Abstract
AL18, an inhibitor of human cytomegalovirus DNA polymerase, was serendipitously found to also block the interaction between the PB1 and PA polymerase subunits of influenza A virus. Furthermore, AL18 effectively inhibited influenza A virus polymerase activity and the overall replication of influenza A and B viruses. A molecular model to explain the binding of AL18 to both cytomegalovirus and influenza targets is proposed. Thus, AL18 represents an interesting lead for the development of new antivirals.
TEXT
Influenza A virus is a major human pathogen responsible for respiratory diseases characterized by high morbidity and significant mortality. Influenza A causes seasonal epidemics affecting millions of people worldwide but can also provoke pandemic outbreaks with higher attack rates and potentially more-severe disease (18).
Currently, two classes of anti-influenza A drugs are available: adamantanes, which block the M2 ion channel and inhibit virus entry, and neuraminidase inhibitors, which prevent the release of virions from the host cell (5). However, they all suffer from limited efficacy, adverse side effects, and emergence of drug resistance. Vaccines also exist, but they must be reformulated annually and they give limited protection. Thus, there is still a considerable need for new anti-influenza drugs.
The influenza A RNA polymerase, a heterotrimer of the PA, PB1, and PB2 subunits, provides an underexploited drug target. The three subunits bind each other and are all essential for viral RNA synthesis (18). Recent crystal structures revealed that the PA-PB1 binding interface consists of an N-terminal helix from PB1 that binds into a groove in the C terminus of PA (7, 17). Since the association of these subunits is essential for viral replication (19), and since the amino acids of both PB1 and PA that are crucial for subunit interaction are highly conserved among influenza A strains (6, 10), this interaction represents an attractive target for antiviral drugs. To this end, we recently developed a PA-PB1 interaction assay and used it to identify compounds able to inhibit the interaction (16).
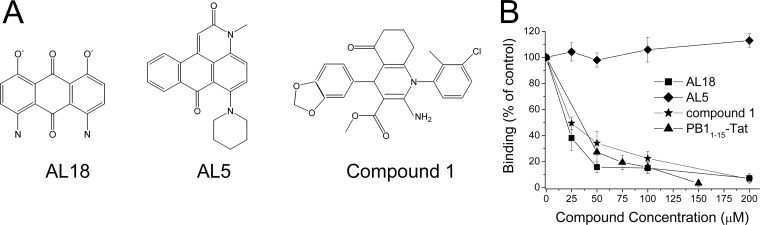
In these influenza A PB1-PA binding assays, microtiter wells coated with 6His-PA239-716, a 6His-tagged form of the PA C-terminal domain, are incubated with glutathione S-transferase (GST)-PB11-25, a GST fusion with the N-terminal 25 residues of PB1 (which are sufficient to bind the PA C-terminal domain [6, 7]), in the absence or the presence of test compounds. Active inhibitors, such as a PB1-derived peptide fused to sequences from HIV Tat (PB11-15-Tat [6]) or the small-molecule “compound 1” inhibitor we recently identified (16), effectively blocked the PA-PB1 interaction as expected (Fig. 1). During the course of these studies, we also tested small molecules able to disrupt the interaction between the UL54 and UL44 subunits of human cytomegalovirus (HCMV) DNA polymerase (13), expecting them to provide negative controls. Unexpectedly, the most active of the HCMV inhibitors, AL18, also inhibited the PA-PB1 interaction with a 50% inhibitory concentration (IC50; calculated by linear regression using the computer program GraphPad Prism version 4.0) of 20.3 ± 2.6 μM (Fig. 1). In contrast, another of the previously characterized UL54-UL44 inhibitors, AL5 (13), did not affect PA-PB1 binding (Fig. 1).
Fig 1.
(A) Chemical structures of AL18, AL5, and compound 1. (B) Effects of AL18 on PA-PB1 interactions. Increasing concentrations of the indicated compounds were added together with 200 ng of GST-PB11-25 to wells coated with 400 ng of 6His-PA239-716. Binding of GST-PB11-25 to 6His-PA239-716 was detected with a horseradish peroxidase (HRP)-conjugated anti-GST antibody, followed by measurement of the absorbance at 450 nm. Data shown are the means ± standard deviations (SDs) of three independent experiments.
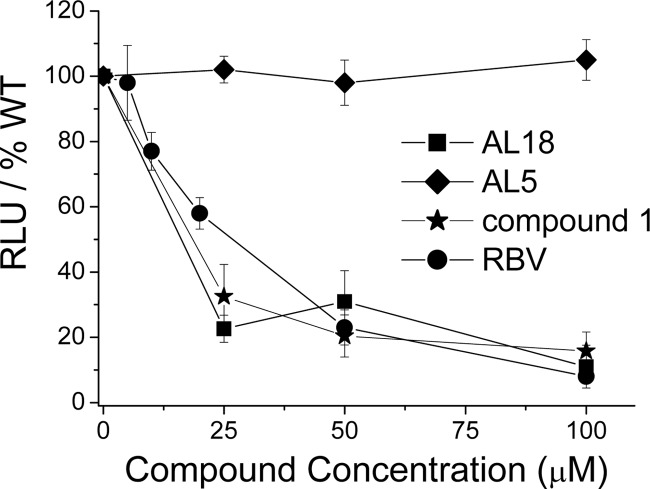
To investigate whether the in vitro inhibitory activity of AL18 on the minimal PA-PB1 binding domains was matched by a corresponding effect on the influenza A transcription machinery in cells, the compound was tested in influenza A minireplicon assays (15, 16). Ribavirin (RBV; from Roche), a known inhibitor of viral RNA polymerases (20), and the anti-PA-PB1 compound 1 served as positive controls. Human embryonic kidney (HEK) 293T cells were cotransfected with plasmids encoding the three polymerase subunits and the viral nucleoprotein (NP) along with a plasmid carrying the firefly luciferase reporter gene flanked by the noncoding sequences of influenza A (A/WSN/33 strain) segment 8 and a plasmid coding for Renilla luciferase (pRL-SV40; Promega) which served to normalize variations in transfection efficiency, and the cells were treated with test or control compounds. The firefly reporter gene expression indicates that a negative-sense RNA is synthesized and is reconstituted intracellularly into functional RNPs in which all four viral proteins are coexpressed and interact with each other. A strong inhibition of firefly luciferase activity, measured at 24 h posttransfection (Fig. 2), was observed upon treatment with AL18 (50% effective concentration [EC50], calculated by linear regression using GraphPad Prism 4.0, of 16.5 ± 3.5 μM) and, as expected, with RBV and compound 1 (EC50s of 26.5 ± 4.8 μM and 18.4 ± 4.5 μM, respectively), whereas AL5 had no effect (EC50 > 100 μM).
Fig 2.
Ability of AL18 to inhibit influenza A polymerase activity in minireplicon assays. HEK 293T cells were cotransfected with plasmids encoding PB1, PB2, PA, and NP along with a plasmid carrying the firefly luciferase reporter gene flanked by the noncoding sequences of influenza A segment 8 and treated with the indicated compounds. The transfection mixtures also contained a plasmid constitutively expressing Renilla luciferase which served to normalize variations in transfection efficiency. Luciferase activity was quantified at 24 h posttransfection. The data are the means ± SDs of three independent experiments plotted relative to the activity seen in the presence of the compound vehicle (dimethyl sulfoxide [DMSO]). Omission of PB2 served as a negative control.
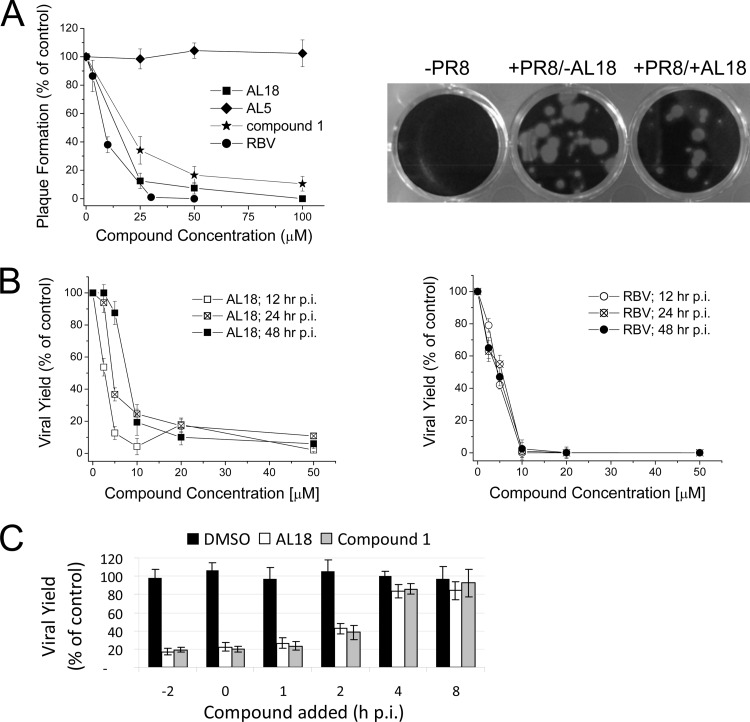
