Abstract
The recent and dramatic rise of antibiotic resistance among bacterial pathogens underlies the fear that standard treatments for infectious disease will soon be largely ineffective. Resistance has evolved against nearly every clinically used antibiotic, and in the near future, we may be hard-pressed to treat bacterial infections previously conquered by “magic bullet” drugs. While traditional antibiotics kill or slow bacterial growth, an important emerging strategy to combat pathogens seeks to block the ability of bacteria to harm the host by inhibiting bacterial virulence factors. One such virulence factor, the type three secretion system (T3SS), is found in over two dozen Gram-negative pathogens and functions by injecting effector proteins directly into the cytosol of host cells. Without T3SSs, many pathogenic bacteria are unable to cause disease, making the T3SS an attractive target for novel antimicrobial drugs. Interdisciplinary efforts between chemists and microbiologists have yielded several T3SS inhibitors, including the relatively well-studied salicylidene acylhydrazides. This review highlights the discovery and characterization of T3SS inhibitors in the primary literature over the past 10 years and discusses the future of these drugs as both research tools and a new class of therapeutic agents.
INTRODUCTION
One of the most pressing threats to the future of human health is the rapid and alarming evolution of antimicrobial resistance by pathogenic bacteria. Since the introduction of the first antibiotics, the development of resistance has dependably followed clinical use, often in as little as 3 years (10). Currently, 70% of hospital-acquired infections are resistant to one or more antibiotics (10). Methicillin-resistant Staphylococcus aureus (MRSA) heads this group and is responsible for more U.S. deaths each year than HIV (42). These substantial concerns are most pressing for Gram-negative bacteria, for which only a single new agent has been approved in the last decade (62).
Despite a clear necessity for the development of new drugs, most large pharmaceutical companies have abandoned the field (13). The prevailing view among corporations like Glaxo SmithKline, Roche, and Eli Lilly is that research dollars are better invested in developing treatments that command high prices and require long courses of therapy (61). As expensive clinical trials and low success rates have made antibiotic research less profitable, Washington lawmakers are considering legislation like the recently passed GAIN Act for installing tax incentives, longer patents, and even federal funding to promote corporate innovation (60). Yet it is unlikely that any new classes of antibiotic drugs will reach the market within the next 10 years (12). Clearly, a renaissance in antimicrobial research is needed to combat the emergence of multidrug-resistant and untreatable pan-resistant bacterial infections.
VIRULENCE BLOCKERS
In the past decade, a significant portion of academic antibiotic research has shifted from bactericidal or bacteriostatic drugs to virulence blockers (37). Unlike established antibiotics, virulence blockers inhibit pathogens by disarming the bacteria and preventing normal infection. These targeted antivirulence drugs inherently have benefits and disadvantages over conventional antibacterials. For example, traditional antibiotics are directed at widespread bacterial structures or processes required for growth. While this approach produces broadly effective drugs, these antibiotics indiscriminately kill both pathogens and members of the microbiota. Disrupting the normal flora of the gut can have harmful side effects, including increased risk of colitis caused by microbiota dysbiosis (9, 29). Additionally, recent research suggests that during antibiotic treatment, resistance arises in the abundant commensal flora, and this antibiotic resistance can then be passed on to more-scarce pathogens in the gut through horizontal gene transfer (37, 42, 64, 65). Since the targets of virulence blockers are found only in a small subset of bacteria, they should apply selective pressure on fewer organisms than established antibiotics and reduce the evolution and spread of antibiotic resistance genes.
Virulence blockers should circumvent several common drug resistance pathways. For instance, some classes of virulence blockers target external processes, thus avoiding the common resistance avenues of drug efflux and diminished permeability (70). Additionally, these drugs may not promote a rapid rise of resistance, as they limit bacterial replication in the host but not in other environments, where antibiotic contamination from agriculture and animal farms can drive the evolution of resistance (37, 46). Though bacterial virulence mechanisms are diverse, anticipated progress in rapid infection diagnosis bolsters the potential for targeted therapeutic strategies (7). Several classes of inhibitors have already been researched or even accepted into the clinic (10).
The most-established virulence blockers are classified as antitoxins and are administered to counteract the secreted toxins of pathogens, including Bacillus anthracis, Corynebacterium diphtheriae, and Clostridium tetani (10, 48, 66, 77). Often in the form of antibodies, these virulence blockers differ from most of the inhibitors currently being developed but have been well studied and used since the late 19th century (32, 63). More recently, distinct molecules inhibiting Vibrio cholerae cholera toxin expression and biofilm formation have been explored (28, 58). Similarly, new work has examined the potential of inhibiting extracellular molecules and receptors involved in quorum sensing. Certain pathogens, including Pseudomonas aeruginosa, require sufficient bacterial numbers before forming biofilms or expressing virulence genes (25, 50, 69). Keeping these bacteria “blind” to their neighbors may be one strategy to allow easier immune clearance.
Targeting bacterial appendages is another potential method to reduce virulence, and specialized secretion systems are an obvious aim for researchers, as they are required for growth in human hosts but not in other environments. While much of this research has focused on type three secretion systems (T3SSs), type two (T2SS) and type four (T4SS) secretion systems are also promising targets for study (5, 37). Both T2SSs and T4SSs translocate virulence factors across the bacterial membrane, but the T4SS mediates the translocation of genetic information as well (5). Thus, inhibiting T4S may play an important role in reducing the spread of antibiotic resistance. In addition to translocating virulence factors, T2SSs are also responsible for assembling adhesive pili on the outer surface of bacteria (5). Many pathogens use their pili to attach to eukaryotic cells, and pilicides have been proposed as one method of reducing adhesion and thereby limiting infection (41, 59, 73).
TYPE THREE SECRETION SYSTEMS
Type three secretion systems (T3SSs) are required by dozens of animal and plant pathogens, including the causative agents of plague, typhoid fever, and pneumonia, to cause disease (16). During mammalian infection, pathogens use the T3SS to inject effector proteins directly into target host cells, disrupting host defense mechanisms and allowing disease progression (8). In contrast, when the T3SS is nonfunctional, most T3SS-expressing bacteria are rendered benign and are easily cleared by the host immune system (8, 81). While not found in commensal bacteria (24), T3SSs of pathogens share many structural features, suggesting that a single class of molecules may inhibit T3S by multiple different bacterial species.
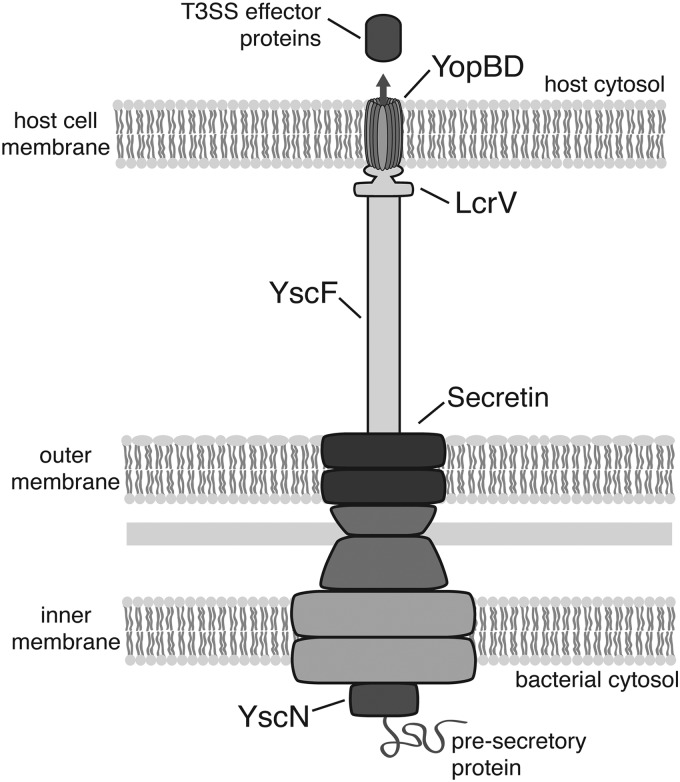
A growing number of studies have explored T3SS inhibition as a therapeutic strategy for novel antibiotics. In a majority of these studies, Yersinia spp. have served as the model organisms due to their well-characterized T3SSs and readily available tools for research. Found only in Gram-negative bacteria, T3SSs span the inner and outer bacterial membranes and share remarkable structural similarity to flagella (15, 16). The membrane-bound portion is known as the basal body and consists of a number of proteins (Fig. 1) (15, 16). Notable basal body proteins include secretin, which forms a ring found in the outer membrane, and the ATPase known as YscN in Yersinia spp. (49). YscN sits at the base of the T3SS and is thought to remove molecular chaperones from effector proteins, facilitating effector protein translocation into target cells (15, 16). The extracellular needle of the Yersinia T3SS consists of a 60-nm-long hollow tube made up of 100 to 150 polymerized YscF subunits (15, 16). The needle tip is formed by LcrV, a hydrophilic protein thought to provide a scaffold for pore formation in host cell membranes (15, 16). The host cell membrane pore is formed by the insertion of two hydrophobic proteins, YopB and YopD, which are known to interact with LcrV (8, 15). Upon host cell contact, various effectors are translocated through the T3SS into the host cell cytosol, where they target host cell signaling pathways (8, 81).
Fig 1.
Yersinia type III secretion system. The T3SS is composed of a basal body, a needle structure, and a needle tip complex. The basal body spans the bacterial inner and outer membranes and is composed of proteins, including the secretin protein YscC and the ATPase YscN (47). Once the basal body is assembled, it becomes a functional secretion system. The YscF needle subunit is then targeted for secretion through the basal body, and secreted YscF subunits assemble to form a hollow extracellular appendage extending 60 nm from the bacterial outer membrane (15). The LcrV needle tip protein is secreted next and assembles into a pentamer at the apex of the needle. Upon host cell contact, the hydrophobic translocator proteins YopB and YopD are secreted through the T3SS. The LcrV tip complex is then thought to insert YopB and YopD into the host membrane, forming a pore. Finally, partially unfolded effector proteins are translocated through the fully assembled T3SS into the host cell's cytoplasm, where they interfere with host defense mechanisms.
As is the case for many T3SS-expressing pathogens (16), expression and activity of the Yersinia T3SS are tightly regulated in response to external factors (19). LcrF is the main transcriptional regulator of T3SS genes in Yersinia and is expressed only at low levels at 26°C (14, 31). However, at 37°C, the temperature inside a mammalian host, translation of LcrF is thermoinduced, leading to increased expression of virulence genes (8, 14, 31). Further regulation is mediated by LcrQ in conjunction with YopD and the YopD chaperone LcrH; these cytoplasmic proteins inhibit translation of effector proteins until host cell contact has occurred (19, 31). LcrQ is then rapidly secreted, derepressing expression of the T3SS effector proteins (20). T3S can be artificially induced in a lab setting by growing Yersinia at 37°C and in medium containing a low level of calcium, which may mimic host cell contact.
While the T3S injectisome is highly conserved among pathogens, the function and number of effector proteins vary between species. In Yersinia pseudotuberculosis, six proteins serve as effectors: Yop proteins E, H, J, M, O, and T (8). YopE, YopH, YopO, and YopT all target proteins involved in the host's actin cytoskeleton, leading to the inhibition of both phagocytosis and the formation of reactive oxygen species (47, 71). As a result, Yersinia can avoid being taken up and killed by professional phagocytes. YopJ downregulates the inflammatory response by blocking the mitogen-activated protein kinase (MAPK), NF-κB, and IRF3 signaling pathways (47, 74). YopM is critical for virulence and modulates immune cell function (1, 36, 72, 85). Collectively, the Yop proteins allow Yersinia to evade innate immune defenses. T3SS-mediated disruptions of phagocytosis and host immune responses are essential for the progression of Yersinia infection. In fact, Yersinia pestis mutants missing the T3SS are avirulent even if injected into a host's bloodstream (81). T3S is also necessary for the virulence of many other human pathogens, including Pseudomonas aeruginosa, Shigella flexneri, enteropathogenic Escherichia coli, and Salmonella enterica serovar Typhimurium (16, 81). Accordingly, antimicrobial strategies that block T3S have become attractive alternatives to traditional bactericidal drugs.
T3SS INHIBITORS: A RECENT HISTORY OF DISCOVERY
Salicylidene acylhydrazides.
One of the earliest accounts of a T3SS inhibitor came in 2003 when a group from Umeå University in Sweden announced the promise of salicylidene acylhydrazides (Fig. 2, compound 1) (Table 1) (33, 34). This initial breakthrough came from an assay designed to measure T3S through a reporter-gene construct (33). In their experimental setup, Kauppi et al. took advantage of the tight coupling of T3S and translation of effector proteins in Yersinia pseudotuberculosis by fusing a luciferase gene to the YopE promoter (33). Compounds that inhibited T3S resulted in lower YopE translation and reduced luminescence. This high-throughput assay was used to successfully screen a synthetic library of 9,400 small molecules purchased from ChemBridge, obtaining three confirmed hits, including compound 1 (Fig. 2), a salicylidene acylhydrazide (33). The authors demonstrated that the inhibitors did not affect bacterial growth in vitro and concluded that they were acting on T3S in some capacity (33). As T3SSs and flagella are closely related, the authors sensibly investigated the effect of T3SS inhibitors on motility, finding that only compound 1 reduced Yersinia motility (33). By screening in a whole-cell model, Kauppi et al. confronted obstacles of cell permeability and metabolism head-on, yet the exact targets and mechanisms of compounds 1 to 3 remained uncertain and required future study (33).
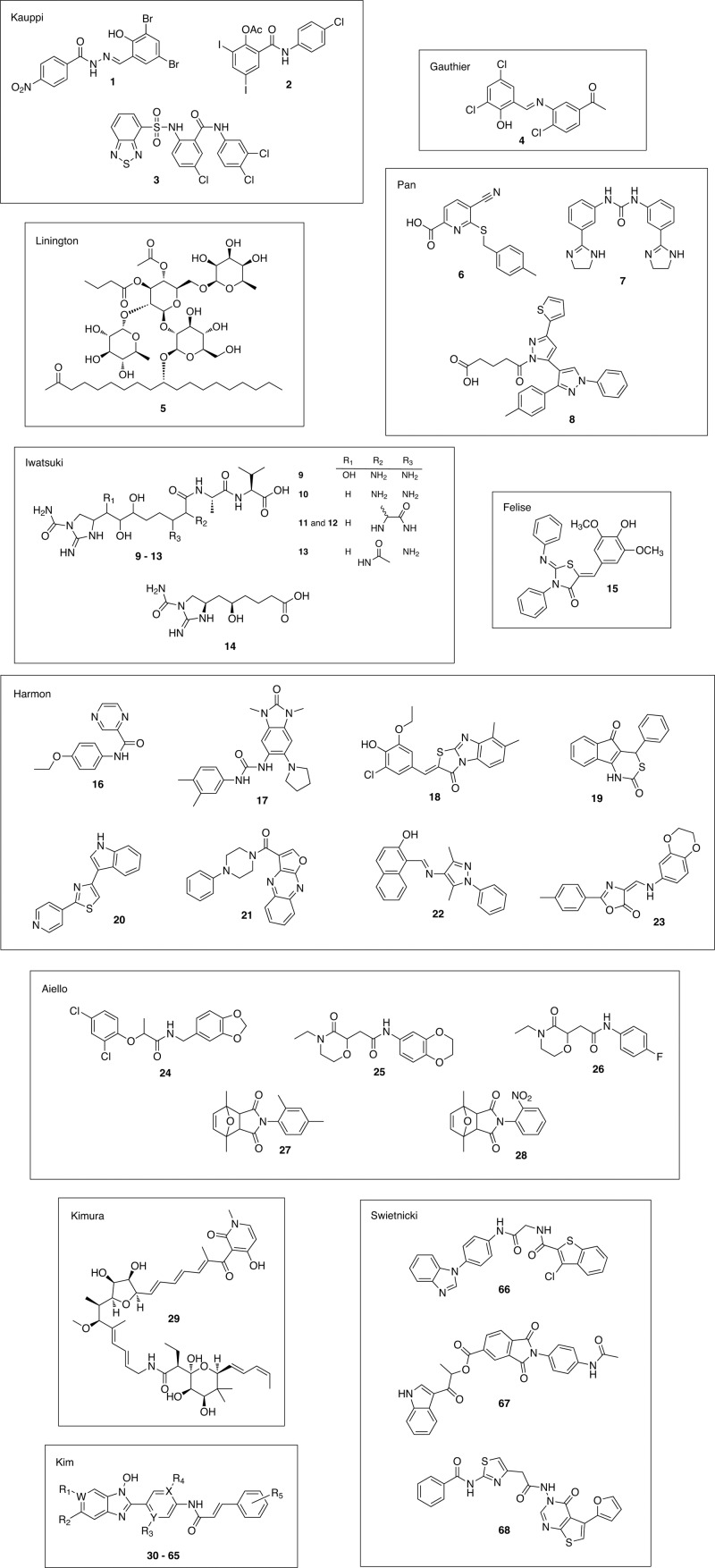
Fig 2.
Chemical structures of identified type three secretion system (T3SS) inhibitors. Each inhibitor is numbered in order of appearance in the text and labeled by the first author of the study that discovered it. The apparent chemical diversity among T3SS inhibitors suggests that there are multiple targets for inhibition.
Table 1.
Published type three secretion system inhibitors
| Compound name(s)/class(es) (compound no. in this study) | Yr | Reference | Molecular target | Source | Effective against: |
|---|---|---|---|---|---|
| Caminoside A (5) | 2002 | Linington et al. (44) | Unknown | Marine sponge | E. coli |
| Salicylidene acylhydrazide (1) | 2003 | Kauppi et al. (33) | Unknown; possibly WrbA, Tpx, and FolX | Synthetic compound library | Yersinia, Chlamydia, Pseudomonas, E. coli, Salmonella, Shigella |
| Clioxanide (2), 2-arylsulfonylamino-benzanilide (3) | 2003 | Kauppi et al. (33) | Unknown | Synthetic compound library | Yersinia |
| Salicylideneanilide (4) | 2005 | Gauthier et al. (24) | Unknown | Natural compound library | E. coli, Pseudomonas |
| Dipropionate (7), compounds 1 and 4 (6, 8) | 2007 | Pan et al. (56) | Unknown | Natural/synthetic compound library | Yersinia |
| Guadinomines A-D (9–13), guadinomic acid (14) | 2008 | Iwatsuki et al. (30) | Unknown | Microbial extracts from soil samples | E. coli |
| Thiazolidinone (15) | 2008 | Felise et al. (21) | Unknown; possibly secretin | Natural/synthetic compound library | Yersinia, Salmonella, Francisella, Pseudomonas |
| N-Hydroxybenzimidazole (30–65) | 2009 | Kim et al. (38) | LcrF (transcriptional activator) | Synthetic compound library | Yersinia |
| Compounds 1–8 (16–23) | 2010 | Harmon et al. (26) | Unknown; possibly a T3SS-host membrane interaction | Natural/synthetic compound library | Yersinia |
| Compounds 1–5 (24–28) | 2010 | Aiello et al. (2) | Unknown | Natural/synthetic compound library | Yersinia, Pseudomonas, Chlamydia |
| Aurodox (29) | 2011 | Kimura et al. (39) | Unknown | Natural compound library | E. coli, Citrobacter rodentium |
| 7086, 7832, 7812 (66–68) | 2011 | Swietnicki et al. (75) | YscN (ATPase) | Synthetic compound library | Yersinia, E. coli |
Two years later, the group published a follow-up paper characterizing the salicylidene acylhydrazides (55). This research sought to elucidate the results presented in their previous study; it had remained unclear whether the inhibitors were acting specifically on the T3SS or reducing effector levels by broadly lowering overall transcription or translation. To make this important distinction, the authors tested whether compound 1 reduced transcription of the LcrQ promoter, which is upregulated at 37°C but not controlled by LcrF (55). The authors found that compound 1 did not affect expression of an LcrQ-lux fusion reporter, suggesting that this compound does not generally inhibit transcription of temperature-inducible promoters (55). However, compound 1 did block secretion of T3SS effector proteins in both wild-type and LcrQ mutant Yersinia strains (55). The authors concluded that compound 1 specifically targets T3S (55).
In 2005, Gauthier et al. independently identified salicylideneanilides, compounds closely related to salicylidene acylhydrazides, in a high-throughput screen designed to measure T3SS inhibition in enteropathogenic E. coli (EPEC) (24). The authors screened a Maybridge library of 20,000 molecules, monitoring secretion of the EPEC effector protein EspB by an enzyme-linked immunosorbent assay (ELISA) (24). Their primary salicylideneanilide (Fig. 2, compound 4) reduced the expression of T3SS-associated virulence proteins (Tir, EspB, EscJ, EscC, and EspC), as demonstrated by a Western blot (24, 82). The group also used transcriptional fusions to confirm that compound 4 inhibited the transcription of virulence-associated promoters, while nonvirulence promoters were unaffected (24). In contrast to findings by Kauppi et al., the compound caused no change in motility or levels of flagellin in EPEC (24).
These breakthroughs were noted by several groups studying Chlamydia trachomatis, an obligate intracellular pathogen that encodes a T3SS but for which there is no genetic system to aid its study. In 2006, Wolf et al. and Muschiol et al. showed that the salicylidene acylhydrazides were able to disrupt normal progression of the Chlamydia infectious cycle (51, 84). Both groups demonstrated that the compound was able to prevent chlamydial differentiation and multiplication within mammalian cells in a dose-dependent manner when given at various stages of infection (51, 84). The authors also saw downregulation of T3SS genes by reverse transcriptase PCR (RT-PCR) (51, 84). Wolf et al. included Coxiella burnetii, an intracellular pathogen possessing a T4SS, in their study and found that this pathogen's infectious cycle was unperturbed by the T3SS inhibitor (84). Additionally, the group observed that the compound's effects on Chlamydia were reversible, further highlighting the utility of T3SS inhibitors as research tools for notoriously hard-to-study intracellular bacteria (84). One year later, a report proving that the same class of T3SS inhibitors was effective against the related pathogen Chlamydia pneumoniae was published (3).
Surprisingly, it was research on Chlamydia spp. that led to one of the more curious discoveries about the salicylidene acylhydrazides—their inhibitory activity's dependence on iron concentration. In 2007, Slepenkin et al. found that by adding iron to HeLa cells infected with C. trachomatis, they were able to reverse the effects of the inhibitors in a dose-dependent manner, while Ca2+, Mn2+, Mg2+, and Zn2+ had no discernible effect (68). Previously, hydrazones were shown to chelate iron, and the authors demonstrated that the salicylidene acylhydrazide they studied, INP0341, and an analogue, INP0406, were able to bind iron to some extent (68). However, since INP0406 had little inhibitory effect on chlamydial intracellular growth, iron depletion caused by INP0341 might not fully explain the compound's mode of inhibition (68). The authors proposed that the salicylidene acylhydrazides at least partially work by altering the intracellular iron available to Chlamydia or the host cell, though it initially remained unclear how or why iron may be necessary for T3S (68).
Two years later, Layton et al. proposed that salicylidene acylhydrazides rely on iron restriction to inhibit T3S in Salmonella enterica serovar Typhimurium (40). Through whole-genome transcriptome analysis, the authors found that in the presence of INP0403, a quarter of all highly upregulated genes were involved in iron acquisition or transport, suggesting that their compound may be an iron chelator (40). The authors confirmed previous work by showing that INP0403 had restricted iron availability, and its inhibitory activity was able to be reversed by the addition of exogenous free iron (40). Despite these findings, it remains unclear what role iron plays in salicylidene acylhydrazide-mediated T3SS inhibition, and further work is necessary to fully appreciate iron's effects on T3S and T3S inhibitors.
In 2007, Hudson et al. first demonstrated the in vitro and in vivo effectiveness of salicylidene acylhydrazides against SPI-1, one of two T3SSs in Salmonella enterica serovar Typhimurium (27). The authors showed that their salicylidene acylhydrazides did not affect Salmonella growth in broth culture yet decreased SPI-1-induced red blood cell lysis by 30 to 60% and reduced SPI-1-mediated invasion of HeLa cells (27). The group was the first to demonstrate that salicylidene acylhydrazides block virulence in vivo, using a bovine intestinal ligated loop model (27). After injecting three Friesian bull calves with Salmonella, the authors found that bacteria preincubated with the compounds induced less enteritis and recruited fewer neutrophils to the infected loops than Salmonella alone (27).
Negrea et al. further explored the salicylidene acylhydrazides' effects on T3S in Salmonella enterica serovar Typhimurium (53). Analyzing nine related compounds, the authors found that most prevented Salmonella intracellular replication and invasion into MDCK cells (53). However, only one was able to prevent effector translocation without also blocking effector protein expression (53). The authors also created LacZ fusion constructs against several notable T3SS proteins: the transcriptional activator protein HilA, the translocon protein PrgH, and the effector protein SipC (53). All three reporters demonstrated the expected high levels of transcription under T3SS-inducing conditions, but this transcription was severely reduced in the presence of the salicylidene acylhydrazides (53). The salicylidene acylhydrazides did not repress transcription of an effector protein, SipB, suggesting that the compounds do not broadly lower all gene expression (53).
Negrea et al. also explored the effects of salicylidene acylhydrazides on motility (53). Two of the nine studied inhibitors reduced bacterial motility, with this finding corroborated by decreased surface expression of flagella observed by immunoblotting (53). Similarly, Layton et al. saw that most Salmonella flagellar genes were repressed 1.5- to 2-fold in the presence of a salicylidene acylhydrazide inhibitor (40). These findings support observations by Kauppi et al. but are in contrast to those of Gauthier et al., who saw no change in the motility of EPEC cells treated with their closely related salicylideneanilide (24, 33).
In 2008, Veenendaal et al. tested salicylidene acylhydrazides on Shigella flexneri, finding the compounds capable of preventing T3S, HeLa cell invasion, and macrophage killing (80). The authors also studied needle assembly by electron microscopy, noting a 30 to 40% reduction in T3SSs per compound-treated bacterium (80). In addition, the authors measured needle length, observing much shorter needles in compound-treated samples (80). This was the first publication to suggest that the salicylidene acylhydrazides functioned by altering needle assembly.
Slepenkin et al. published an account of a salicylidene acylhydrazide protecting against infection in vivo, working with the sexually transmitted pathogen C. trachomatis (67). During the trial, the authors gave 51 mice 10 vaginal treatments with a salicylidene acylhydrazide, INP0341, or diluent alone (67). The mice were infected with C. trachomatis 2 days after the first treatment, and INP0341 or sham control treatments continued for 5 days after infection (67). The mice treated with INP0341 were 60% less likely to have a positive vaginal C. trachomatis culture than control mice during the 4-week observation period (67). INP0341-treated mice also had lower antibody titers to C. trachomatis after 4 weeks, indicating that salicylidene acylhydrazides may have promise as prophylactics for Chlamydia infection (67).
A follow-up study analyzed 58 salicylidene acylhydrazides for inhibition of C. trachomatis growth, pharmacokinetics in mice, and in vivo efficacy against the pathogen (78). One compound, ME0192, demonstrated poor pharmacokinetic properties but had the highest in vitro antichlamydial activity (78). The molecule inhibited C. trachomatis infection in mice after topical administration while leaving the normal vaginal flora unharmed (78).
In an attempt to characterize the effects of salicylidene acylhydrazides on E. coli, Tree et al. completed a whole-genome transcriptome analysis of the bacterium in the presence of the inhibitory compounds (76). They found repression of many virulence genes, including all genes within the locus of enterocyte effacement (LEE), a conserved pathogenicity island (76). They also observed reduced expression of Ler and PchA, regulators known to be involved in coordinate expression of virulence genes (76). Through their findings, the authors propose that this class of inhibitors functions through repression of virulence genes or possibly horizontally acquired genes (76). Interestingly, they also found upregulation of flagellum expression, unlike observations from previous studies in other organisms (76).
Wang et al. reported their findings from a study aimed at identifying the protein targets of a salicylidene acylhydrazide in E. coli (83). Using an affinity column containing the salicylidene acylhydrazide ME0055, the group identified proteins from an E. coli cell lysate that bound the inhibitor (83). Three enzymes were the most likely targets of the compound: WrbA and Tpx, proteins involved in defense against oxidative stress, and FolX, a poorly characterized epimerase (83). To further study these enzyme targets, the researchers deleted each of the three genes in both Yersinia and E. coli and then examined the T3SS and transcriptome of each mutant (83). They found 27 genes affected by the deletions, with most involved in locomotion, motility, and localization of the cell (83). Interestingly, flagellar genes were downregulated while T3SS genes were upregulated (83). This suggests that the inhibitors may actually increase the efficiency of the enzymes to which they bind, allowing them to better inhibit T3S. The group recently published a follow-up study providing insight into the binding of salicylidene acylhydrazides to Tpx from Yersinia pseudotuberculosis (22). The authors identified the most likely binding conformations for their salicylidene acylhydrazides and found the Tpx binding pocket to be mostly hydrophobic (22). The researchers also used far-Western blotting to demonstrate that the compounds bind to Tpx dimers with higher affinity than Tpx monomers (22).
OTHER INHIBITOR CLASSES
The first published account of a T3SS inhibitor came in 2002. Caminoside A (compound 5), a glycolipid from a marine sponge, was found to inhibit secretion by EPEC (44). Though obstacles in compound supply prevented further characterization of the molecule, its discovery signaled the beginning of a promising area of research.
Following the discovery of the salicylidene acylhydrazides, other groups began screening commercial compound libraries for inhibitors of T3S. In 2007, Pan et al. developed a novel screen to identify T3SS inhibitors in Yersinia using natural and synthetic libraries, and 2 years later, they reported new inhibitors discovered using this screen (56, 57). Normally, when Yersinia is induced to express the T3SS by increasing the temperature and dropping extracellular calcium levels, the bacteria enter a virulent stage and cease replication (56). Assuming that T3SS inhibitors would allow bacterial growth during T3SS-inducing conditions, the authors used a green fluorescent protein (GFP) strain of Yersinia to identify the few compounds that would promote increased fluorescence (growth) in this environment (57). Two of the three discovered compounds (Fig. 2, compounds 6 and 7) reduced effector secretion by Yersinia and EPEC, while all three (Fig. 2, compounds 6 to 8) inhibited T3SS-mediated HeLa killing by Yersinia (57). These compounds were structurally distinct from previously reported T3SS inhibitors and spanned a range of structural classes, including thioether bridged bicyclic and polyheteroaromatic systems (57). Interestingly, each compound inhibited secretion of Yop proteins E, D, and M, but with various efficiencies (57). The targets of these compounds remain unknown.
In 2008, Iwatsuki et al. screened for compounds that inhibited red blood cell lysis through T3S by EPEC using a library of microbial extracts isolated from Japanese soil samples (30). The group discovered six unique inhibitors (Fig. 2, compounds 9 to 14) produced by Actinobacteria, including guadinomines A to D and guadinomic acid (30). These compounds were neither antibiotic nor cytotoxic and showed dose-dependent T3SS inhibition but were not further characterized, and their mechanism of action remains unknown (30).
Later that year, Felise et al. reported a new class of inhibitors identified after screening 92,000 small molecules against the Salmonella T3SS (21). The library contained molecules of synthetic and natural origin purchased from Chem Div, Biomol, Maybridge, IF Labs, and Bionet (21). To set up their screen, the authors fused a phospholipase to a Salmonella effector protein (21). They then classified hits as molecules that reduced the enzyme's extracellular activity and, presumably, its secretion (21). While the study found seven hits, only one, a thiazolidinone (compound 15), did not affect transcription of a transcriptional regulator of T3S machinery (21). This molecule was next shown to inhibit formation of needle complexes and block T3S by Yersinia (21). The compound also prevented Francisella from secreting virulence factors through its unusual secretion system related to type four pilus secretion systems (21). Intriguingly, compound 15 (Fig. 2) was able to block T2S as well but had no effect on flagella or motility (21). The authors propose that their inhibitor targets secretin, the only shared protein between Francisella's secretion system, T2, and T3 secretion systems (21). Thiazolidinone was the first T3SS inhibitor shown to be effective against a plant pathogen, Pseudomonas syringae (21), underscoring the potential for T3SS inhibitors to block virulence in a wide range of T3SS-expressing pathogens.
Harmon et al. published an extremely thorough study detailing both the discovery and the characterization of several new inhibitors (26). The authors developed a novel screen by fusing a Yersinia effector to β-lactamase (26). First, target HEp-2 cells were loaded with a fluorescent dye, CCF2-AM, that emits a lower wavelength of light upon cleavage by β-lactamase (26). After infecting the HEp-2 cells with bacteria containing β-lactamase-T3SS effector fusions, the authors measured translocation efficiency by determining the ratio of blue/green fluorescence within the HEp-2 cells (26). After screening 100,000 small molecules, the authors found eight hits (Fig. 2, compounds 16 to 23) that were not cytotoxic to HEp-2 cells or generally antibiotic toward Yersinia (26). To determine whether the compounds affected T3SS needle formation, the group examined compound-treated bacteria by immunohistochemistry (26). All compound-treated samples showed some fluorescent staining of both YscF and LcrV, though compounds 20 and 23 showed slightly reduced staining of the T3SS needle (26). General chemical cross-linking analysis of YscF indicated that only one of the compounds (Fig. 2, compound 17) caused minor changes in needle structure (26). Interestingly, under T3SS-inducing conditions, none of the compounds were able to prevent Yop synthesis or secretion into culture supernatants (26). Six of the eight (Fig. 2, compounds 18 to 23) were, however, able to prevent YopE translocation into HEp-2 cells, while all eight (Fig. 2, compounds 16 to 23) increased Yop leakage into the surrounding supernatant during HEp-2 infection (26). One compound (Fig. 2, compound 22) greatly reduced Yersinia adherence to HEp-2 cells, and five (Fig. 2, compounds 17, 18, 20, 22, and 23) inhibited effector translocation by Pseudomonas (26). These findings suggest that the compounds function by altering pore formation on target cells or by disrupting hydrophobic interactions occurring between the membranes of Yersinia and the host cell (26).
The same year, Aiello et al. screened 80,000 compounds against the Pseudomonas T3SS, finding several confirmed hits (2). As in Yersinia, reduced T3S capability in Pseudomonas causes decreased expression of all T3SS operons (2). Accordingly, the authors fused the luxCDABE operon to a Pseudomonas effector gene, classifying screened compounds as hits if they caused reduced bacterial bioluminescence (2). Five compounds (Fig. 2, compounds 24 to 28) were confirmed as inhibitors using distinct effector secretion assays (2). However, the phenoxyacetamide (Fig. 2, compound 24) was the most promising, as it was the only compound to rescue CHO cells from T3SS-mediated cell death by Pseudomonas (2). The compound was also effective against Chlamydia and Yersinia, but with reduced potency (2). While the molecular target of compound 24 is not known, the results suggest that this phenoxyacetamide acts directly on the T3SS apparatus, as T2S was unaffected, general protein expression was not blocked, and the authors observed inhibition of T3S even when the compounds were added after T3SS assembly (2).
In 2011, Kimura et al. reported that their T3SS inhibitor protected against in vivo infection (39). The authors screened a diverse collection of natural products, including actinobacterial, fungal, and plant extracts for inhibition of red blood cell lysis by EPEC (39). Among the 10 original hits, aurodox (Fig. 2, compound 29) was selected due to its potent inhibition and history of previous study (39). In an animal experiment, none of the control mice infected with Citrobacter rodentium, including those treated with tetracycline, survived past day 18 (39). Yet all mice treated with aurodox 1 day following infection survived the challenge and demonstrated reduced incidence of colitis (39). Kimura et al. observed that aurodox did not affect expression of the housekeeping gene groEL; however, earlier work has suggested that the molecule may affect bacterial translation by binding to the ribosomal elongation factor Tu (39). The target of aurodox remains unclear, and further study is warranted to determine whether aurodox inhibits T3S via an indirect mechanism.
TARGETED SCREENS IN YERSINIA
Unlike early T3SS inhibitor screens, a few recent studies have targeted specific proteins necessary for regulation of needle formation or effector translocation. The Yersinia virulence regulator LcrF is essential for T3SS formation, and LcrF mutants are severely attenuated in mouse infection models (38). In 2009, Kim et al. identified an N-hydroxybenzimidazole as an inhibitor of LcrF-DNA binding (38). The following year, the same group published a more thorough exploration of their LcrF inhibitors (Fig. 2, compounds 30 to 65) (23). In addition to preventing DNA binding, the authors found the compounds able to reduce Yersinia virulence against macrophages and in vivo in mouse lungs, although their compound was administered to the mice 1 day before infection (23).
Another recent study identified inhibitors of YscN, a Yersinia ATPase responsible for removing chaperones from effector proteins and energizing the translocation process through the T3SS (75). The group confirmed that YscN mutants were fully attenuated in a mouse infection model and developed YscN inhibitors by using computational screening of a virtual three-dimensional (3D) database of small molecules against a model of the active site of YscN (75). Thirty-seven promising compounds were used in biological assays, and the researchers found three (Fig. 2, compounds 66 to 68) able to inhibit both YscN ATPase activity and secretion of YopE into bacterial culture (75). However, in an infection assay, the authors were unable to find significant inhibition, as Yersinia still caused HeLa cell rounding in the presence of the compounds (75). Although the authors were unable to discover an effective YscN inhibitor, their concept shows promise for future research. While bacterial ATPases have long been dismissed as potential antibiotic targets, the authors propose that their seemingly low 25% homology to related human enzymes should alleviate concerns of drug cross-reactivity (75). Additionally, the group's computer-aided approach to compound screening is an unexplored avenue that may bear fruit in future T3SS inhibitor studies.
STRUCTURE-ACTIVITY RELATIONSHIP STUDIES
High-throughput screening using live bacteria has led to several promising classes of T3SS inhibitors. However, the modes of action and biologically active functional groups must be explored afterwards. To systematically identify these biologically active sites, researchers often perform quantitative structure-activity relationship (QSAR) studies by synthesizing focused libraries of molecules that are structurally similar to one or more of the lead compounds. Selecting compounds for these focused libraries by statistical molecular design ensures chemical diversity and enhances the information yield from QSAR studies.
Kauppi et al. used statistical molecular design to create compounds based on a 2-arylsulfonylamino-benzanilide (Fig. 2, compound 3) identified in their original screen and were able to improve inhibitory activity from 68% to 91% (35). Similar studies on salicylanilides and salicylidene acylhydrazides showed strong correlations between theoretical QSAR models and experimental T3SS inhibition (17, 18). This work demonstrated that statistical molecular design coupled with QSAR and validation can be a successful strategy for lead optimization of T3SS inhibitors (18).
CHALLENGES AND PROMISE FOR THERAPEUTIC USE
As a new class of antibacterial compounds, T3SS inhibitors face unique obstacles for reaching the clinic and gaining widespread use. T3SS inhibitors do not affect bacterial replication outside a host, so standard MIC measurements will not be useful for assaying comparative drug activity. Instead, the effective dose of inhibitor will need to be established early in animal models to predict appropriate dosing for humans (6). This strategy is already used for new antitoxins (43) and will likely be the primary measure to compare the activities of antivirulence drugs.
Oral bioavailability is a common parameter that must be considered when developing a new drug for clinical use. Defined as the fraction of an orally administered dose of unchanged drug that reaches systemic circulation, oral bioavailability is important both to allow reasonable dosage of new drugs and to promote patient compliance (45, 79). However, many T3SS-expressing pathogens utilize their T3SS to colonize and/or persist at the mucosal surface or inside host cells within the epithelial barrier, obviating the need to deliver T3SS inhibitors into systemic circulation. Exceptions to this include enteropathogenic Yersinia, which requires T3S to survive in deeper tissues, such as the spleen (4). Therefore, the oral bioavailability of T3SS inhibitors may not be an important consideration for treatment of many infections caused by T3SS-expressing pathogens. However, given the number of pathogens that require a T3SS to grow inside mammalian cells (37), the ability of T3SS inhibitors to access the host cell cytosol and vacuole remains an important consideration.
Researchers will also need to determine the best applications of T3SS inhibitors in the clinic. The majority of in vivo studies reviewed here have employed the drugs as prophylactics, with only aurodox resolving an ongoing infection (39). T3SS inhibitors are less likely to generate resistance, making them prime candidates for use in prophylaxis. The inhibitors might be used to prevent enteric disease outbreaks following flooding, and it is easy to envision prophylactic applications for agricultural farming of plants and animals (37). However, as mentioned above, some pathogens require T3S throughout infection and would be well suited to antivirulence therapy following disease onset.
Since T3SS inhibitors target a specific subset of pathogenic bacteria, accurate diagnosis of infection will be a prerequisite for treatment. Point-of-care nucleic acid testing is beginning to enter the clinic, enabling the rapid and precise identification of infectious agents (7, 11, 54). This long-awaited shift in hospital policy should promote the development of T3SS inhibitors and other antivirulence drugs alike. However, one class of inhibitor can be used more broadly by physicians, since salicylidene acylhydrazides and thiazolidinones have proven effective against several bacterial genera. T3SS inhibitors may even be applied in concert with traditional antibiotics to improve their efficacy.
CONCLUSIONS AND FUTURE DIRECTIONS
The emerging threat of multidrug-resistant bacteria has led researchers to explore virulence blockers as new classes of antibiotics. Much research has focused on inhibitors of the T3SS, which is appropriate given the lack of new antibiotics in development to combat Gram-negative infection and the ubiquity of T3SSs in Gram-negative pathogens. Discovered T3SS inhibitors have been chemically diverse, suggesting that there are many different targets for T3SS inhibition. Consequently, T3SS inhibitors also have enormous potential as research tools, capable of helping microbiologists explore host-pathogen interactions involving the T3SS. Despite an increased interest in T3SS inhibitors, only the salicylidene acylhydrazides have truly begun to be biologically characterized for their mechanism of action. If any of this class of virulence blockers are to reach the clinic in the foreseeable future, more work is needed to identify and optimize the drugs' functions.
Further high-throughput screening to find new inhibitors is certainly necessary but must be undertaken with thought and reflection prioritized over brute force. David Payne of Glaxo SmithKline observed that antibacterial high-throughput screens are five times less likely to yield inhibitors than any other therapeutic targets, emphasizing a need for rethinking the process (62). Whole-cell screening has proven more successful than target-based in vitro approaches, as chemists have struggled to transform hits from target-based screening into active agents in whole-cell systems (62). Though the majority of current FDA-approved antibiotics are derived from natural products, few T3SS inhibitor screens have taken advantage of this resource. Many groups have instead examined extensively overlapping chemical libraries selected for their drug-like qualities (52). Additionally, previous studies have utilized less than 0.1% of the information provided by large screens; ideally, “hit” compounds should be compared with structurally similar but inactive compounds from the original screen in preliminary structure-activity relationship studies to learn why certain compounds are not biologically active (52).
As the rise of antibiotic resistance coincides with a shortage of antibiotic research, unique drugs and methods of treating bacterial infections must be considered. T3SS inhibitors should function not only as research tools but as novel antibiotics that do not promote the rapid evolution of resistance. The development of these antivirulence compounds is a complex goal that requires the resources of both academia and the pharmaceutical industry but, when achieved, should bear invaluable rewards for worldwide human health.
ACKNOWLEDGMENT
The authors acknowledge the California Institute for Quantitative Biosciences for support (to M.C.D. and V.A.).
Footnotes
Published ahead of print 30 July 2012
REFERENCES
- 1. Aepfelbacher M, Trasak C, Ruckdeschel K. 2007. Effector functions of pathogenic Yersinia species. Thromb. Haemost. 98:521–529 [PubMed] [Google Scholar]
- 2. Aiello D, et al. 2010. Discovery and characterization of inhibitors of Pseudomonas aeruginosa type III secretion. Antimicrob. Agents Chemother. 54:1988–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bailey L, et al. 2007. Small molecule inhibitors of type III secretion in Yersinia block the Chlamydia pneumoniae infection cycle. FEBS Lett. 581:587–595 [DOI] [PubMed] [Google Scholar]
- 4. Balada-Llasat J, Panilaitis B, Kaplan D, Mecsas J. 2007. Oral inoculation with type III secretion mutants of Yersinia pseudotuberculosis provides protection from oral, intraperitoneal, or intranasal challenge with virulent Yersinia. Vaccine 25:1526–1533 [DOI] [PubMed] [Google Scholar]
- 5. Baron C. 2010. Antivirulence drugs to target bacterial secretion systems. Curr. Opin. Microbiol. 13:100–105 [DOI] [PubMed] [Google Scholar]
- 6. Bergman KL. 2009. The animal rule and emerging infections: the role of clinical pharmacology in determining an effective dose. Clin. Pharmacol. Ther. 86:328–331 [DOI] [PubMed] [Google Scholar]
- 7. Bissonnette L, Bergeron MG. 2010. Diagnosing infections—current and anticipated technologies for point-of-care diagnostics and home-based testing. Clin. Microbiol. Infect. 16:1044–1053 [DOI] [PubMed] [Google Scholar]
- 8. Bleves S, Cornelis GR. 2000. How to survive in the host: the Yersinia lesson. Microbes Infect. 2:1451–1460 [DOI] [PubMed] [Google Scholar]
- 9. Chang JY, et al. 2008. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 197:435–438 [DOI] [PubMed] [Google Scholar]
- 10. Clatworthy AE, Pierson E, Hung DT. 2007. Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3:541–548 [DOI] [PubMed] [Google Scholar]
- 11. Clerc O, Greub G. 2010. Routine use of point-of-care tests: usefulness and application in clinical microbiology. Clin. Microbiol. Infect. 16:1054–1061 [DOI] [PubMed] [Google Scholar]
- 12. Coates ARM, Halls G, Hu Y. 2011. Novel classes of antibiotics or more of the same? Br. J. Pharmacol. 163:184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cooper M, Shlaes D. 2011. Fix the antibiotics pipeline. Nature 472:32. [DOI] [PubMed] [Google Scholar]
- 14. Cornelis GR, Wolf-Watz H. 1997. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol. Microbiol. 23:861–867 [DOI] [PubMed] [Google Scholar]
- 15. Cornelis GR. 2010. The type III secretion injectisome, a complex nanomachine for intracellular “toxin” delivery. Biol. Chem. 391:745–751 [DOI] [PubMed] [Google Scholar]
- 16. Cornelis GR. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811–825 [DOI] [PubMed] [Google Scholar]
- 17. Dahlgren MK, Kauppi AM, Olsson I, Linusson A, Elofsson M. 2007. Design, synthesis, and multivariate quantitative structure-activity relationship of salicylanilides—potent inhibitors of type III secretion in Yersinia. J. Med. Chem. 50:6177–6188 [DOI] [PubMed] [Google Scholar]
- 18. Dahlgren MK, Zetterström CE, Gylfe SA, Linusson A, Elofsson M. 2010. Statistical molecular design of a focused salicylidene acylhydrazide library and multivariate QSAR of inhibition of type III secretion in the Gram-negative bacterium Yersinia. Bioorg. Med. Chem. 18:2686–2703 [DOI] [PubMed] [Google Scholar]
- 19. Deane JE, Abrusci P, Johnson S, Lea SM. 2010. Timing is everything: the regulation of type III secretion. Cell. Mol. Life Sci. 67:1065–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edqvist PJ. 2007. Multiple twists in the molecular tales of YopD and LcrH in type III secretion by Yersinia pseudotuberculosis. Ph.D. dissertation Umeå University, Umeå, Sweden [Google Scholar]
- 21. Felise HB, et al. 2008. An inhibitor of gram-negative bacterial virulence protein secretion. Cell Host Microbe 4:325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gabrielsen M, et al. 2012. Structural characterisation of Tpx from Yersinia pseudotuberculosis reveals insights into the binding of salicylidene acylhydrazide compounds. PLoS One 7:e32217 doi:10.1371/journal.pone.0032217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garrity-Ryan LK, et al. 2010. Small molecule inhibitors of LcrF, a Yersinia pseudotuberculosis transcription factor, attenuate virulence and limit infection in a murine pneumonia model. Infect. Immun. 78:4683–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gauthier A, et al. 2005. Transcriptional inhibitor of virulence factors in enteropathogenic Escherichia coli. Antimicrob. Agents Chemother. 49:4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Geske GD, Wezeman RJ, Siegel AP, Blackwell HE. 2005. Small molecule inhibitors of bacterial quorum sensing and biofilm formation. J. Am. Chem. Soc. 127:12762–12763 [DOI] [PubMed] [Google Scholar]
- 26. Harmon DE, Davis AJ, Castillo C, Mecsas J. 2010. Identification and characterization of small-molecule inhibitors of Yop translocation in Yersinia pseudotuberculosis. Antimicrob. Agents Chemother. 54:3241–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hudson DL, et al. 2007. Inhibition of type III secretion in Salmonella enterica serovar Typhimurium by small-molecule inhibitors. Antimicrob. Agents Chemother. 51:2631–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hung DT, Shakhnovich E, Pierson E, Mekalanos JJ. 2005. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science 310:670–674 [DOI] [PubMed] [Google Scholar]
- 29. Hviid A, Svanström H, Frisch M. 2011. Antibiotic use and inflammatory bowel diseases in childhood. Gut 60:49–54 [DOI] [PubMed] [Google Scholar]
- 30. Iwatsuki M, et al. 2008. Guadinomines, type III secretion system inhibitors, produced by Streptomyces sp. K01-0509. I. Taxonomy, fermentation, isolation and biological properties. J. Antibiot. (Tokyo) 61:222–229 [DOI] [PubMed] [Google Scholar]
- 31. Izoré T, Job V, Dessen A. 2011. Biogenesis, regulation, and targeting of the type III secretion system. Structure 19:603–612 [DOI] [PubMed] [Google Scholar]
- 32. Kantha SS. 1991. A centennial review; the 1890 tetanus antitoxin paper of von Behring and Kitasato and the related developments. Keio J. Med. 40:35–39 [DOI] [PubMed] [Google Scholar]
- 33. Kauppi AM, Nordfelth R, Uvell H, Wolf-Watz H, Elofsson M. 2003. Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem. Biol. 10:241–249 [DOI] [PubMed] [Google Scholar]
- 34. Kauppi A, Nordfelth R, Hägglund U, Wolf-Watz H, Elofsson M. 2003. Salicylanilides are potent inhibitors of type III secretion in Yersinia. Adv. Exp. Med. Biol. 529:97–100 [DOI] [PubMed] [Google Scholar]
- 35. Kauppi AM, et al. 2007. Inhibitors of type III secretion in Yersinia: design, synthesis and multivariate QSAR of 2-arylsulfonylamino-benzanilides. Bioorg. Med. Chem. 15:6994–7011 [DOI] [PubMed] [Google Scholar]
- 36. Kerschen EJ, Cohen DA, Kaplan AM, Straley SC. 2004. The plague virulence protein YopM targets the innate immune response by causing a global depletion of NK cells. Infect. Immun. 72:4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Keyser P, Elofsson M, Rosell S, Wolf-Watz H. 2008. Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against Gram-negative bacteria. J. Intern. Med. 264:17–29 [DOI] [PubMed] [Google Scholar]
- 38. Kim OK, et al. 2009. N-hydroxybenzimidazole inhibitors of the transcription factor LcrF in Yersinia: novel antivirulence agents. J. Med. Chem. 52:5626–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kimura K, et al. 2011. A small-molecule inhibitor of the bacterial type III secretion system protects against in vivo infection with Citrobacter rodentium. J. Antibiot. (Tokyo) 64:197–203 [DOI] [PubMed] [Google Scholar]
- 40. Layton AN, et al. 2010. Salicylidene acylhydrazide-mediated inhibition of type III secretion system-1 in Salmonella enterica serovar Typhimurium is associated with iron restriction and can be reversed by free iron. FEMS Microbiol. Lett. 302:114–122 [DOI] [PubMed] [Google Scholar]
- 41. Leo JC, Skurnik M. 2011. Adhesins of human pathogens from the genus Yersinia. Adv. Exp. Med. Biol. 715:1–15 [DOI] [PubMed] [Google Scholar]
- 42. Levy SB, Marshall B. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10:S122–S129 [DOI] [PubMed] [Google Scholar]
- 43. Li D, Mattoo P, Keller JE. 2012. New equine antitoxins to botulinum neurotoxins serotypes A and B. Biologicals 40:240–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Linington RG, et al. 2002. Caminoside A, an antimicrobial glycolipid isolated from the marine sponge Caminus sphaeroconia. Org. Lett. 4:4089–4092 [DOI] [PubMed] [Google Scholar]
- 45. Lipinski C, Lombardo F, Dominy BW, Feeney PJ. 2001. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46:3–26 [DOI] [PubMed] [Google Scholar]
- 46. Martínez JL. 2008. Antibiotics and antibiotic resistance genes in natural environments. Science 321:365–367 [DOI] [PubMed] [Google Scholar]
- 47. Matsumoto H, Young GM. 2009. Translocated effectors of Yersinia. Curr. Opin. Microbiol. 12:94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moayeri M, Wiggins JF, Lindeman RE, Leppla SH. 2006. Cisplatin inhibition of anthrax lethal toxin. Antimicrob. Agents Chemother. 50:2658–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moraes TF, Spreter T, Strynadka NCJ. 2008. Piecing together the type III injectisome of bacterial pathogens. Curr. Opin. Struct. Biol. 18:258–266 [DOI] [PubMed] [Google Scholar]
- 50. Müh U, et al. 2006. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob. Agents Chemother. 50:3674–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Muschiol S, et al. 2006. A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U. S. A. 103:14566–14571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nathan C. 2011. Making space for anti-infective drug discovery. Cell Host Microbe 9:343–348 [DOI] [PubMed] [Google Scholar]
- 53. Negrea A, et al. 2007. Salicylidene acylhydrazides that affect type III protein secretion in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 51:2867–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Niemz A, Ferguson TM, Boyle DS. 2011. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 29:240–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nordfelth R, Kauppi AM, Norberg HA, Wolf-Watz H, Elofsson M. 2005. Small-molecule inhibitors specifically targeting type III secretion. Infect. Immun. 73:3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pan N, Lee C, Goguen J. 2007. High throughput screening for small-molecule inhibitors of type III secretion in Yersinia pestis. Adv. Exp. Med. Biol. 603:367–375 [DOI] [PubMed] [Google Scholar]
- 57. Pan NJ, Brady MJ, Leong JM, Goguen JD. 2009. Targeting type III secretion in Yersinia pestis. Antimicrob. Agents Chemother. 53:385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Peach KC, et al. 2011. An image-based 384-well high-throughput screening method for the discovery of biofilm inhibitors in Vibrio cholerae. Mol. Biosyst. 7:1176–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pinkner JS, et al. 2006. Rationally designed small compounds inhibit pilus biogenesis in uropathogenic bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:17897–17902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pollack A. 5 November2010. Antibiotics research subsidies weighed by U.S. The New York Times, New York, NY [Google Scholar]
- 61. Projan SJ. 2003. Why is big Pharma getting out of antibacterial drug discovery? Curr. Opin. Microbiol. 6:427–430 [DOI] [PubMed] [Google Scholar]
- 62. Projan SJ, Bradford P. 2007. Late stage antibacterial drugs in the clinical pipeline. Curr. Opin. Microbiol. 10:441–446 [DOI] [PubMed] [Google Scholar]
- 63. Rainey GJ, Young J. 2004. Antitoxins: novel strategies to target agents of bioterrorism. Nat. Rev. Microbiol. 2:721–726 [DOI] [PubMed] [Google Scholar]
- 64. Schjørring S, Krogfelt K. 2011. Assessment of bacterial antibiotic resistance transfer in the gut. Int. J. Microbiol. 2011:312956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schjørring S, Struve C, Krogfelt KA. 2008. Transfer of antimicrobial resistance plasmids from Klebsiella pneumoniae to Escherichia coli in the mouse intestine. J. Antimicrob. Chemother. 62:1086–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shoop WL, et al. 2005. Anthrax lethal factor inhibition. Proc. Natl. Acad. Sci. U. S. A. 102:7958–7963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Slepenkin A, Chu H, Elofsson M, Keyser P, Peterson EM. 2011. Protection of mice from a Chlamydia trachomatis vaginal infection using a salicylidene acylhydrazide, a potential microbicide. J. Infect. Dis. 204:1313–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Slepenkin A, et al. 2007. Reversal of the antichlamydial activity of putative type III secretion inhibitors by iron. Infect. Immun. 75:3478–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Smith KM, Bu Y, Suga H. 2003. Induction and inhibition of Pseudomonas aeruginosa quorum sensing by synthetic autoinducer analogs. Chem. Biol. 10:81–89 [DOI] [PubMed] [Google Scholar]
- 70. Smith PA, Romesberg FE. 2007. Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Nat. Chem. Biol. 3:549–556 [DOI] [PubMed] [Google Scholar]
- 71. Songsungthong W, Higgins MC, Rolán HG, Murphy JL, Mecsas J. 2010. ROS-inhibitory activity of YopE is required for full virulence of Yersinia in mice. Cell. Microbiol. 12:988–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Soundararajan V, Patel N, Subramanian V, Sasisekharan V, Sasisekharan R. 2011. The many faces of the YopM effector from plague causative bacterium Yersinia pestis and its implications for host immune modulation. Innate Immun. 17:548–557 [DOI] [PubMed] [Google Scholar]
- 73. Svensson A, et al. 2001. Design and evaluation of pilicides: potential novel antibacterial agents directed against uropathogenic Escherichia coli. Chembiochem 2:915–918 [DOI] [PubMed] [Google Scholar]
- 74. Sweet CR, Conlon J, Golenbock DT, Goguen J, Silverman N. 2007. YopJ targets TRAF proteins to inhibit TLR-mediated NF-κB, MAPK and IRF3 signal transduction. Cell. Microbiol. 9:2700–2715 [DOI] [PubMed] [Google Scholar]
- 75. Swietnicki W, et al. 2011. Identification of small-molecule inhibitors of Yersinia pestis type III secretion system YscN ATPase. PLoS One 6:e19716 doi:10.1371/journal.pone.0019716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tree JJ, et al. 2009. Characterization of the effects of salicylidene acylhydrazide compounds on type III secretion in Escherichia coli O157:H7. Infect. Immun. 77:4209–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Turk BE, et al. 2004. The structural basis for substrate and inhibitor selectivity of the anthrax lethal factor. Nat. Struct. Mol. Biol. 11:60–66 [DOI] [PubMed] [Google Scholar]
- 78. Ur-Rehman T, et al. 6 June 2012, posting date Pre-clinical pharmacokinetics and anti-chlamydial activity of salicylidene acylhydrazide inhibitors of bacterial type III secretion. J. Antibiot. (Tokyo) doi:10.1038/ja.2012.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Veber DF, et al. 2002. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 45:2615–2623 [DOI] [PubMed] [Google Scholar]
- 80. Veenendaal AKJ, Sundin C, Blocker AJ. 2009. Small-molecule type III secretion system inhibitors block assembly of the Shigella type III secreton. J. Bacteriol. 191:563–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Viboud GI, Bliska JB. 2005. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu. Rev. Microbiol. 59:69–89 [DOI] [PubMed] [Google Scholar]
- 82. Vidal JE, Navarro-García F. 2008. EspC translocation into epithelial cells by enteropathogenic Escherichia coli requires a concerted participation of type V and III secretion systems. Cell. Microbiol. 10:1975–1986 [DOI] [PubMed] [Google Scholar]
- 83. Wang D, et al. 2011. Identification of bacterial target proteins for the salicylidene acylhydrazide class of virulence blocking compounds. J. Biol. Chem. 286:29922–29931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wolf K, et al. 2006. Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol. Microbiol. 61:1543–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ye Z, et al. 2011. Distinct CCR2+ Gr1+ cells control growth of the Yersinia pestis ΔyopM mutant in liver and spleen during systemic plague. Infect. