Abstract
L-forms are cell wall-deficient bacteria that can grow and proliferate in osmotically stabilizing media. Recently, a strain of the Gram-positive model bacterium Bacillus subtilis was constructed that allowed controlled switching between rod-shaped wild-type cells and corresponding L-forms. Both states can be stably maintained under suitable culture conditions. Because of the absence of a cell wall, L-forms are known to be insensitive to β-lactam antibiotics, but reports on the susceptibility of L-forms to other antibiotics that interfere with membrane-anchored steps of cell wall biosynthesis are sparse, conflicting, and strongly influenced by strain background and method of L-form generation. Here we investigated the response of B. subtilis to the presence of cell envelope antibiotics, with regard to both antibiotic resistance and the induction of the known LiaRS- and BceRS-dependent cell envelope stress biosensors. Our results show that B. subtilis L-forms are resistant to antibiotics that interfere with the bactoprenol cycle, such as bacitracin, vancomycin, and mersacidin, but are hypersensitive to nisin and daptomycin, which both affect membrane integrity. Moreover, we established a lacZ-based reporter gene assay for L-forms and provide evidence that LiaRS senses its inducers indirectly (damage sensing), while the Bce module detects its inducers directly (drug sensing).
INTRODUCTION
L-forms are cell wall-deficient bacteria that have been described and analyzed in many bacterial species (15, 16, 21, 37, 76). In several studies, a spontaneous conversion of wild-type cells to L-forms under exposure to cell wall-affecting substances was demonstrated (36, 37, 43). In most cases, L-forms were generated by inhibiting cell wall biosynthesis through β-lactam treatment or by degradation of the intact cell wall using lysozyme (33, 60). The resulting L-forms are very osmotically sensitive and need osmotic stabilizers (e.g., KCl salt or sucrose) in the medium to survive (31, 51). L-forms are very diverse in their morphology but are generally resistant to β-lactam antibiotics because they lack a cell wall. Clinically, L-forms of pathogens (e.g., Mycobacterium tuberculosis, Listeria monocytogenes, Staphylococcus aureus, and Helicobacter pylori) exhibit resistance to a wide panel of commercially used cell wall antibiotics, including vancomycin, streptomycin, and penicillin (18, 70, 71). This feature has raised much concern regarding the persistence of pathogenic L-forms capable of evading classical antibiotic therapy (43).
In this study, we used the rod-shaped Gram-positive model bacterium Bacillus subtilis and derived L-forms, which had already been well characterized (9, 24, 75, 76). It is known that B. subtilis can grow and proliferate after a permanent loss of the cell wall as stable nonreverting L-forms (1, 72). Recently, a defined B. subtilis strain was constructed that can easily be converted from rod-shaped to L-form cells (19, 34). In this strain, the expression of the murE operon is under the control of a xylose-inducible promoter. The murE operon consists of four genes—murE, mraY, murD, and spoVD—which encode enzymes involved in essential steps of cell wall precursor biosynthesis (14). In the absence of xylose, the precursor UDP-N-acetylmuramyl (MurNAc) pentapeptide cannot be assembled and loaded on the lipid carrier, undecaprenol phosphate (UP) (4, 20). Under such conditions, the cells are therefore unable to synthesize the essential cell wall precursors, including lipid I and lipid II. Thus, L-forms can be conveniently generated in the absence of xylose in osmotically stabilized medium. Addition of xylose to the medium leads to normal expression of the murE operon, resulting in correct cell wall biosynthesis and hence rod-shaped cells (19, 34).
In B. subtilis and other Gram-positive bacteria, the cell wall represents the first and major line of defense against environmental threats, including cell wall antibiotics. To ensure its integrity, B. subtilis cells harbor a complex regulatory network for permanently monitoring the state of cell envelope integrity (29). This cell envelope stress response (CESR) network consists of at least four alternative σ-factors of the extracytoplasmic function (ECF) protein family (65) and a similar number of two-component systems (TCS) (29).
The TCS LiaRS is strongly induced by a diverse range of antibiotics that target the bactoprenol cycle of cell wall biosynthesis, e.g., the nonribosomally synthesized cyclic dodecylpeptide antibiotic bacitracin, the lantibiotic nisin, the glycolipodepsipeptide ramoplanin, and the glycopeptide antibiotic vancomycin (39, 40, 56, 74). It is also induced by some antimicrobial compounds directly interfering with membrane integrity, such as the antimicrobial (lipo)peptides daptomycin and LL-37 (40, 53, 73). Although the physiological role of the Lia system in B. subtilis is not fully understood, the target promoter PliaI has been developed as a biosensor and also adapted for high-throughput screens, based on its strong and specific induction by a variety of peptide antibiotics interfering with cell envelope integrity (8, 40, 64).
The BceRS system belongs to a second type of cell envelope stress-sensing TCS in B. subtilis. It is functionally associated with a peptide antibiotic-specific detoxification pump, the ABC transporter BceAB (2, 55). The BceRS TCS plays a central role in the bacitracin stress response, and BceAB represents the most efficient bacitracin resistance determinant in B. subtilis (39, 49, 55). More recently, it was shown that this module also strongly responds to some additional, chemically unrelated peptide antibiotics, such as plectasin, and the lantibiotics mersacidin and actagardine, while conferring only weak or no resistance against them (58, 64). The specific range of inducers for the BceRSAB module led to the development of a biosensor, based on the respective target promoter PbceA (64).
While the LiaRS and BceRS TCS respond exclusively to antibiotics interfering with the membrane-associated steps of cell wall synthesis, the specificity by which they sense and distinguish individual peptide antibiotics is unclear, with regard to both the chemical nature of these compounds and their mechanism of action (MOA).
This functional and chemical diversity of antibiotics acting as inducers provokes the question of the exact nature of the stimuli sensed by the CESR systems LiaRS and BceRS. Are the two systems able to sense these very different compounds directly, i.e., by binding them (drug sensing)? Or is it rather some aspect of the damage caused by these compounds on the cell envelope that is ultimately detected (damage sensing)? We decided to employ B. subtilis L-forms to address these questions and also gain some deeper insight into the MOA of these antibiotics.
The genetically well-defined background of the B. subtilis strain described above should allow us to separate these effects by using identical strains, propagated either as rod-shaped cells with cell walls or as osmotically stabilized L-forms lacking the murein sacculus. Hence, this strain and the derived biosensors, based on PbceA and PliaI, should provide powerful tools for studying the mechanism of action of novel compounds targeting the cell envelope in vivo both directly, by assessing their antimicrobial activity, and indirectly, by monitoring the induction of biosensors specifically responding to peptide antibiotics that interfere with cell envelope integrity.