Next, the effect of AL18 on the replication of influenza A (A/PR/8/34 strain, from the Department of Pathology, University of Cambridge, United Kingdom) in Madin-Darby canine kidney (MDCK) cells was evaluated by plaque reduction assays (PRAs) as described previously (16). AL18 inhibited influenza A plaque formation in a dose-dependent manner (Fig. 3A), with EC50s similar to those of compound 1 (EC50s were 14.5 ± 2.1 μM and 19.4 ± 3.2 μM, respectively) (Fig. 3A and Table 1), while AL5 did not show significant activity (EC50 > 100 μM) (Fig. 3A). In addition, AL18 showed antiviral activity against several influenza A strains, including pandemic swine-originated influenza virus strains (from C. Salata, University of Padua, Italy) and an oseltamivir-resistant clinical isolate (A/Parma/24/09; from I. Donatelli, Istituto Superiore di Sanità, Rome, Italy), with EC50s ranging from 13.5 to 27.8 μM. In parallel, we tested the cytotoxicity of the compound both by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT; Sigma) assays (16) and by using a bioluminescence-based ATP determination kit (ATPlite; PerkinElmer). AL18 did not exhibit cytotoxicity in MDCK cells up to at least 250 μM (Table 1). We also tested the activity of AL18 on viral yield. MDCK cells were infected with A/PR/8/34 at a multiplicity of infection (MOI) of 0.01 and then treated with AL18 or RBV as a control, and the viral titers in compound-treated and control cells were measured at 12, 24, and 48 h postinfection (p.i.). AL18 inhibited virus yield with an EC50 of 2.5 ± 0.9 μM at 12 h, an EC50 of 4.5 ± 1.5 μM at 24 h, and an EC50 of 7.5 ± 2.6 μM at 48 h p.i. (Fig. 3B). As expected, RBV also effectively reduced viral titers (Fig. 3B). In time-of-addition experiments, performed as described in reference 9, inhibition was observed when AL18, or compound 1 for comparison, was added preinfection or up to 2 h postinfection but not at later time points (Fig. 3C). Thus, in contrast to M2 and neuraminidase inhibitors, which block virus replication at earlier and later stages, respectively (8, 21), both AL18 and compound 1 appear to act at early to middle stages, consistent with the observation of inhibition of the viral polymerase activity in minireplicon assays (Fig. 2).
Fig 3.
Activity of AL18 against influenza A replication. (A) Effects of the indicated compounds on plaque formation by A/PR/8/34 virus in MDCK cells. Representative pictures of mock-infected (−PR8), A/PR/8/34-infected untreated (+PR8/−AL18), and A/PR/8/34-infected, AL18-treated (+PR8/+AL18) cells are shown on the right. (B) Effects of AL18 (left) and of RBV as a control (right) on the yield of A/PR/8/34 virus following low-MOI infections of MDCK cells, determined at 12, 24, and 48 h p.i. (C) Inhibition of A/PR/8/34 virus replication by AL18 and by compound 1 added at different times before or during the viral life cycle. Data shown in panels A, B, and C are the means ± SDs of two or three independent experiments in duplicate.
Table 1.
Activity of AL18 against a panel of DNA and RNA virusesa
| Virus (strain) | Family (subfamily) | Genome | Activity of: |
||||||
|---|---|---|---|---|---|---|---|---|---|
| AL18 |
Controlb |
||||||||
| EC50c (μM) | MTT assay CC50d (μM) | ATP assay CC50d (μM) | SIe | EC50 (μM) | MTT assay CC50 (μM) | SI | |||
| HSV-1 (F) | Herpesviridae (α) | dsDNA | >50 | >250 | >5 | 1.6 ± 0.7 | >250 | >156 | |
| HCMV (AD169) | Herpesviridae (β) | dsDNA | 0.9 ± 0.2 | 250 ± 58 | 232 ± 24 | 278 | 1.3 ± 0.3 | 320 ± 82 | 246 |
| HHV-6A (GS) | Herpesviridae (β) | dsDNA | 5.8 ± 1.2 | 35.3 ± 3.7 | 22.4 ± 4.8 | 6 | 6.1 ± 2.5 | 770 ± 132 | 126 |
| HHV-6B (Z29) | Herpesviridae (β) | dsDNA | 6.1 ± 2.6 | 38.0 ± 6.7 | 25.6 ± 5.7 | 6 | 3.7 ± 1.9 | 980 ± 144 | 265 |
| MCMV (Smith) | Herpesviridae (β) | dsDNA | >30 | >100 | >3 | 0.7 ± 0.2 | 278 ± 12 | 397 | |
| HHV-8 | Herpesviridae (γ) | dsDNA | >50 | >250 | >5 | 5.8 ± 1.3 | 312 ± 80 | 54 | |
| AdV | Adenoviridae | dsDNA | >50 | >250 | >5 | 25.6 ± 3.2 | >250 | >10 | |
| Influenza A (A/PR/8/34) | Orthomyxoviridae | (−) ssRNA | 14.5 ± 2.1 | >250 | >250 | >17 | 8.5 ± 1.8 | >250 | >29 |
| Influenza B (B/Lee/40) | Orthomyxoviridae | (−) ssRNA | 8.3 ± 1.9 | >250 | >250 | >30 | 18.3 ± 2.6 | >250 | >14 |
| MV | Paramyxoviridae | (−) ssRNA | >50 | >250 | >5 | 85.4 ± 5.8 | >250 | >3 | |
| RSV | Paramyxoviridae | (−) ssRNA | >50 | >250 | >5 | 21.6 ± 3.6 | >250 | >12 | |
| VSV | Rhabdoviridae | (−) ssRNA | >50 | >250 | >5 | 10.3 ± 2.9 | >250 | >24 | |
| COX B1 | Picornaviridae | (+) ssRNA | >50 | >250 | >5 | ND | |||
dsDNA, double-stranded DNA; ND, not determined; ssRNA, single-stranded RNA.
Control compounds were GCV for HSV-1, HCMV, and HHV-8, FOS for HHV-6A and HHV-6B, CDV for MCMV and AdV, and RBV for all RNA viruses except COX B1.
EC50, the concentration of compound that inhibits 50% of virus replication, was determined by PRAs for HCMV, HSV-1, MCMV, AdV, influenza A, influenza B, MV, RSV, and VSV, by quantitative real-time PCR for HHV-6 and HHV-8, and by estimation of cytopathic effect for COX B1.
CC50, the concentration of compound that inhibits 50% of cell growth, was determined in different cell lines by MTT assays and by ATP assays and was calculated by linear regression using GraphPad Prism 4.0. Cell lines were Vero for HSV-1, MV, and COX B1, HFF for HCMV, HSB-2 for HHV-6A, MOLT-3 for HHV-6B, NIH 3T3 for MCMV, BC-3 for HHV-8, A549 for AdV, MDCK for influenza A and influenza B, L929 for VSV, and HEp-2 for RSV.
SI, the selectivity index as determined by the ratio between the CC50 determined by MTT assays and the EC50. CC50 and EC50 were determined under the same cell culture conditions.
To further evaluate the therapeutic potential and the antiviral selectivity of AL18, we tested its effects on the replication of a panel of other DNA and RNA viruses (Table 1): herpes simplex virus type 1 (HSV-1; from the American Type Culture Collection [ATCC], Manassas, VA), HCMV (from ATCC), human herpesvirus 6 type A and type B (HHV-6A and HHV-6B; from L. Naesens, Rega Institute for Medical Research, Leuven, Belgium), murine cytomegalovirus (MCMV; from D. Lembo, University of Turin, Italy), human herpesvirus 8 (HHV-8; from C. Salata, University of Padua, Italy), adenovirus (AdV; from R. Cusinato, Padua University Hospital, Italy), influenza B virus (B/Lee/40 strain), vesicular stomatitis virus (VSV), respiratory syncytial virus (RSV), measles virus (MV), and coxsackie virus B1 (COX B1) (all RNA viruses except influenza B, which was from W. S. Barclay, Imperial College, London, United Kingdom, were from R. Cusinato). The ability of AL18 to block virus growth was evaluated by PRAs (for HSV-1, HCMV, MCMV, AdV, influenza B, VSV, RSV, and MV), by quantification of viral DNA by quantitative real-time PCR (for HHV6-A, HHV6-B, and HHV-8), or by estimation of the cytopathic effect (for COX B1) as described previously (13, 14, 16). The antiviral drugs ganciclovir (GCV; from Sigma), foscarnet (FOS; from Sigma), cidofovir (CDV; from Pfizer), and RBV were used as reference compounds.
As previously reported (13), AL18 showed potent antiviral activity against HCMV with a selectivity index (SI) comparable to that of GCV (Table 1). Conversely, it was inactive against other herpesviruses, such as HSV-1, HHV-8, or MCMV, but showed some inhibitory activity against HHV-6A and HHV-6B (Table 1). However, the latter activity may have been due in part to cytotoxicity in the cell lines used to grow the HHV-6 viruses, as the SI values were poor (Table 1). No significant antiviral activity was seen against another DNA virus (adenovirus) or RNA viruses other than influenza A and influenza B (Table 1). Remarkably, AL18 exhibited an EC50 against influenza B (8.3 ± 1.9 μM) that was comparable to that against influenza A (Table 1). Thus, AL18 has dual specificity, acting as a potent inhibitor of one or more members from two evolutionarily distinct virus families: HCMV and influenza A and B.
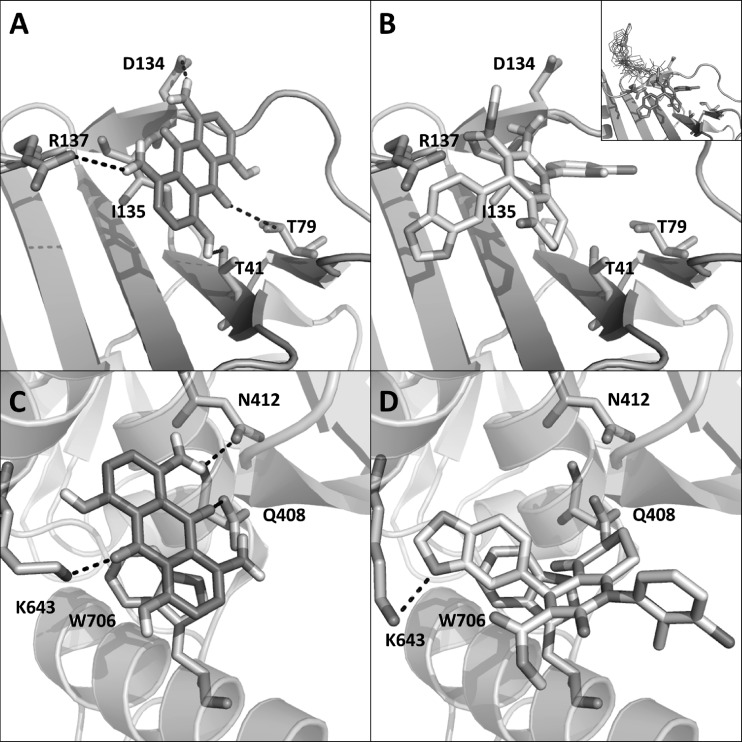
The activity of AL18 against both HCMV DNA polymerase (13) and influenza A RNA polymerase was surprising and would not have been predicted on the basis of any obvious sequence similarity of the polymerase interaction domains. Indeed, the lack of specific antiviral activity of AL18 against herpesviruses other than HCMV was not completely unexpected. Previous studies have shown that the molecular details of the HCMV UL54-UL44 interaction are quite different from those of the interaction between the counterparts from other herpesviruses (1, 2, 4, 11, 12, 22). Nevertheless, knowing the predicted target sites of the drug in both systems allowed us to use molecular modeling to investigate whether a plausible explanation for the dual specificity was able to be found. In fact, by analyzing docked poses of AL18 with the UL44 protein of HCMV DNA polymerase and with the PA subunit of influenza A RNA polymerase (Fig. 4) using the FLAP (Fingerprints for Ligands And Proteins) software (3), possible molecular explanations for the dual anti-HCMV and anti-influenza activities of the compound were able to be proposed. According to the FLAP docking poses with HCMV UL44 (Fig. 4A), AL18 strongly interacts with three residues in the central part of the so-called “connector loop,” a region of UL44 shown to be crucial for UL54 binding (1, 12). In particular, the two anilinic groups of AL18 form hydrogen bonds with D134 and R137. Additionally, the side chain of I135 is positioned as a hydrophobic anchor underneath the hydrogen-bonding network of AL18, mimicking the behavior already observed for UL54 binding by UL44 (1). Furthermore, a second set of weaker hydrogen-bonding interactions might occur between the carbonyl and hydroxyl groups of AL18 at positions 10 and 4 and the T41 and T79 residues of UL44, respectively. Inspecting the top 10 docking solutions, eight out of 10 poses of AL18 in UL44 are in agreement with the binding mode reported in Fig. 4A. Compound 1 was reported to not inhibit HCMV replication (16). Nine out of the 10 best FLAP poses of compound 1 in UL44 (Fig. 4B) are located in a more external binding region (inset in Fig. 4B), and thus, being highly exposed at the UL44 surface, compound 1 may be easily displaced. The only pose predicted at the same AL18 and UL54 binding regions is driven by hydrophobic interactions only, while H-bond interactions do not occur.
