LETTER
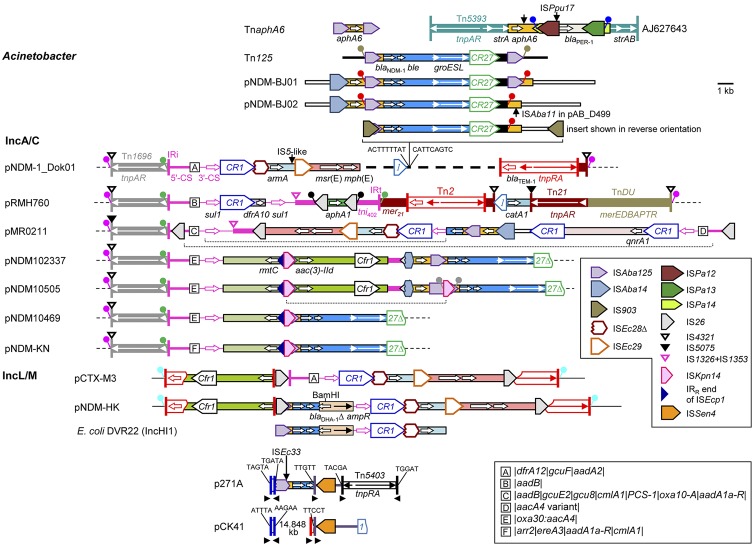
The blaNDM-1 gene, first reported in 2009 (24), has received considerable attention, and the sequences of several plasmids/regions carrying blaNDM-1 are now available. In all cases blaNDM-1 lies downstream of an intact or truncated ISAba125 element (Fig. 1) which provides the −35 region of a promoter (17, 19). Using the corrected definition of the ends of ISAba125 (http://www-is.biotoul.fr/is.html), 93 bp separate the right-hand inverted repeat (IRR) of ISAba125 from the start codon of blaNDM-1. An ISCR3-like element, ISCR27 (called ISCR21 in reference 16), found downstream of blaNDM-1, may have been responsible for initial gene capture (16, 22). However, as ISCR elements can progressively acquire DNA segments at their terIS end and the mosaic ISCR3-like elements (11) apparently derive from an ancestor already associated with Xanthomonas-like groES-groEL genes (23), the region between blaNDM-1 and ISCR27 may be made up of segments with different origins (5).
Fig 1.
Known wider contexts of blaNDM-1. Extents and orientations of selected resistance and other genes are shown by arrows labeled with the gene name. Different types of horizontal lines indicate plasmid backbone or the A. baumannii chromosome. IS are shown as boxes labeled with the name/number of the IS or shown in the key with the pointed end corresponding to the IRR (or oriIS for ISCR elements). IS flanked by direct repeats (DRs) have generally been omitted, with vertical arrows indicating their locations. Tall bars represent Tn3-like 38-bp IR or IRi of class 1 In/Tn, as indicated. Boxes containing letters represent different cassette arrays (see key). DRs are shown by filled circles of the same color on short stalks, and nonmatching sequences of expected DR length are given. The vertical arrow on the AJ627643 diagram indicates the site of insertion of Tn125 in pNDM-BJ01. The dotted lines below the pMR0211 structure indicate possible inversion events due to recombination between fragments of class 1 integrons, and 6.564 kb of the IncA/C backbone is missing at the right-hand end compared with pNDM-1_Dok01. The pNDM10505 structure could be derived from the pNDM102337 structure by insertion of ISKpn14 in ISAba125, the pNDM10469 structure by recombination between these two ISKpn14 to delete the intervening segment (dotted line), and the pNDM-HK structure by switching of the cassette array. In these four plasmids, 3.473 kb of the IncA/C backbone is missing at the right-hand end compared with pNDM-1_Dok01. The BamHI site shown on the pNDM-HK structure marks the end of the blaDHA-1 fragment in the K. pneumoniae 05-506/pKpANDM-1 (untyped plasmid; GenBank accession number FN396876) context (24). Several other partial blaNDM-1 contexts are illustrated in Fig. 2 of reference 18 and Fig. 3 of reference 10, but in both of these figures ISAba125 is shown in the wrong orientation for E. coli 271 and potentially in other structures (the sequences are not available) and the ISAba125Δ-blaNDM-1-ble portion of the K. pneumoniae 05-506 structure is also shown in the wrong orientation; i.e., IS26 should truncate ISAba125. Diagrams were drawn from the following GenBank accession numbers: TnaphA6, JF343537; Tn125, HQ857107; pNDM-BJ01, JQ001791; pNDM-BJ02, JQ060896; pAB_D499, AGFH01000030; pNDM-1_Dok01, AP012208; pRMH760, AY123253; pMR0211, JN687470; pNDM102337, JF714412; pNDM10505, JF503991; pNDM10469, JN861072; pNDM-KN, JN157804; pCTX-M3, AF550415; pNDM-HK, HQ451074; E. coli DVR22, JF922606; p271A, JF785549; pCK41, HQ332785.
blaNDM-1 was recently identified as a chimera (22), with the sequence from IRR of ISAba125 to nucleotide 20 of blaNDM-1 matching the region between ISAba125 and aphA6 in the ISAba125-flanked composite transposon TnaphA6 found in Acinetobacter baumannii (9). This could have occurred by placement of the fragment mobilized by ISCR27 so that the blaNDM-1 precursor fused in frame with aphA6 or by a deletion (22). However, the situation appears more complex, as in pNDM-BJ01 from Acinetobacter lwoffii (5), ISAba125 upstream of blaNDM-1 appears to be part of an ISAba125-mediated composite transposon (Tn125), flanked by 3-bp direct repeats (DRs), that has been inserted downstream of aphA6 in the context found in Tn5393d from Alcaligenes faecalis (6). In the closely related plasmids pNDM-BJ02 from A. lwoffii (5) and pAB_D499 from A. pittii (2), only a small fragment of ISAba125 remains downstream of blaNDM-1, possibly due to recombination between two copies of a repeated short sequence. Tn125 flanked by different DRs has also been found on the A. baumannii chromosome (15, 16).
In six IncA/C plasmids, segments apparently derived from the Acinetobacter plasmid structures are inserted within large multiresistance regions (MRRs) in the same location as, and presumably derived from, the MRR of pRMH760 from Klebsiella pneumoniae (11, 12) (Fig. 1). The pNDM-1_Dok01 (20) MRR includes a segment probably mobilized as an IS903-mediated composite transposon, while pMR0211 (7) contains a different segment that includes plasmid backbone, ISAba14, and aphA6. The pNDM102337 MRR includes a segment in which ISAba14 and ISCR27 are both truncated. The MRR of the remaining three plasmids could have been progressively derived from this by insertion of ISKpn14 (pNDM10505), deletion of the region between two ISKpn14 (pNDM10469), and switching of the cassette array (pNDM-KN) (1) (Fig. 1).
In the IncL/M plasmid pNDM-HK (4), a short blaNDM-1-containing segment and part of the blaDHA-1-ampR region replace part of the MRR of the closely related pCTX-M3 (3). This segment may be derived from a partially mapped plasmid with a similar configuration but an intact ISAba125 (21). The first reported context of blaNDM-1 (24) is similar to that in pNDM-HK, but blaDHA-1 is truncated and the adjacent region, carrying a gene encoding a putative efflux protein, matches the Klebsiella pneumoniae chromosome. A BamHI site at the boundary of these two regions (Fig. 1) suggests accidental cloning of a noncontiguous fragment not present in the native plasmid.
p271A from Escherichia coli (17) includes most of ISAba125, interrupted by ISEc33 and truncated by a region consisting of two Tn3-like 38-bp IR separated by 181 bp (13). Similar IR-(181-bp)-IR structures are found on other plasmids, e.g., close to Tn4401 carrying blaKPC (8) and on pCK41 from Edwardsiella tarda. The region downstream of blaNDM-1, containing another putative 38-bp IR and ISSen4, also matches pCK41 and may be part of a transposon truncated by Tn5403. This configuration could have resulted from insertion of a pregenerated hybrid transposon bounded by a 38-bp IR or by progressive insertions with accompanying deletions (13).
blaNDM-1 may have been created by initial capture of a precursor gene by ISCR27 and then fusion to the aphA6/ISAba125 promoter region in Acinetobacter. Different mobile elements then seem to have moved segments that contain blaNDM-1 into existing MRRs on different plasmids found in Enterobacteriaceae. This variety of wider contexts provides an interesting and potentially informative contrast with the globally dominant blaCTX-M-15 gene, generally found in variants of the same MRR on IncF plasmids (14).
