 4°C for 10 min. Five hundred microliters of lysis buffer (3 mM EDTA, 200 mM NaCl) was added to suspend the pellets. The cell suspension was transferred to a screw-cap tube containing 500 μl phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol) with glass beads (diameter, 0.1 mm; Sartorius) and immediately mechanically disrupted by using a ribolyser at 6.5 m/s for 30 s. The mixture was centrifuged at 13,000 × g at 4°C for 10 min. The supernatant was extracted again with 600 μl phenol-alcohol-isoamyl alcohol. The upper phase was extracted twice with chloroform-isoamyl alcohol (25:1, vol/vol). RNA was precipitated with a 0.5× volume of absolute ethanol containing a 0.1× volume of 3 M sodium acetate at −80°C for 3 h. After centrifugation, the pellet was washed with 70% ethanol and resuspended in deionized water. RNA integrity was confirmed by agarose gel electrophoresis, and photometric measurements were made using a NanoDrop apparatus.
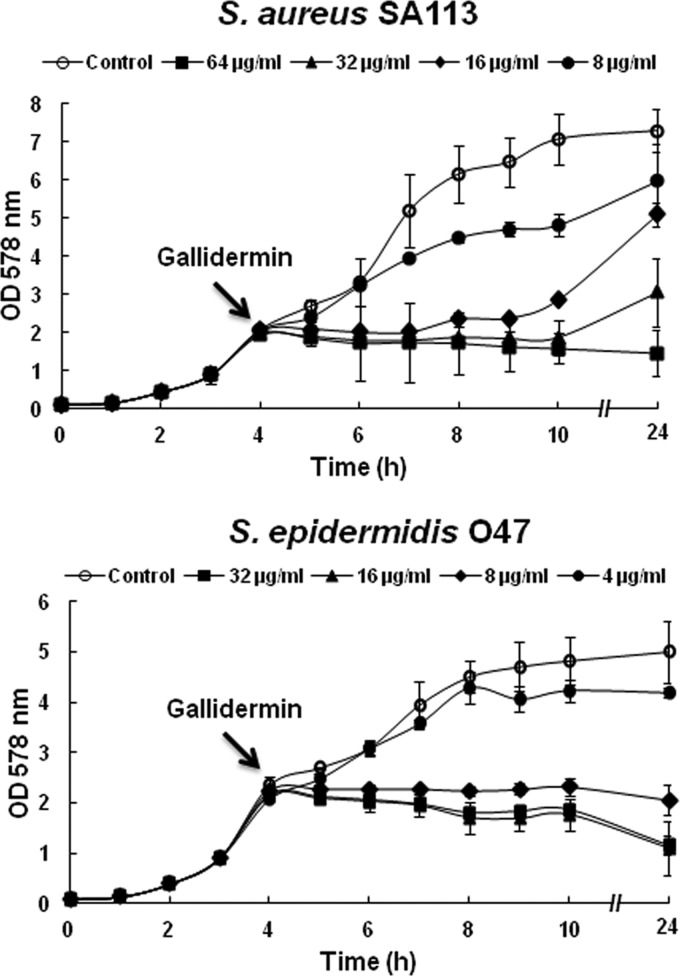
Fig 1.
Growth curves of S. aureus SA113 and S. epidermidis O47 after treatment with gallidermin at 1×, 2×, 4×, and 8× MIC of gallidermin. Gallidermin was added at the mid-exponential growth phase, after 4 h of cultivation. The MIC values of gallidermin for S. aureus SA113 and S. epidermidis O47 were 8 and 4 μg/ml, respectively.
RNA probe preparation.
Digoxigenin-labeled RNA probes of the corresponding genes were prepared by transcription with T7 RNA polymerase by using a PCR fragment as the template (16) that was generated from chromosomal DNA of S. aureus SA113 and S. epidermidis O47 and the respective oligonucleotides (Table 1). The reverse oligonucleotides contained a T7 RNA polymerase recognition site sequence at the 5′ end.
Table 1.
Oligonucleotide used in this study
| Genea | Gene namea | Direction | Primer sequence (5′–3′)b |
|---|---|---|---|
| SA0905 | atlA | Forward | GCTAATCAATCAGCGACTAC |
| Reverse | CTAATACGACTCACTATAGGGAGACAAATGCATGTACGAATGCG | ||
| SERP0636 | atlE | Forward | GGGACGTCCTGAAGGTATCG |
| Reverse | CTAATACGACTCACTATAGGGAGAGATGTTGTGCCCCAAGGTGC | ||
| SA2459 | icaA | Forward | GCCAACGCACTCAATCAAGG |
| Reverse | CTAATACGACTCACTATAGGGAGACTTCCAAAGACCTCCCAATG | ||
| SERP2293 | icaA | Forward | GAGGGAATCAAACAAGCATC |
| Reverse | CTAATACGACTCACTATAGGGAGACCATCGAACCCTTTGTTTCC |
Based on KEGG classification (www.genome.jp/kegg/).
The underlined sequence portions at the 5′ ends represent the recognition sequence for the T7 RNA polymerase.
Northern blot analyses.
Ten micrograms of total RNA was loaded into each well. The RNA was separated under denaturing conditions in a 1% agarose gel containing 20 mM morpholinepropanesulfonic acid, 5 mM sodium acetate, 1 mM EDTA, 1.85% formaldehyde. The gel was blotted onto a nylon membrane with 20× SSPE (3 M NaCl, 0.2 M NaH2PO4, 0.02 M EDTA; pH 7.4) by using a vacuum blotter for 4 h. The RNA was cross-linked to the membrane by UV for 1 min and stained with methylene blue. 16S and 23S rRNA bands on the membrane indicated the efficiency of blotting and served as a control for the quantity of RNA in each lane. RNA probes were used to detect gene-specific hybridization. Transcriptional signals were detected by using ChemiDoc (Bio-Rad).
Effect of gallidermin on autolysin activity in a zymogram.
A 20-ml aliquot of a culture at an OD578 of 0.1 of S. aureus SA113 or S. epidermidis O47 was cultured in TSB at 37°C with shaking at 150 rpm until mid-logarithmic phase was reached (after approximately 4 h). At that time point, gallidermin was added at a final concentration of 4× MIC and cultivated for another 4 h. Then, cells were centrifuged, washed in Tris-HCl (pH 6.8), and centrifuged at 13,000 rpm for 5 min. The cell pellet was treated with sample buffer (SDS with β-mercaptoethanol), incubated at 65°C for 3 min to remove cell wall-attached proteins, and put on ice for 5 min. After centrifugation, the supernatant was electrophoresed in a 12% SDS-PAGE gel containing heat-killed Micrococcus luteus cells as a substrate. After electrophoresis, the PAGE gel was washed with deionized water and then incubated in regeneration buffer overnight at 37°C. The gel was stained with 0.1% methylene blue and destained with deionized water. Proteins with autolysin activity were observed as clear bands in the gel.
Statistical analysis.
Statistical analysis was performed using analysis of variance (ANOVA). Comparisons between means were carried out according to Duncan's test. Differences were considered significant at a P level of <0.05.
RESULTS
Dose-dependent inhibition of staphylococcal growth by gallidermin.
The activities of gallidermin on planktonic cells of S. aureus and S. epidermidis were investigated by determination of MIC and MBC values in MHB medium. Gallidermin inhibited the growth of both S. aureus and S. epidermidis, and the MICs were in the range between 4 and 8 μg/ml, irrespective of the resistance of strains to methicillin, such as USA300. The biofilm-positive test strains used here were S. aureus SA113 and S. epidermidis O47; they showed MIC values of 8 and 4 μg/ml, respectively, if gallidermin was present from the beginning of growth (data not shown). However, when gallidermin was added to a mid-exponential-phase culture, higher doses (on average, 2× MIC) were necessary to completely inhibit growth (Fig. 1). In contrast to the O47 strain, SA113 was not only less sensitive to gallidermin but also showed a tendency to resume growth at 1× and 2× MIC after 10 h; only at 4× and 8× MIC was growth arrested over the entire period.
Gallidermin inhibits biofilm formation.