Fig 4.
Molecular basis of the interaction of AL18 and compound 1 with the UL44 subunit of HCMV DNA polymerase and the PA subunit of influenza A RNA polymerase. (A and B) FLAP best pose for AL18 (A) and compound 1 (B) in HCMV UL44. The inset in panel B shows the 10 best poses for compound 1 in HCMV UL44: nine out of 10 poses (in wireframe style) are clustered in a more external binding region. (C and D) FLAP best pose for AL18 (C) and compound 1 (D) in influenza A PA. Flexibility of the K643 side chain in influenza A PA was considered. In all panels, the residues of the protein target that interacts with AL18 or compound 1 are reported in sticks mode. The indicated AL18-binding residues of influenza A PA are conserved in influenza B.
By analyzing FLAP docked poses with influenza A PA, AL18 (Fig. 4C) appears to be docked in the same binding region of compound 1 (Fig. 4D) in the PA cavity (16). Looking at the best 10 poses in terms of FLAP similarity scores, all of them display the same binding region. Seven poses out of 10 are oriented as reported in Fig. 4C. The aromatic moiety of AL18 is involved in a π-π stacking with W706. Furthermore, AL18 forms a hydrogen-bonding network with the N412, Q408, and K643 residues. All the mentioned AL18-interacting residues of PA are highly conserved among influenza A virus strains (6), suggesting that AL18 will likely have broad-spectrum activity against influenza A viruses of both human and animal origin. Intriguingly, these residues of influenza A PA are also conserved in influenza B and match well upon structural alignment (16). Thus, the FLAP docking pose is also in agreement with the experimentally observed inhibition of both influenza A and influenza B replication by AL18. The remaining three poses of AL18 in influenza A PA overlap the other poses, but the structures are oppositely oriented, having the hydroxyl moiety pointed toward the N412 residue. Due to the specific structure of AL18, the hydrogen-bonding network, as well as the hydrophobic interaction with W706, is largely retained also in the second binding mode.
In contrast to AL18 and compound 1, AL5 docking into the influenza A PA cavity seems to be driven mainly by hydrophobic interactions involving the W706 residue (data not shown). All the best 10 poses for AL5 are perfectly matching. The lack of an H-bond network can be responsible for a weak interaction of AL5 with PA, which may explain the lack of anti-influenza activity of AL5. Mutagenesis studies to confirm the molecular modeling predictions are planned.
In conclusion, the AL18 compound exhibits anti-influenza A and anti-influenza B activity with a potency comparable to that of compound 1, a recently identified antiviral agent that acts by the same mechanism (16). Unlike compound 1, however, AL18 also possesses potent anti-HCMV activity. Thus, these data suggest that AL18 merits further consideration as a starting point for the development of new antiviral agents.
ACKNOWLEDGMENTS
We thank W. S. Barclay, R. Cusinato, I. Donatelli, D. Lembo, L. Naesens, and C. Salata for kindly providing viruses. We gratefully acknowledge Donald M. Coen for critical reading of the manuscript.
This work was supported by MURST EX60% and PRIN 2008 (grant no. 20085FF4J4 to A.L.), by the Italian Ministry of Health and Istituto Superiore Sanità, Progetto finalizzato 2009 “Studio e sviluppo di nuovi farmaci antivirali contro infezioni da virus influenzale A-H1N1” (to A.L. and G.P.), and by the Medical Research Council (grant no. G0801931 to P.D.).
Footnotes
Published ahead of print 20 August 2012
REFERENCES
- 1. Appleton BA, et al. 2006. Crystal structure of the cytomegalovirus DNA polymerase subunit UL44 in complex with the C terminus from the catalytic subunit: differences in structure and function relative to unliganded UL44. J. Biol. Chem. 281:5224–5232 [DOI] [PubMed] [Google Scholar]
- 2. Baltz JL, et al. 2009. The crystal structure of PF-8, the DNA polymerase accessory subunit from Kaposi's sarcoma-associated herpesvirus. J. Virol. 83:12215–12228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baroni M, Cruciani G, Sciabola S, Perruccio F, Mason JS. 2007. A common reference framework for analyzing/comparing proteins and ligands. Fingerprints for Ligands and Proteins (FLAP): theory and application. J. Chem. Infect. Model 47:279–294 [DOI] [PubMed] [Google Scholar]
- 4. Bridges KG, Chow CS, Coen DM. 2001. Identification of crucial hydrogen-bonding residues for the interaction of herpes simplex virus DNA polymerase subunits via peptide display, mutational, and calorimetric approaches. J. Virol. 75:4990–4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Clercq E. 2006. Antiviral agents active against influenza A viruses. Nat. Rev. Drug Discov. 5:1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ghanem A, et al. 2007. Peptide-mediated interference with influenza A virus polymerase. J. Virol. 81:7801–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. He X, et al. 2008. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 454:1123–1126 [DOI] [PubMed] [Google Scholar]
- 8. Hoffmann CE, Neumayer EM, Haff RF, Goldsby RA. 1965. Mode of action of the antiviral activity of amantadine in tissue culture. J. Bacteriol. 90:623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoffmann HH, Kunz A, Simon VA, Palese P, Shaw ML. 2011. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 108:5777–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu H, Yao X. 2010. Molecular basis of the interaction for an essential subunit PA-PB1 in influenza virus RNA polymerase: insights from molecular dynamics simulation and free energy calculation. Mol. Pharm. 7:75–85 [DOI] [PubMed] [Google Scholar]
- 11. Loregian A, Appleton BA, Hogle JM, Coen DM. 2004. Residues of human cytomegalovirus DNA polymerase catalytic subunit UL54 that are necessary and sufficient for interaction with the accessory protein UL44. J. Virol. 78:158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Loregian A, Appleton BA, Hogle JM, Coen DM. 2004. Specific residues in the connector loop of the human cytomegalovirus DNA polymerase accessory protein UL44 are crucial for interaction with the UL54 catalytic subunit. J. Virol. 78:9084–9092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Loregian A, Coen DM. 2006. Selective anti-cytomegalovirus compounds discovered by screening for inhibitors of subunit interactions of the viral polymerase. Chem. Biol. 13:191–200 [DOI] [PubMed] [Google Scholar]
- 14. Mercorelli B, et al. 2009. A 6-aminoquinolone compound, WC5, with potent and selective anti-human cytomegalovirus activity. Antimicrob. Agents Chemother. 53:312–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mullin AE, Dalton RM, Amorim MJ, Elton D, Digard P. 2004. Increased amounts of the influenza virus nucleoprotein do not promote higher levels of viral genome replication. J. Gen. Virol. 85:3689–3698 [DOI] [PubMed] [Google Scholar]
- 16. Muratore G, et al. 2012. Small molecule inhibitors of influenza A and B viruses that act by disrupting subunit interactions of the viral polymerase. Proc. Natl. Acad. Sci. U. S. A. 109:6247–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Obayashi E, et al. 2008. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454:1127–1131 [DOI] [PubMed] [Google Scholar]
- 18. Palese P, Shaw ML. 2007. Orthomyxoviridae: the viruses and their replication, p 1648–1698 In Knipe DM, et al. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins Co., Philadelphia, PA [Google Scholar]
- 19. Perez DR, Donis RO. 2001. Functional analysis of PA binding by influenza A virus PB1: effects on polymerase activity and viral infectivity. J. Virol. 75:8127–8136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sidwell RW, et al. 1972. Broad-spectrum antiviral activity of virazole: 1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science 177:705–706 [DOI] [PubMed] [Google Scholar]
- 21. Smee DF, Huffman JH, Morrison AC, Barnard DL, Sidwell RW. 2001. Cyclopentane neuraminidase inhibitors with potent in vitro anti-influenza virus activities. Antimicrob. Agents Chemother. 45:743–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zuccola HJ, Filman DJ, Coen DM, Hogle JM. 2000. The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol. Cell 5:267–278 [DOI] [PubMed] [Google Scholar]