Our results not only establish B. subtilis L-forms as a powerful tool for MOA studies of antibiotics that target the cell envelope but also provide an insight into the nature of the stimuli sensed by the LiaRS and BceRS CESR modules.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used in this study (Table 1) are derivatives of strain PDC204 and can therefore be converted from rod-shaped cells to cell wall-deficient L-forms simply through cultivation on osmotically stabilized medium. The strain PDC134 was used for quantitative real-time PCR (qRT-PCR). TMB1075 and TMB1077 were used for determination of total protein concentration. TMB1080 and TMB1082 were used for gradient plates and β-galactosidase assays. To cultivate rod-shaped cells, strains were inoculated in selective nutrient broth (NB) or streaked onto selective nutrient agar (NA) supplemented with 0.5% xylose. The concentrations of antibiotics used in this study were as follows: erythromycin, 1 μg ml−1; chloramphenicol, 5 μg ml−1; and spectinomycin, 100 μg ml−1. L-forms were induced and propagated under nonselective conditions in the absence of xylose in NA medium or on NB plates supplemented with 20 mM MgCl2 and 0.5 M sucrose and buffered with 20 mM maleic acid (MSM medium). All strains were incubated at 30°C. In liquid culture, only rod-shaped cells were inoculated with aeration at 200 rpm.
Table 1.
B. subtilis strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| PDC134 | W168 trpC2 Ω spoVD::Cm Pxyl-murE Ω amyE::xylR | 19 |
| PDC204 | W168 trpC2 Ω spoVD::Erm Pxyl-murE Ω | 19 |
| RM29 | W168 trpC2 Ω spoVD::Cm Pxyl-murE Ω amyE::xylR sacB::Spec | R. Mercier |
| TMB1075 | PDC204 Ω spoVD::Erm Pxyl-murE Ω amyE::pTM1 | This study |
| TMB1077 | PDC204 Ω spoVD::Erm Pxyl-murE Ω amyE::pER603 | This study |
| TMB1080 | PDC204 Ω spoVD::Erm Pxyl-murE Ω amyE::pTM1, sacB::Spec | This study |
| TMB1082 | PDC204 Ω spoVD::Erm Pxyl-murE Ω amyE::pER603, sacB::Spec | This study |
| Plasmids | ||
| pAC6 | Bla amyE′ lacZ Cat ′amyE (integrative promoter probe vector) | 69 |
| pTM1 | pAC6 PliaI-lacZ | 30 |
| pER603 | pAC6 PbceA-lacZ | 55 |
| Oligonucleotides | ||
| liaI-fwd(RT) | GGTATCGGAGCCATTATGCTC | This study |
| liaI-rev(RT) | CCCATTCGTCATCAAAGTGAG | This study |
| bceA-fwd(RT) | CGTCAGTATTATGGGTGCTTC | This study |
| bceA-rev(RT) | GAATGGTTCCGTGACTGACCTG | This study |
Resistance cassettes: Bla, ampicillin; Cat, chloramphenicol; Spec, spectinomycin; Erm, erythromycin.
Strain construction.
Plasmids pTM1 (30) and pER603 (55) (Table 1) were used to generate promoter-lacZ reporter strains. PDC204 was transformed with ScaI-linearized plasmids (pTM1 and pER603) and chromosomal DNA of strain RM29 (sacB::spec) with chloramphenicol and spectinomycin selection, resulting in the strains TMB1080 and TMB1082, respectively. Transformation and preparation of chromosomal DNA was carried out as described elsewhere (27).
Response to antibiotics and resistance experiments.
Gradient plates (41, 50) were used to compare the response of promoter-lacZ fusions as well as the antibiotic resistance of rod-shaped cells and L-forms. Mid-log-phase (optical density at 600 nm [OD600] ∼ 0.4 to 0.6) cultures of rod-shaped cells were spotted on NA supplemented with 0.5% xylose. L-form cells were spotted on NA-MSM. The plates contained gradients from 0 μg ml−1 to the following maximal antibiotic concentrations: for both PbceA and PliaI, bacitracin at 50 μg ml−1 and mersacidin at 5 μg ml−1; additionally for PliaI, vancomycin at 0.0625 μg ml−1, nisin at 10 μg ml−1, and daptomycin at 0.15 μg ml−1. Additionally, all gradient plates were supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 100 μg ml−1) to visualize the promoter activity of PliaI and PbceA.
The strains used for gradient plate experiments are PDC204 derivatives that carry an additional deletion of the sacB gene, which encodes levansucrase (Table 1). This extracellular enzyme is expressed in the presence of sucrose and is involved in the degradation of carbohydrates (61). During this study, we observed that L-form colonies growing on agar supplemented with sucrose are not well defined and show similarities to water droplets. Since cells with this colony morphology are very hard to handle on agar plates, we decided to knock out sacB by allelic replacement with a spectinomycin resistance cassette. This deletion resulted in the formation of well-defined colonies on agar plates.
Measurement of promoter induction by β-galactosidase assay.
For rod-shaped cells, the assay was performed as described previously (44). For L-forms, a modified procedure was developed in the course of this study. The OD600 of mid-logarithmic-phase L-form cells of strains TMB1080 and TMB1082 was measured, the cultures were split, and one half was treated with bacitracin (50 μg ml−1) and mersacidin (5 μg ml−1). The other half of the cultures served as uninduced controls. After additional incubation for 4 h at 30°C with gentle shaking, 800 μl of each sample was harvested. The cell pellets were resuspended in 800 μl working buffer (60 mM Na2HPO4 · 2H2O, 40 mM NaH2PO4 · H2O, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol). This treatment reliably disrupted the L-form cells, and the samples could directly be used for the β-galactosidase assays. Nevertheless, lysozyme (which is necessary for the lysis of rod-shaped cells) was also added in this assay in order to directly compare the results for both cell types.
Quantitative real-time RT-PCR (qRT-PCR).
Strain PDC134 and liaI- and bceA-specific primers (Table 1) were used to perform qRT-PCR on total RNA, prepared from mid-logarithmic-phase rod-shaped and L-form cells after 30 min incubation in the presence of bacitracin and mersacidin, respectively (concentrations as listed above), as described previously (55). A reference value for the liaI and bceA transcription was obtained using the same strain without antibiotic treatment.
RESULTS
The initial goal of this work was to evaluate the use of inducible L-forms of B. subtilis, based on the depletion of cell wall precursors by the repression of the murE operon in strain PDC204, as a tool for studying the MOA of cell wall antibiotics both at the level of their biological activity and at the level of induction of the established B. subtilis biosensors PliaI and PbceA. This analysis was initiated by introducing the PliaI-lacZ and PbceA-lacZ fusions into the amyE locus of strain PDC204, resulting in the reporter strains TMB1080 (PliaI) and TMB1082 (PbceA), respectively.
The L-form state dramatically alters the antibiotic sensitivity profile.
As a first test, we used antibiotic gradient diffusion plates (see Materials and Methods for details) to assess the inhibitory actions of selected cell wall antibiotics and their effects on the two biosensors, both before (rod-shaped cells) and after conversion of the strains into the L-form state. Mid-log-phase cultures of rod-shaped cells and L-forms were spotted in a line across the antibiotic gradient (Fig. 1). After incubation, the plates were visually inspected to determine the relative inhibitory effect of the tested antibiotics on the different strains, as indicated by the point of growth inhibition on the gradient. The addition of X-Gal to these plates also provided an indication for the induction of the reporter constructs in each strain by the development of a blue coloration of the patch of bacterial growth (Fig. 1). Since it is not possible to ensure an identical cell number's being initially spotted for the L-forms, this assay cannot be used as a quantitative measure of inhibitory action, at least for this cell type. Nevertheless, this easy-to-perform assay provided a first qualitative readout for the antimicrobial action of these compounds, due to the dramatic differences in sensitivities and regulatory response between the two strains.
Fig 1.
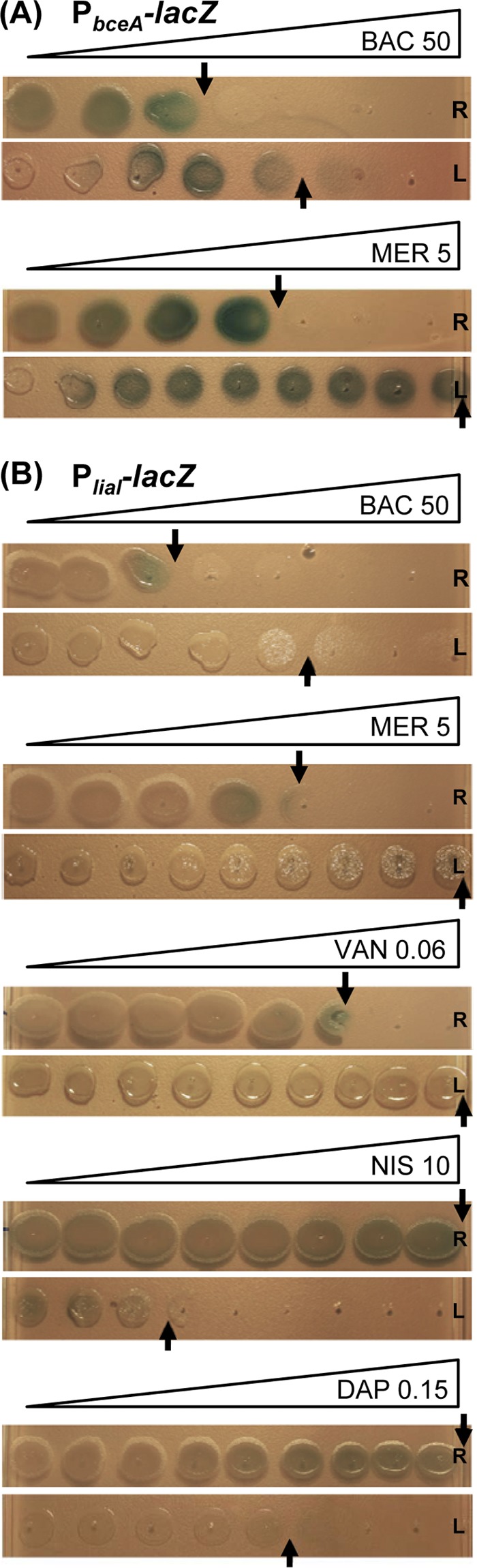
Spot assay on antibiotic gradient plates. Mid-logarithmic-phase rod-shaped cells (R) and the corresponding L-forms (L) were spotted on NA- or NB-MSM agar plates, respectively, and incubated overnight or for 2 days, respectively, at 30°C. All plates were supplemented with X-Gal (100 μg ml−1) and a gradient overlay of the antibiotics to be tested at the following final concentrations: bacitracin, 50 μg ml−1; mersacidin, 5 μg ml−1; vancomycin, 0.0625 μg ml−1; nisin, 10 μg ml−1; and daptomycin, 0.15 μg ml−1. (A) Induction of PbceA (TMB1082); (B) induction of PliaI (TMB1080).
While resistance against β-lactam antibiotics is well documented for L-forms, their response to antibiotics interfering with the membrane-anchored steps of cell wall biosynthesis, such as those used in this study, is less well understood. In the case of mersacidin and vancomycin, L-form colonies are fully viable, even at the highest antibiotic concentration applied (Fig. 1). While only moderate concentrations of vancomycin (maximum, 0.06 μg ml−1) could be used in gradient plates and still allow growth of the highly sensitivity rod-shaped wild type, we verified that L-form cultures could survive in the presence of over 10 μg ml−1 vancomycin, demonstrating a >100-fold-increased MIC for L-forms relative to the MIC for the wild type (data not shown). This dramatically increased resistance in the absence of a cell wall indicates that the target structure for both compounds is lacking or of no consequence to viability in L-forms, which is in good agreement with the respective MOA. In contrast, L-forms are dramatically more susceptible to daptomycin and nisin (Fig. 1), which both disrupt the integrity of the cytoplasmic membrane.
While L-forms are more resistant to bacitracin then their corresponding wild-type cells, they nevertheless retain a certain degree of susceptibility at higher antibiotic concentrations (Fig. 1). The primary inhibitory effect is a specific binding of bacitracin to undecaprenol pyrophosphate, thereby preventing its recycling to the monophosphate form, UP (66, 67). Hence, the cellular pool of UP, an essential precursor of the lipid II cycle, is rapidly depleted, thereby ultimately stopping cell wall biosynthesis. While this step is no longer essential in L-forms, our observation is in agreement with earlier reports demonstrating that bacitracin can also interfere with membrane integrity, i.e., by disrupting protoplasts (summarized in reference 46). However, the exact mechanism for this interference has not yet been elucidated.
In contrast, no significantly altered sensitivity was observed for the translational inhibitor kanamycin or the DNA-intercalating agent phleomycin (data not shown), demonstrating that the observed effects are indeed cell wall specific, as expected.
Taken together, our results demonstrate that the susceptibility of B. subtilis against antibiotics that inhibit membrane-anchored steps of cell wall biosynthesis is strongly affected. The hypersensitivity obtained in L-forms treated with nisin and daptomycin also indicates that L-forms represent a useful tool to distinguish between cell wall and membrane perturbation and to identify potential secondary MOAs. Moreover, our results suggest that both nisin and daptomycin might be potent drugs against persistent infections with pathogenic L-forms.
The response of cell wall antibiotic biosensors is altered in L-forms.
Since classical disk diffusion assays (10, 64) could not be successfully adapted for L-forms (data not shown), we instead modified the gradient plates described above for this purpose. Addition of the chromogenic β-galactosidase substrate X-Gal allowed direct assessment of the response of the LiaRS- and BceRS-dependent promoters in both the rod-shaped and L-form states as a function of the antibiotic concentration. In analogy to the disk diffusion assay, the strongest response of our wild-type biosensors was observed in colonies spotted at the highest tolerated concentrations (Fig. 1). Unfortunately, the intensity of the blue color was significantly weaker on gradient plates than in disk diffusion assays. Moreover, the strength of coloration does not directly correlate with induction strength but is instead reporter strain dependent, as noted in previous studies (11, 40, 64). Nevertheless, we observed a clear blue color for the L-forms containing PbceA in the presence of suitable amounts of both bacitracin and mersacidin (Fig. 1A), which are known inducers of the Bce system (64). The induction of the PliaI-derived reporter, which already provides a much weaker readout in disk diffusion assays (our unpublished data), is harder to detect on gradient plates. Nevertheless, a reproducible formation of light blue colonies at the highest tolerated concentrations can be observed for rod-shaped wild-type colonies in the presence of four of the five known inducers tested (Fig. 1B). In the case of daptomycin, the lack of a signal correlates with a lack of inhibition of rod-shaped cells even at the highest concentrations, indicative of insufficient antibiotic concentration. However, higher concentrations could not be used, since they completely prevented the growth of the corresponding L-form colonies (data not shown).
The BceR-dependent reporter strain TMB1082 showed comparable promoter activities in both wild-type and the corresponding L-form colonies in response to bacitracin and mersacidin (Fig. 1A). Interestingly, the minimal inducing concentrations for the two cell types were identical, irrespective of the significant differences in sensitivity, as described above. In contrast, no induction could be observed in L-form colonies of the LiaRS-dependent biosensor TMB1080 (Fig. 1B).
Our results therefore demonstrate that gradient plate experiments using the established biosensors PbceA and PliaI can be applied as a first comparative and qualitative screen for antibiotic induction, both in rod-shaped wild-type and in L-form colonies. Importantly, these initial results indicate that the response of the PbceA-derived reporter strain is unaffected by the absence of a cell wall and therefore the damage caused by these antibiotics, indicative of a more direct sensing of the two compounds. In contrast, the PliaI-based reporter no longer seems to respond to its known inducers in the L-forms, which can be interpreted as a more indirect, damage-induced sensing of antibiotic action. However, the low sensitivity in case of PliaI of the assay and the narrow range of concentrations that can be tested are significant limitations of this method. Hence, a more sensitive and quantitative measure is necessary to support these initial observations.
Promoter activity in L-forms can be quantified by β-galactosidase assay.
B. subtilis strains carrying fusions of reporter genes to PbceA and PliaI have proven to be valuable biosensors for the study of antibiotics that interfere with the membrane-anchored steps of cell wall biosynthesis (8, 40, 64). The gradient plate experiments described above indicated that the classical lacZ-based reporter gene fusions can also be used in L-forms (Fig. 1). We therefore next aimed at establishing the classical β-galactosidase assay for liquid cultures of B. subtilis L forms to further develop and evaluate the use of promoter-based biosensors in this genetic background. This type of assay has traditionally been normalized by expressing the enzyme activities relative to cell density (44). Hence, we first verified that measuring the OD600 is also a reliable measure for liquid L-form cultures by demonstrating a linear correlation of total protein concentration with optical density (data not shown).
Next, we investigated if β-galactosidase assays can be applied to L-forms to quantitatively monitor gene expression in these cell wall-deficient bacteria. For this purpose, we measured the promoter activity in strains TMB1080 (PliaI) and TMB1082 (PbceA) in both rod-shaped cells and their corresponding L-forms. As inducers, only bacitracin and mersacidin were chosen for these in-depth studies, since they are the only two compounds that activate both systems in wild-type cells (64). Samples of mid-logarithmic-phase cultures were split into three aliquots: one was an uninduced reference sample, and the other two were induced, one with bacitracin (50 μg ml−1) and one with mersacidin (5 μg ml−1). Because of the significantly reduced growth (and hence also protein production) rate of L-forms, it was critical to adjust the incubation time postinduction to 4 h (instead of the usual 30 min for wild-type cultures). This incubation time did not affect the basal promoter activity but resulted in significantly increased activities after antibiotic induction (data not shown). After 30 min or 4 h, the cells of wild-type or L-form cultures, respectively, were harvested and subjected to β-galactosidase assays as described in Materials and Methods. The results are shown in Fig. 2A. Rod-shaped wild type cells containing either PbceA-lacZ (TMB1082) or PliaI-lacZ (TMB1080) responded strongly to the presence of bacitracin and mersacidin, as expected (Fig. 2A) (64). In the corresponding L forms, the overall background activity was higher than in the wild type for both biosensors (Fig. 2A). No induction of PliaI activity was observed in strain TMB1080 after 4 h of incubation in the presence of bacitracin and mersacidin. In contrast, TMB1082 (PbceA-lacZ) showed five- and sevenfold-increased β-galactosidase activities for bacitracin and mersacidin, respectively (Fig. 2A). These data are in good agreement with the results obtained with gradient plates (Fig. 1). Inhibitors of translation did not induce either of the biosensors, as expected, thereby demonstrating the specificity of the responses in both rods and L-forms (data not shown).
Fig 2.
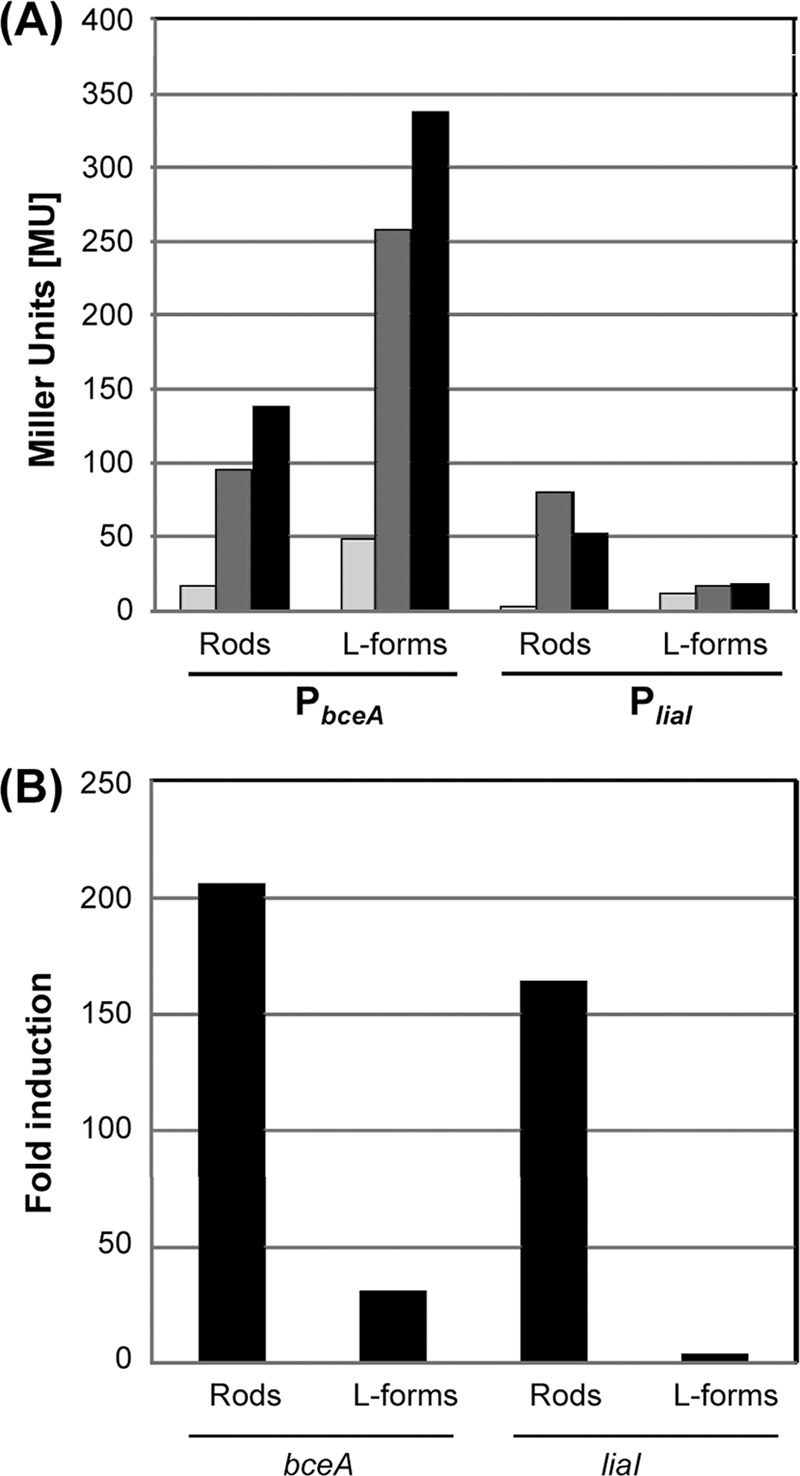
Induction of PbceA (TMB1082) and PliaI (TMB1080) by bacitracin (dark gray bars) and mersacidin (black bars) in rod-shaped wild-type cells and the corresponding L-forms. (A) Mid-log-phase cultures of TMB1080 and TMB1082 from both cell types were split and induced with bacitracin (50 μg ml−1) and mersacidin (5 μg ml−1). An uninduced sample served as a control (light gray bars). Cells were harvested after 30 min (wild type) and 4 h (L-forms), and samples were analyzed by β-galactosidase assay for PliaI (TMB1080) and PbceA (TMB1082) activity as described previously (44). Promoter activity is expressed in Miller units (MU). (B) Induction of bceA and liaI monitored by quantitative real-time RT-PCR. RNA was prepared from cells of strain PDC134 harvested at mid-logarithmic growth phase from cultures of rod-shaped cells or L forms that had been induced for 30 min with bacitracin (50 μg ml−1). RT-PCR with gene-specific primers (Table 1) and calculation of induction ratios (x-fold changes; black bars) of liaI or bceA expression relative to an uninduced reference sample were performed as described in Materials and Methods.
To verify our data, we next analyzed the transcription of liaI and bceA directly by quantitative real-time RT-PCR, using RNA prepared from wild-type and L-form cultures of strain PDC134 after treatment of the cells for 30 min with bacitracin. RNA from an uninduced culture was prepared in parallel as a reference. Bacitracin strongly induced the expression of both genes in wild-type cells (>150- to 200-fold relative to the basal expression level) (Fig. 2B), as expected. Moreover, a clear induction of bceA expression was also observed in the L-form sample (∼40-fold). In contrast, no significant induction of liaI can be detected in L-forms containing PliaI-lacZ. For TMB1080, we also tested the activity of PliaI in the presence of other known inducers, such as vancomycin and nisin. Again, PliaI responded to both antibiotics in the wild type, as described previously (40), while no induction of the liaI promoter was observed in the derived L-forms (data not shown). The lower induction ratio for bceA in the L-form samples was due to an overall higher basal signal, as was observed by β-galactosidase assay (Fig. 2A). The overall higher dynamic range determined by quantitative real-time RT-PCR is commonly observed, due to the higher sensitivity of this assay. Thus, the quantitative real-time RT-PCR results are in perfect agreement with the observations from both gradient plates and liquid β-galactosidase assays.
DISCUSSION
In this study, we used cell wall-less L-forms of B. subtilis to investigate their sensitivity against antibiotics that interfere with membrane-anchored steps of cell wall biosynthesis. Moreover, we have established β-galactosidase assays for these cell types and investigated the response of two established biosensors for cell wall antibiotics, PbceA-lacZ (TMB1082) and PliaI-lacZ (TMB1080), by comparing their response in cell wall-containing wild-type and derived cell wall-less L-form cultures. Our results demonstrated a strong hypersensitivity of L-forms to nisin and daptomycin. Moreover, they shed some light on the nature of the inducing stimulus for our two biosensors, as discussed below.
L-forms are hypersensitive to the lantibiotic nisin and the lipopeptide antibiotic daptomycin.
The hypersensitivity of L-forms to nisin seems somewhat surprising. Despite its known pore-forming bactericidal activity, nisin is known to require the pyrophosphate moiety of lipid II as a docking molecule to initiate efficient pore formation at low antibiotic concentrations (5, 7, 35), a molecule that is absent in our L-forms. On the other hand, lipid II-independent pore formation has also been observed at higher nisin concentrations and the presence of anionic lipids in the target membrane (5, 6, 22). Moreover, it was recently shown that nisin can also bind the precursor of wall teichoic acid biosynthesis (47). This molecule is also loaded onto the lipid carrier undecaprenol phosphate, formerly termed lipid IV, and flipped to the outer surface of the cytoplasmic membrane. While formation of wall teichoic acid cannot occur in the absence of peptidoglycan, a specific target molecule of nisin is therefore still present in our murE deletion strain.
Daptomycin has not been reported to require a component of the cell wall biosynthesis machinery for its inhibitory action (57, 62, 68). But both the exact MOA and hence the reason for the hypersusceptibility of B. subtilis L-forms is less clear. Our results seem to argue that the local daptomycin concentration on the outer surface of the cytoplasmic membrane is significantly increased in the absence of a cell wall to allow disruption of the cytoplasmic membrane even at very low external antibiotic concentrations. We therefore postulate that the Gram-positive cell wall seems to function as a molecular sieve that keeps the local daptomycin concentration low at its site of action.
An alternative explanation for the dramatic difference in daptomycin sensitivity between rod-shaped cells and L-forms could be a difference in membrane composition between the two cell types. This idea is based on three independent observations. First, phosphatidylglycerol levels in membranes have been identified as important determinants of the daptomycin susceptibility of B. subtilis (26). Second, it was shown previously that removing the cell wall of S. aureus in order to generate protoplasts significantly alters the membrane composition (77). Third, a very recent report on B. subtilis L-forms—based on the same strain that was used in this study—showed that a number of membrane lipids and their precursors, such as cardiolipin, phosphatidylethanolamine, unsaturated fatty acids, and lysyl-phosphatidylglycerol, are not essential for proliferation of cells in the L-form state. On the other hand, membrane fluidity determined by the incorporation of branched-chain fatty acids does play a crucial role in this process (42).
In order to determine the reason for the observed hypersensitivity, it might be worthwhile for a future study to determine the membrane composition of L-forms and their respective wild types. Both potential mechanisms described for daptomycin might of course also contribute to the observed hypersensitivity against nisin.
Does daptomycin also interfere with cell wall biosynthesis?
Daptomycin is the only compound for which no clear experimental evidence has so far been provided that links its activity to interference with cell wall biosynthesis. While such an activity has repeatedly been discussed (12, 48), our results can be viewed as the first, albeit indirect, evidence for such a MOA. It is generally accepted that the primary bactericidal activity of daptomycin is its interference with membrane integrity (12, 26, 48). Accordingly, L-forms show a significantly increased susceptibility in the absence of a protecting cell wall (Fig. 1B). Strong induction of liaIH expression by daptomycin, through activation of the LiaRS-dependent promoter PliaI, was well documented by two independent studies (25, 73). But despite maintaining its biological potency, daptomycin fails to induce the PliaI-lacZ biosensor in L-forms (Fig. 1B and data not shown). Therefore, the primary killing mechanism of daptomycin does not act as the stimulus sensed by the LiaRS system. Instead, this again requires an intact cell wall biosynthesis machinery. This observation strongly suggests that daptomycin—in addition to its primary MOA—also somehow interferes with (most likely some membrane-anchored step of) cell wall biosynthesis. This hypothesis is strongly supported by a recent report demonstrating that daptomycin distorts the cytoplasmic membrane in B. subtilis. This results in mislocalization of membrane-anchored proteins of the cell wall biosynthesis machinery, thereby also affecting cell wall biosynthesis and integrity (54). These data not only provide a convincing explanation for the observed induction of the cell envelope stress response in B. subtilis described above but also highlight the potential and predictive power of using the PliaI- and PbceA-derived biosensors in both the wild-type and L-forms of B. subtilis.
The response of PliaI and PbceA in L-forms sheds light on the mechanism of antibiotic sensing.
Taken together, our results reveal a fundamental difference in the mechanism of sensing cell wall antibiotics by two well-established biosensors, which are derived from the cell envelope stress-responsive promoters PliaI and PbceA of B. subtilis (40, 64). These new findings are summarized in a graphic model (Fig. 3). In rod-shaped wild-type cells that contain an intact cell wall, all five antibiotics exhibit their respective inhibitory activity, which directly or indirectly also interferes with the lipid II cycle of cell wall synthesis. In the case of mersacidin and bacitracin, this cycle is directly affected (3, 45) while vancomycin prevents the release of undecaprenol pyrophosphate by blocking the incorporation of new cell wall material into the existing peptidoglycan network (13, 63). Nisin uses lipid II as a docking molecule and thereby also interferes with this molecule (3). Daptomycin, on the other hand, primarily affects the integrity of the cell membrane and hence does not require cell wall biosynthesis for exhibiting its primary inhibitory function (57, 62, 68). All five antibiotics studied are known inducers of PliaI activity in wild-type cells (40, 64). In L-forms, mersacidin and vancomycin completely lose their inhibitory activity, while only a reduced activity is seen for bacitracin. In contrast, nisin and daptomycin—which both perturb the cytoplasmic membrane—show a strongly increased potency (Fig. 1). Nevertheless, none of these five antibiotics is able to induce PliaI activity in L-forms (Fig. 1B and data not shown). Hence, the compounds themselves cannot represent the true stimuli sensed by the LiaRS system. Instead, we conclude that this TCS requires the presence of a functional cell wall biosynthesis machinery to sense the five antibiotics addressed in this study. Our collective data strongly suggest that the true stimulus sensed by the LiaRS system is some aspect of interference with cell wall biosynthesis by those antibiotics. LiaRS is therefore a damage-sensing signal-transducing system. This hypothesis is supported by the wide and varying range of cell wall antibiotics sensed by the homologous systems in other members of the Firmicutes, such as VraSR of Staphylococcus aureus and CesSR of Lactococcus lactis. In addition to some of the compounds used in this study, these systems also respond to β-lactams or cationic antimicrobial peptides of the immune system (32, 38, 52). This rather general induction mechanism of LiaRS-like TCS is also reflected by their biological function: especially in Gram-positive cocci, these systems represent the primary but also rather general cell envelope stress response. They control large regulons that seem to provide general layers of protection in the presence of adverse conditions affecting the cell envelope (29, 59).
Fig 3.
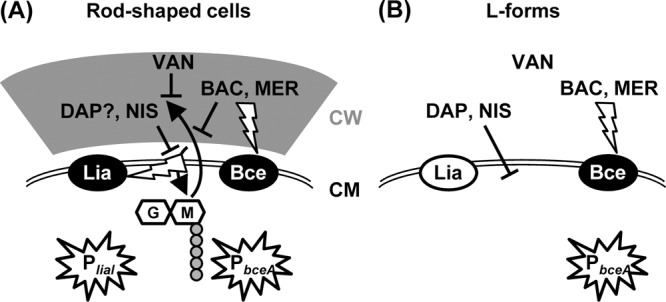
Graphic model illustrating the inhibitory action (T-shaped lines) of the antibiotics (BAC, bacitracin; DAP, daptomycin; MER, mersacidin; NIS, nisin; VAN, vancomycin) used in this study and the mechanism of stimulus perception (flash-shaped arrows) resulting in activation of the biosensors PbceA and PliaI (stars) in both rod-shaped cells (A) and L-forms (B). CM, cytoplasmic membrane; CW, cell wall. The lipid II cycle and incorporation of cell wall building blocks are indicated by the two black bent arrows. Activation of the LiaRS (Lia) or BceRS (Bce) TCS is indicated by white letters on a black background, while an inactive system is indicated by black letters on a white background. See the text for details.
In contrast, the BceRS-dependent promoter PbceA responds to its known inducers irrespective of the presence or absence of a cell wall, despite the fact that mersacidin and bacitracin have no and very little inhibitory activity in L-forms, respectively. The promoter responds in a concentration-dependent manner, and the regulatory sensitivity is unaffected by the lack of a cell wall in the L-forms and the resulting differences in resistance (Fig. 1A). This strongly argues for a direct drug-sensing mechanism of stimulus perception. This observation is in good agreement with the known mechanism of signal transduction for Bce-like detoxification modules. In contrast to classical TCS, the histidine kinase BceS and its homologs do not function as sensor proteins themselves. Instead, they recruit the transporter BceAB for this purpose (17, 23, 55). Since these ABC transporters therefore act as both sensors and highly efficient resistance determinants against peptide antibiotics (17, 23, 28, 64), such a direct sensing of the respective compound to be transported makes perfect sense.
Summary and outlook.
The goal of this study was to establish β-galactosidase assays for the analysis of lacZ-based biosensors in B. subtilis L-forms and to evaluate such biosensors for cell wall antibiotics in the genetic background of strain PDC134. In this strain, the murE operon is under the transcriptional control of a xylose-dependent promoter. Using an osmotically stabilizing medium allows the conversion of rod-shaped (wild-type) cells into L-forms simply by omitting xylose and thereby depleting the cells of essential steps in cell wall biosynthesis (34).
This strain therefore allows a direct comparison of the response of cell wall-containing rod-shaped cells with that of cell wall-deficient L-forms when both types of cells are exposed to antibiotics that interfere with cell wall biosynthesis and the integrity of the cell envelope. By establishing β-galactosidase assays using these L-forms, it is now possible to analyze both the physiological response of the cells and the transcriptional activation of biosensors in parallel and directly compare the results obtained with wild-type and L-forms. This approach allows an unprecedented insight into antibiotic action and sensing at the same time, as demonstrated for a number of antibiotics interfering with the membrane-anchored steps of cell wall biosynthesis or membrane integrity. We believe that the PDC134-derived biosensors for cell wall antibiotics constitute a very useful addition to the toolbox currently available both for initial screening and subsequent MOA studies of novel antibiotics that target the cell envelope.
Moreover, our study highlights the potential of using daptomycin or type A lantibiotics such as nisin to combat persistent infections by bacterial L-forms, which have repeatedly been reported (43) and which are insensitive to many classical antibiotics such as the β-lactams or vancomycin.
ACKNOWLEDGMENTS
We thank Romain Mercier (University of Newcastle upon Tyne) for strain RM29, Gabriele Bierbaum for the generous gift of mersacidin, and Tanja Schneider for helpful discussions.
This work was supported by DFG grant MA2873/1-3 (to T.M.) and a UK Biotechnology and Biological Sciences Research Council grant (to R.D.). The work of Patricia Domínguez-Cuevas was supported by a Marie Curie Intra-European Fellowship. Diana Wolf was supported by a Marie Curie Short Term Fellowship during her stay at the University of Newcastle upon Tyne (United Kingdom).
Footnotes
Published ahead of print 10 September 2012
REFERENCES
- 1. Allan EJ. 1991. Induction and cultivation of a stable L form of Bacillus subtilis. J. Appl. Bacteriol. 70:339–343 [DOI] [PubMed] [Google Scholar]
- 2. Bernard R, Guiseppi A, Chippaux M, Foglino M, Denizot F. 2007. Resistance to bacitracin in Bacillus subtilis: Unexpected requirement of the BceAB ABC transporter in the control of expression of its own structural genes. J. Bacteriol. 189:8636–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bierbaum G, Sahl HG. 2009. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 10:2–18 [DOI] [PubMed] [Google Scholar]
- 4. Bouhss A, Crouvoisier M, Blanot D, Mengin-Lecreulx D. 2004. Purification and characterization of the bacterial MraY translocase catalyzing the first membrane step of peptidoglycan biosynthesis. J. Biol. Chem. 279:29974–29980 [DOI] [PubMed] [Google Scholar]
- 5. Breukink E, de Kruijff B. 2006. Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 5:321–332 [DOI] [PubMed] [Google Scholar]
- 6. Breukink E, et al. 1997. The C-terminal region of nisin is responsible for the initial interaction of nisin with the target membrane. Biochemistry 36:6968–6976 [DOI] [PubMed] [Google Scholar]
- 7. Breukink E, et al. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361–2364 [DOI] [PubMed] [Google Scholar]
- 8. Burkard M, Stein T. 2008. Microtiter plate bioassay to monitor the interference of antibiotics with the lipid II cycle essential for peptidoglycan biosynthesis. J. Microbiol. Methods 75:70–74 [DOI] [PubMed] [Google Scholar]
- 9. Burmeister HR, Hesseltine CW. 1968. Induction and propagation of a Bacillus subtilis L form in natural and synthetic media. J. Bacteriol. 95:1857–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cao M, et al. 2002. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443–457 [DOI] [PubMed] [Google Scholar]
- 11. Cao M, Wang T, Ye R, Helmann JD. 2002. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σW and σM regulons. Mol. Microbiol. 45:1267–1276 [DOI] [PubMed] [Google Scholar]
- 12. Cotroneo N, Harris R, Perlmutter N, Beveridge T, Silverman JA. 2008. Daptomycin exerts bactericidal activity without lysis of Staphylococcus aureus. Antimicrob. Agents Chemother. 52:2223–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Courvalin P. 2006. Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 42(Suppl 1):S25–S34 [DOI] [PubMed] [Google Scholar]
- 14. Daniel RA, Errington J. 1993. DNA sequence of the murE-murD region of Bacillus subtilis 168. J. Gen. Microbiol. 139:361–370 [DOI] [PubMed] [Google Scholar]
- 15. Dell'Era S, et al. 2009. Listeria monocytogenes L forms respond to cell wall deficiency by modifying gene expression and the mode of division. Mol. Microbiol. 73:306–322 [DOI] [PubMed] [Google Scholar]
- 16. Dienes L, Weinberger HJ. 1951. The L forms of bacteria. Bacteriol. Rev. 15:245–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dintner S, et al. 2011. Co-evolution of ABC-transporters and two-component regulatory systems as resistance modules against antimicrobial peptides in Firmicutes bacteria. J. Bacteriol. 193:3851–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Domingue GJ, Sr, Woody HB. 1997. Bacterial persistence and expression of disease. Clin. Microbiol. Rev. 10:320–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dominguez-Cuevas P, Mercier R, Leaver M, Kawai Y, Errington J. 2012. The rod to L form transition of Bacillus subtilis is limited by a requirement for the protoplast to escape from the cell wall sacculus. Mol. Microbiol. 83:52–66 [DOI] [PubMed] [Google Scholar]
- 20. Foster SJ, Popham DL. 2002. Structure and synthesis of cell wall, spore cortex, teichoic acid, S-layers and capsules, p 21–41 In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC [Google Scholar]
- 21. Fuller E, et al. 2005. Beta-lactam resistance in Staphylococcus aureus cells that do not require a cell wall for integrity. Antimicrob. Agents Chemother. 49:5075–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garcera MJ, Elferink MG, Driessen AJ, Konings WN. 1993. In vitro pore-forming activity of the lantibiotic nisin. Role of protonmotive force and lipid composition. Eur. J. Biochem. 212:417–422 [DOI] [PubMed] [Google Scholar]
- 23. Gebhard S, Mascher T. 2011. Antimicrobial peptide sensing and detoxification modules: unraveling the regulatory circuitry of Staphylococcus aureus. Mol. Microbiol. 81:581–587 [DOI] [PubMed] [Google Scholar]
- 24. Gilpin RW, Young FE, Chatterjee AN. 1973. Characterization of a stable L form of Bacillus subtilis 168. J. Bacteriol. 113:486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hachmann A-B, Angert ER, Helmann JD. 2009. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob. Agents Chemother. 53:1598–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hachmann A-B, et al. 2011. Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtilis. Antimicrob. Agents Chemother. 55:4326–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harwood CR, Cutting SM. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 28. Hiron A, Falord M, Valle J, Debarbouille M, Msadek T. 2011. Bacitracin and nisin resistance in Staphylococcus aureus: a novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol. Microbiol. 81:602–622 [DOI] [PubMed] [Google Scholar]
- 29. Jordan S, Hutchings MI, Mascher T. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107–146 [DOI] [PubMed] [Google Scholar]
- 30. Jordan S, Junker A, Helmann JD, Mascher T. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: Identification of inhibitor proteins, regulator binding sites and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 188:5153–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. King JR. 1986. L forms of group D Streptococcus, p 43–58 In Madoff S. (ed), The bacterial L forms. Marcel Dekker Inc., New York, NY [Google Scholar]
- 32. Kuroda M, et al. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807–821 [DOI] [PubMed] [Google Scholar]
- 33. Landman OE, Halle S. 1963. Enzymically and physically induced inheritance changes in Bacillus subtilis. J. Mol. Biol. 7:721–738 [DOI] [PubMed] [Google Scholar]
- 34. Leaver M, Dominguez-Cuevas P, Coxhead JM, Daniel RA, Errington J. 2009. Life without a wall or division machine in Bacillus subtilis. Nature 457:849–853 [DOI] [PubMed] [Google Scholar]
- 35. Linnett PE, Strominger JL. 1973. Additional antibiotic inhibitors of peptidoglycan synthesis. Antimicrob. Agents Chemother. 4:231–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maisnier-Patin S, Richard J. 1996. Cell wall changes in nisin-resistant variants of Listeria innocua grown in the presence of high nisin concentrations. FEMS Microbiol. Lett. 140:29–35 [DOI] [PubMed] [Google Scholar]
- 37. Markova N, Slavchev G, Michailova L, Jourdanova M. 2010. Survival of Escherichia coli under lethal heat stress by L form conversion. Int. J. Biol. Sci. 6:303–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martinez B, Zomer AL, Rodriguez A, Kok J, Kuipers OP. 2007. Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol. Microbiol. 64:473–486 [DOI] [PubMed] [Google Scholar]
- 39. Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50:1591–1604 [DOI] [PubMed] [Google Scholar]
- 40. Mascher T, Zimmer SL, Smith TA, Helmann JD. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48:2888–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McCallum N, Meier PS, Heusser R, Berger-Bachi B. 2011. Mutational analyses of open reading frames within the vraSR operon and their roles in the cell wall stress response of Staphylococcus aureus. Antimicrob. Agents Chemother. 55:1391–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mercier R, Dominguez-Cuevas P, Errington J. 2012. Crucial role for membrane fluidity in proliferation of primitive cells. Cell Rep. 1:417–423 [DOI] [PubMed] [Google Scholar]
- 43. Michailova L, Kussovsky V, Radoucheva T, Jordanova M, Markova N. 2007. Persistence of Staphylococcus aureus L form during experimental lung infection in rats. FEMS Microbiol. Lett. 268:88–97 [DOI] [PubMed] [Google Scholar]
- 44. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 45. Ming L-J, Epperson JD. 2002. Metal binding and structure-activity relationship of the metalloantibiotic peptide bacitracin. J. Inorg. Biochem. 91:46–58 [DOI] [PubMed] [Google Scholar]
- 46. Ming L-J. 2003. Structure and function of “metalloantibiotics.” Med. Res. Rev. 23:697–762 [DOI] [PubMed] [Google Scholar]
- 47. Müller A, Ulm H, Reder-Christ K, Sahl HG, Schneider T. 2012. Interaction of type A lantibiotics with undecaprenol-bound cell envelope precursors. Microb. Drug Resist. 18:261–270 [DOI] [PubMed] [Google Scholar]
- 48. Muraih JK, Pearson A, Silverman J, Palmer M. 2011. Oligomerization of daptomycin on membranes. Biochim. Biophys. Acta 1808:1154–1160 [DOI] [PubMed] [Google Scholar]
- 49. Ohki R, Giyanto, et al. 2003. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49:1135–1144 [DOI] [PubMed] [Google Scholar]
- 50. O'Leary JO, et al. 2004. Effects of sarA inactivation on the intrinsic multidrug resistance mechanism of Staphylococcus aureus. FEMS Microbiol. Lett. 237:297–302 [DOI] [PubMed] [Google Scholar]
- 51. Onoda T, Oshima A, Fukunaga N, Nakatani A. 1992. Effect of Ca2+ and K+ on the intracellular pH of an Escherichia coli L form. J. Gen. Microbiol. 138:1265–1270 [DOI] [PubMed] [Google Scholar]
- 52. Pietiäinen M, et al. 2009. Transcriptome analysis of the responses of Staphylococcus aureus to antimicrobial peptides and characterization of the roles of vraDE and vraSR in antimicrobial resistance. BMC Genomics 10:429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pietiäinen M, et al. 2005. Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology 151:1577–1592 [DOI] [PubMed] [Google Scholar]
- 54. Pogliano J, Pogliano N, Silverman JA. 2012. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J. Bacteriol. 194:4494–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rietkötter E, Hoyer D, Mascher T. 2008. Bacitracin sensing in Bacillus subtilis. Mol. Microbiol. 68:768–785 [DOI] [PubMed] [Google Scholar]
- 56. Salzberg LI, Luo Y, Hachmann A-B, Mascher T, Helmann JD. 2011. The Bacillus subtilis GntR family repressor YtrA responds to cell wall antibiotics. J. Bacteriol. 193:5793–5801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schneider T, et al. 2009. The lipopeptide antibiotic friulimicin B inhibits cell wall biosynthesis through complex formation with bactoprenol phosphate. Antimicrob. Agents Chemother. 53:1610–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schneider T, et al. 2010. Plectasin, a fungal defensin, targets the bacterial cell wall precursor lipid II. Science 328:1168–1172 [DOI] [PubMed] [Google Scholar]
- 59. Schrecke K, Staron A, Mascher T. 2012. Two-component signaling in the Gram-positive envelope stress response: intramembrane-sensing histidine kinases and accessory membrane proteins, p 199–229 In Gross R, Beier D. (ed), Two component systems in bacteria. Horizon Scientific Press, Hethersett, United Kingdom [Google Scholar]
- 60. Sharp JT. 1954. L colonies from hemolytic streptococci: new technic in the study of L forms of bacteria. Proc. Soc. Exp. Biol. Med. 87:94–97 [DOI] [PubMed] [Google Scholar]
- 61. Shimotsu H, Henner DJ. 1986. Modulation of Bacillus subtilis levansucrase gene expression by sucrose and regulation of the steady-state mRNA level by sacU and sacQ genes. J. Bacteriol. 168:380–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Silverman JA, Perlmutter NG, Shapiro HM. 2003. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:2538–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Smyth EG, Pallett AP. 1988. Vancomycin. Br. J. Hosp. Med. 39:308–312 [PubMed] [Google Scholar]
- 64. Staroń A, Finkeisen DE, Mascher T. 2011. Peptide antibiotic sensing and detoxification modules of Bacillus subtilis. Antimicrob. Agents Chemother. 55:515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Staroń A, et al. 2009. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) σ factor protein family. Mol. Microbiol. 74:557–581 [DOI] [PubMed] [Google Scholar]
- 66. Stone KJ, Strominger JL. 1971. Mechanism of action of bacitracin: complexation with metal ion and C 55-isoprenyl pyrophosphate. Proc. Natl. Acad. Sci. U. S. A. 68:3223–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Storm DR, Strominger JL. 1973. Complex formation between bacitracin peptides and isoprenyl pyrophosphates. The specificity of lipid-peptide interactions. J. Biol. Chem. 248:3940–3945 [PubMed] [Google Scholar]
- 68. Straus SK, Hancock RE. 2006. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biochim. Biophys. Acta 1758:1215–1223 [DOI] [PubMed] [Google Scholar]
- 69. Stülke J, et al. 1997. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol. 25:65–78 [DOI] [PubMed] [Google Scholar]
- 70. Wang D, Chen Z, Hu Z, Yuan W, Rao X. 2011. A recombinant antimicrobial peptide inhibits the growth of oxacillin-induced L forms of Staphylococcus aureus. Int. J. Antimicrob. Agents 38:177–178 [DOI] [PubMed] [Google Scholar]
- 71. Wang KX, Chen L. 2004. Helicobacter pylori L form and patients with chronic gastritis. World J. Gastroenterol. 10:1306–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ward JB. 1975. Peptidoglycan synthesis in L-phase variants of Bacillus licheniformis and Bacillus subtilis. J. Bacteriol. 124:668–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wecke T, et al. 2009. Daptomycin versus friulimicin B: in-depth profiling of Bacillus subtilis cell envelope stress responses. Antimicrob. Agents Chemother. 53:1619–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wolf D, et al. 2010. In-depth profiling of the LiaR response of Bacillus subtilis. J. Bacteriol. 192:4680–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wyrick PB, McConnell M, Rogers HJ. 1973. Genetic transfer of the stable L form state to intact bacterial cells. Nature 244:505–507 [DOI] [PubMed] [Google Scholar]
- 76. Wyrick PB, Rogers HJ. 1973. Isolation and characterization of cell wall-defective variants of Bacillus subtilis and Bacillus licheniformis. J. Bacteriol. 116:456–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Xiong YQ, Mukhopadhyay K, Yeaman MR, Adler-Moore J, Bayer AS. 2005. Functional interrelationships between cell membrane and cell wall in antimicrobial peptide-mediated killing of Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3114–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]