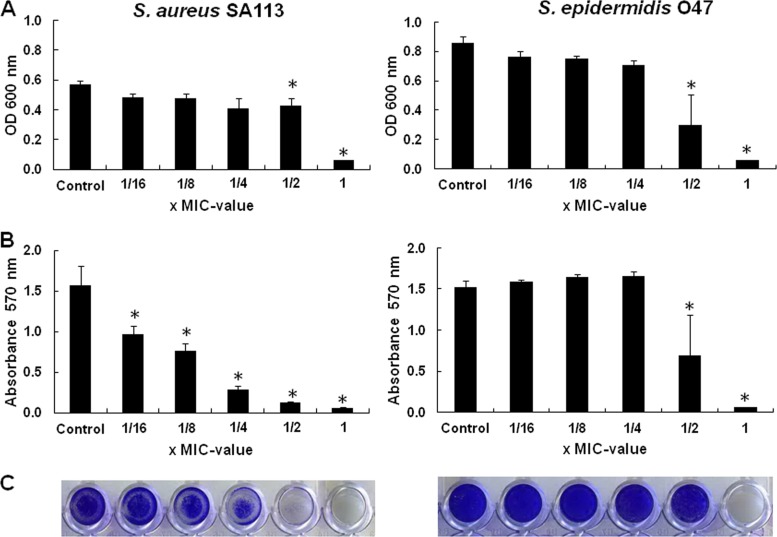
For determining the effect of gallidermin on biofilm formation, we used the quantitative microtiter plate method (7). The bacteria were cultured in microtiter plates for 24 h in the presence of subinhibitory concentrations of gallidermin (0.16× to 1× MIC), and the formed biofilm was stained with crystal violet. At 1× MIC of gallidermin, growth of S. aureus SA113 and S. epidermidis O47 was completely arrested; the O47 strain was arrested even at 0.5× MIC (Fig. 2A). However, biofilm formation, particularly of S. aureus SA113, was already inhibited at sublethal concentrations of gallidermin (0.16× MIC [0.5 μg/ml]; P < 0.05) (Fig. 2B and C). This result indicated that gallidermin already exerts an inhibiting effect on biofilm formation long before it inhibits growth and viability, probably by repression of biofilm-related genes (see below). In S. epidermidis O47, the effect was less pronounced than in S. aureus SA113; 0.5× MIC (2 μg/ml) affected both growth and biofilm formation.
Fig 2.
Biofilm formation of S. aureus SA113 and S. epidermidis O47 in the presence of subinhibitory concentrations of gallidermin. Growth and biofilm formation occurred in microtiter plates. Shown are the turbidity (the OD600) of the cell culture (A), the absorbance (A570) of stained cells attached to the surface after washing (B), and photos of biofilm cells attached to microtiter wells after crystal violet staining (C). Differences were considered significant at P < 0.05.
Effect of gallidermin on 1- and 5-day-old preformed biofilms.
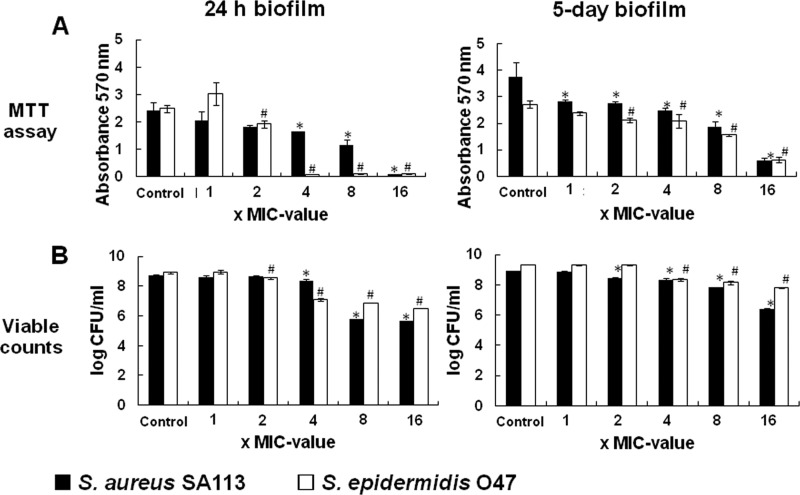
Once a biofilm has been established, the cells are extremely robust against all kinds of antibiotics. We therefore studied the effect of gallidermin on cell viability in 24-h and 5-day-old biofilms. For viability testing, an indirect method (the MTT assay) and direct method (viable counting) were used. The MTT assay is based on the activities of MTT-reducing enzymes (frequently NADH-dependent dehydrogenases) as an indication of live cells. Increasing concentrations (1× to 16× MIC) of gallidermin led to a decrease in formazan production, which was more rapid in the 24-h biofilm than in the 5-day biofilms (Fig. 3A), indicating that in an old biofilm, cells are more robust and protected.
Fig 3.
Gallidermin and viability of bacterial cells in 24-h and 5-day established biofilms of S. aureus SA113 and S. epidermidis O47. The viability of bacterial cells in 24-h and 5-day biofilms was detected by MTT assay (A) or viable count (B). Differences were considered significant at P < 0.05.
The MTT assay results correlated quite well with the viable count method results. Here too, gallidermin caused a dose-dependent (1× to 16× MIC) decrease of 3 logs in the 24-h biofilm and of 1 to 2 logs in the 5-day biofilms (Fig. 3B). S. epidermidis O47 survived slightly better than S. aureus SA113. While the bactericidal effect of gallidermin in planktonic cultures was very high (7- to 8-log reduction in CFU), it was low in biofilms, with only 1- to 3-log reductions of viable cells, depending on the age of the biofilm.
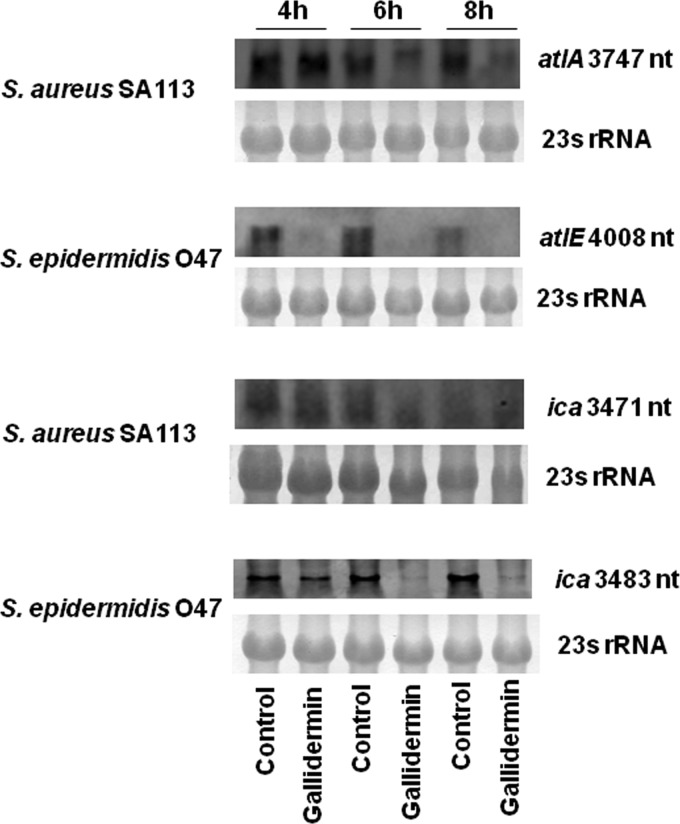
Gallidermin inhibits transcription of atl and ica genes.
As shown in Fig. 2B and C, biofilm formation was inhibited already at sublethal concentrations of gallidermin, suggesting that the expression of some biofilm-related genes was affected. We therefore carried out transcription analysis (Northern blotting) for atl and ica genes. atl encodes the major autolysin, which is involved in attachment to a surface, one of the first steps in biofilm formation (21); ica (intercellular adhesin) encodes polysaccharide intercellular adhesin (PIA), an exopolysaccharide composed of β-1,6-linked glucosaminoglycan (15, 22, 33). Staphylococci were grown to mid-exponential phase (4 h), gallidermin (4× MIC) was added, and RNA was isolated at 4, 6, and 8 h for Northern blot analysis. Indeed, in the presence of gallidermin, atl and ica transcripts were markedly reduced in both S. aureus and S. epidermidis strains (Fig. 4). The inhibiting effect of gallidermin on atl and ica transcription was more pronounced in S. epidermidis than in S. aureus.
Fig 4.
Effects of gallidermin on atl and ica transcription. RNA from S. aureus SA113 and S. epidermidis O47 was taken from 4-, 6-, and 8-h cultures (with or without 4× MIC gallidermin treatment) and analyzed by Nothern blotting. 23S rRNA was used as a control.
Major autolysin activity is decreased after treatment with gallidermin.
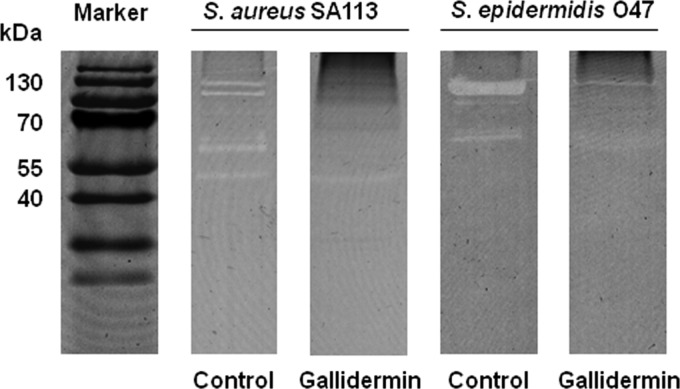
To verify whether the inhibition of atl transcription correlated with the major autolysin activity, we analyzed the major autolysin activity pattern in a zymogram with heat-inactivated Micrococcus luteus cells as the substrate. Indeed, the intensities of the autolysin bands were markedly reduced in the presence of gallidermin for both strains (Fig. 5). For S. aureus, the characteristic lytic bands (48) with molecular masses of 138 kDa (proautolysin), 113 kDa (autolysin), and 62 kDa (amidase) almost disappeared; only the 52-kDa lytic band (most likely the glucosaminidase) was only slightly decreased compared to the untreated culture.
Fig 5.
Autolysin zymogram profiles. S. aureus SA113 and S. epidermidis O47 were treated with or without 4× MIC of gallidermin, and the cell wall-associated proteins were separated by SDS-PAGE with heat-inactivated Micrococcus luteus cells as the substrate. Proteins with autolysin activity were visualized as clear (white) bands.
DISCUSSION
It has been known for some time that the lanthionine-containing peptide antibiotics epidermin and gallidermin have high bactericidal activities against many Gram-positive bacteria, including Bacteroides fragilis, Micrococcus luteus, Propionibacterium acnes, S. aureus, S epidermidis, Streptococcus pneumoniae, and Streptococcus pyogenes (25). Even the multidrug-resistant S. aureus (including MRSA) and oxacillin-resistant S. aureus (ORSA) strains, which are resistant to β-lactam antibiotics, including penicillins (methicillin, dicloxacillin, nafcillin, and oxacillin) and cephalosporins, were as susceptible as the nonresistant strains. The susceptibilities of the various staphylococcal species were quite diverse. While S. aureus and S. epidermidis showed MIC values in the range of 4 to 8 μg/ml (2 to 4 μM), some other staphylococcal species, such as nonpathogenic S. carnosus or S. simulans, are much more susceptible to gallidermin, with MIC values of approximately 0.3 μg/ml (0.15 μM) (4). This indicates that S. aureus and S. epidermidis have acquired some resistance traits that are lacking in the nonpathogenic species. There are several ways to improve tolerance against cationic antimicrobial peptides (CAMPs), including not only defensins but also many lantibiotics. It has been shown that modifications of the cell envelope that contribute to an increased positive charge render bacteria less susceptible to CAMPs. For example, mutants of S. aureus and other Gram-positive species that lack d-alanine (dlt mutants) in their teichoic acids (44), or tagO mutants that lack wall teichoic acid (2, 27, 55), or the mprF mutants that are unable to lysinylate lipids (10), are significantly more susceptible than the wild-type strains. As S. aureus and the skin-colonizing S. epidermidis are exposed to defensins, it is likely that they have acquired functions that increase tolerance to defensins. However, although S. aureus and S. epidermidis were less susceptible to gallidermin than the nonpathogenic species representatives, they were still sufficiently susceptible to consider gallidermin an antimicrobial therapeutic agent.
A major issue of this study was gallidermin activity against biofilms and biofilm formation. As test bacteria, we chose the well-studied biofilm-forming S. aureus SA113 (8) and S. epidermidis O47 (21). We found that gallidermin was able to completely inhibit biofilm formation at 1× MIC. For S. aureus, a biofilm-inhibiting effect was already observed in the sub-MIC range (Fig. 2), suggesting that biofilm-relevant functions were already affected at lower concentrations. Indeed, the transcription levels of two genes, one involved in primary adhesion (the major autolysin, atl) and one involved in exopolysaccharide production (the intercellular adhesin, ica), were significantly decreased in the presence of gallidermin (Fig. 4). The inhibition of transcription by gallidermin was also reflected by a decrease in autolytic protein bands in the zymogram (Fig. 5).
While gallidermin is a good drug for inhibiting the onset of biofilm formation, the killing activity on 24-h and 5-day-old preformed biofilms was comparatively low. Significant effects were only seen at ≥4× MIC. The number of CFU of biofilm-associated staphylococci treated with 8× MIC of gallidermin was decreased by 3 logs in the 24-h biofilms and 1 to 2 logs in the 5-day biofilms. This means that 0.1 to 1% of biofilm-associated staphylococcal cells survive gallidermin treatment. In this respect, gallidermin behaves like many other antibiotics that show good activity with planktonic cells but low activity with already-established biofilms. There are many reports on the activities of natural compounds on staphylococcal biofilm formation. Hydroxypropyltrimethyl ammonium chloride chitosan inhibits biofilm formation as well as expression of the icaA gene (40). Providone iodine also inhibits biofilm formation and decreases transcription of the ica operon (38). The effects of natural antibacterial agents, such as berberine, carvacrol, farnesol, oregano, rhodomyrtone, and thymol, against staphylococcal biofilms have been frequently reported (24, 37, 47, 54). Oregano, carvacrol, and thymol exhibit antistaphylococcal activity in biofilms. They apparently disseminate through the biofilm matrix and lead to cell damage (37). In addition, 1% tea tree oil is able to inhibit the metabolism of biofilms formed by all test isolates of S. aureus (32). Those authors suggested that biofilm destruction by tea tree oil was due not only to bacteria killing but also to destruction of the extracellular matrix and clearing of the biofilms from the surface (32). However, little is known about the side effects of these natural substances.
Maintenance of a high dose of antibiofilm drugs appears crucial, at least for some antibiotics. It has been reported that subinhibitory concentrations of various antibiotics even enhance staphylococcal biofilm formation, such as erythromycin (53), furanone (28), nafcillin (11), quinupristin-dalfopristin and tetracycline (46), and vancomycin (23). Gallidermin does not fall into this class of antibiotics, as biofilm formation is inhibited by concentrations that are subinhibitory for growth.
One of the future tasks will be the characterization of the genotype and phenotype of the gallidermin survivors. Such survivors, also called persisters, are found with almost every type of antibiotic tested so far, and this is therefore a general problem (26). Little is known about how these persisters arise and about their physiological state. Do these persisters comprise dormant cells or small-colony variants (52)? Are they genetically altered, or so they pause in a certain physiological state? More investigation into these persister cells is necessary in order to be able to develop appropriate antibiotic treatments against them.
ACKNOWLEDGMENTS
We appreciate the expert technical help of Vera Augsburger, Regine Stemmler, and Daniel Kühner.
The work was supported by Deutsche Forschungsgemeinschaft SFB766 and EuroSYNBIO.
Footnotes
Published ahead of print 27 August 2012
REFERENCES
- 1. Augustin J, et al. 1992. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur. J. Biochem. 204:1149–1154 [DOI] [PubMed] [Google Scholar]
- 2. Bera A, et al. 2007. Influence of wall teichoic acid on lysozyme resistance in Staphylococcus aureus. J. Bacteriol. 189:280–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bierbaum G, Brötz H, Koller KP, Sahl HG. 1995. Cloning, sequencing and production of the lantibiotic mersacidin. FEMS Microbiol. Lett. 127:121–126 [DOI] [PubMed] [Google Scholar]
- 4. Bonelli RR, Schneider T, Sahl HG, Wiedemann I. 2006. Insights into in vivo activities of lantibiotics from gallidermin and epidermin mode-of-action studies. Antimicrob. Agents Chemother. 50:1449–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brötz H, et al. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 30:317–327 [DOI] [PubMed] [Google Scholar]
- 6. Clinical and Laboratory Standards Institute 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, CLSI document M7-A7, 7th ed Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7. Cramton SE, Gerke C, Götz F. 2001. In vitro methods to study staphylococcal biofilm formation. Methods Enzymol. 336:239–255 [DOI] [PubMed] [Google Scholar]
- 8. Cramton SE, Gerke C, Schnell NF, Nichols WW, Götz F. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Diep BA, et al. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367:731–739 [DOI] [PubMed] [Google Scholar]
- 10. Ernst CM, et al. 2009. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 5:e1000660 doi:10.1371/journal.ppat.1000660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frank KL, Reichert EJ, Piper KE, Patel R. 2007. In vitro effects of antimicrobial agents on planktonic and biofilm forms of Staphylococcus lugdunensis clinical isolates. Antimicrob. Agents Chemother. 51:888–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Freund S, et al. 1991. The solution structure of the lantibiotic gallidermin. Biopolymers 31:803–811 [DOI] [PubMed] [Google Scholar]
- 13. Fuchs S, Pane-Farre J, Kohler C, Hecker M, Engelmann S. 2007. Anaerobic gene expression in Staphylococcus aureus. J. Bacteriol. 189:4275–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geissler S, Götz F, Kupke T. 1996. Serine protease EpiP from Staphylococcus epidermidis catalyzes the processing of the epidermin precursor peptide. J. Bacteriol. 178:284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gerke C, Kraft A, Sussmuth R, Schweitzer O, Götz F. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273:18586–18593 [DOI] [PubMed] [Google Scholar]
- 16. Gertz S, et al. 1999. Regulation of σB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 261:558–566 [DOI] [PubMed] [Google Scholar]
- 17. Götz F. 2004. Staphylococci in colonization and disease: prospective targets for drugs and vaccines. Curr. Opin. Microbiol. 7:477–487 [DOI] [PubMed] [Google Scholar]
- 18. Götz F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367–1378 [DOI] [PubMed] [Google Scholar]
- 19. Götz F, Peters G. 2000. Colonization of medical devices by coagulase-negative staphylococci, p 55–88 In Waldvogel FA, Bisno AL. (ed), Infections associated with indwelling medical devices, 3rd ed ASM Press, Washington, DC [Google Scholar]
- 20. Gould IM, et al. 2012. New insights into methicillin-resistant Staphylococcus aureus (MRSA) pathogenesis, treatment and resistance. Int. J. Antimicrob. Agents 39:96–104 [DOI] [PubMed] [Google Scholar]
- 21. Heilmann C, Hussain M, Peters G, Götz F. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013–1024 [DOI] [PubMed] [Google Scholar]
- 22. Heilmann C, et al. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083–1091 [DOI] [PubMed] [Google Scholar]
- 23. Hsu CY, et al. 2011. Vancomycin promotes the bacterial autolysis, release of extracellular DNA, and biofilm formation in vancomycin-non-susceptible Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 63:236–247 [DOI] [PubMed] [Google Scholar]
- 24. Jabra-Rizk MA, Meiller TF, James CE, Shirtliff ME. 2006. Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob. Agents Chemother. 50:1463–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kellner R, et al. 1988. Gallidermin: a new lanthionine-containing polypeptide antibiotic. Eur. J. Biochem. 177:53–59 [DOI] [PubMed] [Google Scholar]
- 26. Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13–18 [DOI] [PubMed] [Google Scholar]
- 27. Köhler T, Weidenmaier C, Peschel A. 2009. Wall teichoic acid protects Staphylococcus aureus against antimicrobial fatty acids from human skin. J. Bacteriol. 191:4482–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuehl R, et al. 2009. Furanone at subinhibitory concentrations enhances staphylococcal biofilm formation by luxS repression. Antimicrob. Agents Chemother. 53:4159–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuipers OP, Beerthuyzen MM, Siezen RJ, De Vos WM. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281–291 [DOI] [PubMed] [Google Scholar]
- 30. Kupke T, Götz F. 1996. Post-translational modifications of lantibiotics. Antonie van Leeuwenhoek 69:139–150 [DOI] [PubMed] [Google Scholar]
- 31. Kupke T, Kempter C, Gnau V, Jung G, Götz F. 1994. Mass spectroscopic analysis of a novel enzymatic reaction. Oxidative decarboxylation of the lantibiotic precursor peptide EpiA catalyzed by the flavoprotein EpiD. J. Biol. Chem. 269:5653–5659 [PubMed] [Google Scholar]
- 32. Kwiecinski J, Eick S, Wojcik K. 2009. Effects of tea tree (Melaleuca alternifolia) oil on Staphylococcus aureus in biofilms and stationary growth phase. Int. J. Antimicrob. Agents 33:343–347 [DOI] [PubMed] [Google Scholar]
- 33. Mack D, et al. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Majumdar D, Avissar YJ, Wyche JH. 1991. Simultaneous and rapid isolation of bacterial and eukaryotic DNA and RNA: a new approach for isolating DNA. Biotechniques 11:94–101 [PubMed] [Google Scholar]
- 35. Müller A, Ulm H, Reder-Christ K, Sahl HG, Schneider T. 2012. Interaction of type A lantibiotics with undecaprenol-bound cell envelope precursors. Microb. Drug Resist. 18:261–270 [DOI] [PubMed] [Google Scholar]
- 36. Niu WW, Neu HC. 1991. Activity of mersacidin, a novel peptide, compared with that of vancomycin, teicoplanin, and daptomycin. Antimicrob. Agents Chemother. 35:998–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nostro A, et al. 2007. Effects of oregano, carvacrol and thymol on Staphylococcus aureus and Staphylococcus epidermidis biofilms. J. Med. Microbiol. 56:519–523 [DOI] [PubMed] [Google Scholar]
- 38. Oduwole KO, et al. 2010. Anti-biofilm activity of sub-inhibitory povidone iodine concentrations against Staphylococcus epidermidis and Staphylococcus aureus. J. Orthop. Res. 28:1252–1256 [DOI] [PubMed] [Google Scholar]
- 39. Ottenwälder B, et al. 1995. Isolation and characterization of genetically engineered gallidermin and epidermin analogs. Appl. Environ. Microbiol. 61:3894–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peng ZX, et al. 2011. Quaternized chitosan inhibits icaA transcription and biofilm formation by Staphylococcus on a titanium surface. Antimicrob. Agents Chemother. 55:860–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peschel A, Augustin J, Kupke T, Stevanovic S, Götz F. 1993. Regulation of epidermin biosynthetic genes by EpiQ. Mol. Microbiol. 9:31–39 [DOI] [PubMed] [Google Scholar]
- 42. Peschel A, Götz F. 1996. Analysis of the Staphylococcus epidermidis genes epiF, -E, and -G involved in epidermin immunity. J. Bacteriol. 178:531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Peschel A, Ottenwälder B, Götz F. 1996. Inducible production and cellular location of the epidermin biosynthetic enzyme EpiB using an improved staphylococcal expression system. FEMS Microbiol. Lett. 137:279–284 [DOI] [PubMed] [Google Scholar]
- 44. Peschel A, et al. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405–8410 [DOI] [PubMed] [Google Scholar]
- 45. Peschel A, Schnell N, Hille M, Entian KD, Götz F. 1997. Secretion of the lantibiotics epidermin and gallidermin: sequence analysis of the genes gdmT and gdmH, their influence on epidermin production and their regulation by EpiQ. Mol. Gen. Genet. 254:312–318 [DOI] [PubMed] [Google Scholar]
- 46. Rachid S, Ohlsen K, Witte W, Hacker J, Ziebuhr W. 2000. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44:3357–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Saising J, Ongsakul M, Voravuthikunchai SP. 2011. Rhodomyrtus tomentosa (Aiton) Hassk ethanol extract and rhodomyrtone: a potential strategy for the treatment of biofilm-forming staphylococci. J. Med. Microbiol. 60:1793–1800 [DOI] [PubMed] [Google Scholar]
- 48. Schlag M, et al. 2010. Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol. Microbiol. 75:864–873 [DOI] [PubMed] [Google Scholar]
- 49. Schnell N, et al. 1992. Analysis of genes involved in the biosynthesis of lantibiotic epidermin. Eur. J. Biochem. 204:57–68 [DOI] [PubMed] [Google Scholar]
- 50. Schnell N, et al. 1991. The operon-like organisation of lantibiotic epidermin biosynthesis genes, p 269–276 In Jung G, Sahl H-G. (ed), Nisin and novel lantibiotics. ESCOM, Leiden, The Netherlands [Google Scholar]
- 51. Schnell N, et al. 1988. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature 333:276–278 [DOI] [PubMed] [Google Scholar]
- 52. von Eiff C, et al. 1997. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 179:4706–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang Q, et al. 2010. Enhancement of biofilm formation by subinhibitory concentrations of macrolides in icaADBC-positive and -negative clinical isolates of Staphylococcus epidermidis. Antimicrob. Agents Chemother. 54:2707–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang XQ, et al. 2009. Effect of berberine on Staphylococcus epidermidis biofilm formation. Int. J. Antimicrob. Agents 34:60–66 [DOI] [PubMed] [Google Scholar]
- 55. Weidenmaier C, et al. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10:243–245 [DOI] [PubMed] [Google Scholar]