REFERENCES
- 1. Carattoli A, Villa L, Poirel L, Bonnin RA, Nordmann P. 2012. Evolution of IncA/C blaCMY-2 carrying plasmids by acquisition of the blaNDM-1 carbapenemase gene. Antimicrob. Agents Chemother. 56:783–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen Y, et al. 2012. Draft genome sequence of an Acinetobacter genomic species 3 strain harboring a blaNDM-1 gene. J. Bacteriol. 194:204–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gołêbiewski M, et al. 2007. Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum β-lactamase gene blaCTX-M-3. Antimicrob. Agents Chemother. 51:3789–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ho PL, et al. 2011. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One 6:e17989 doi:10.1371/journal.pone.0017989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hu H, et al. 2012. A novel plasmid and its variant harboring both blaNDM-1 gene and T4SS in clinical isolates of Acinetobacter lwoffii. Antimicrob. Agents Chemother. 56:1698–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mantengoli E, Rossolini GM. 2005. Tn5393d, a complex Tn5393 derivative carrying the PER-1 extended-spectrum β-lactamase gene and other resistance determinants. Antimicrob. Agents Chemother. 49:3289–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McGann P, et al. 2012. Complete sequence of a novel 178-kilobase plasmid carrying blaNDM-1 in a Providencia stuartii strain isolated in Afghanistan. Antimicrob. Agents Chemother. 56:1673–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Naas T, et al. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob. Agents Chemother. 52:1257–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nigro SJ, Post V, Hall RM. 2011. Aminoglycoside resistance in multiply antibiotic-resistant Acinetobacter baumannii belonging to global clone 2 from Australian hospitals. J. Antimicrob. Chemother. 66:1504–1509 [DOI] [PubMed] [Google Scholar]
- 10. Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol. 19:588–595 [DOI] [PubMed] [Google Scholar]
- 11. Partridge SR. 2011. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol. Rev. 35:820–855 [DOI] [PubMed] [Google Scholar]
- 12. Partridge SR, Hall RM. 2004. Complex multiple antibiotic and mercury resistance region derived from the r-det of NR1 (R100). Antimicrob. Agents Chemother. 48:4250–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Partridge SR, Paulsen IT, Iredell JR. 2012. pJIE137 carrying blaCTX-M-62 is closely related to p271A carrying blaNDM-1. Antimicrob. Agents Chemother. 56:2166–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Partridge SR, Zong Z, Iredell JR. 2011. Recombination in IS26 and Tn2 in the evolution of multiresistance regions carrying blaCTX-M-15 on conjugative IncF plasmids from Escherichia coli. Antimicrob. Agents Chemother. 55:4971–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pfeifer Y, et al. 2011. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J. Antimicrob. Chemother. 66:1998–2001 [DOI] [PubMed] [Google Scholar]
- 16. Poirel L, et al. 2012. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56:1087–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Poirel L, Bonnin RA, Nordmann P. 2011. Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli by high-throughput genome sequencing. Antimicrob. Agents Chemother. 55:4224–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 55:5403–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poirel L, Lagrutta E, Taylor P, Pham J, Nordmann P. 2010. Emergence of metallo-β-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob. Agents Chemother. 54:4914–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sekizuka T, et al. 2011. Complete sequencing of the blaNDM-1-positive IncA/C plasmid from Escherichia coli ST38 isolate suggests a possible origin from plant pathogens. PLoS One 6:e25334 doi:10.1371/journal.pone.0025334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Solé M, et al. 2011. First description of an Escherichia coli strain producing NDM-1 carbapenemase in Spain. Antimicrob. Agents Chemother. 55:4402–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Toleman M, Spencer J, Jones L, Walsh TR. 2012. blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56:2773–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Toleman MA, Walsh TR. 2008. Evolution of the ISCR3 group of ISCR elements. Antimicrob. Agents Chemother. 52:3789–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yong D, et al. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]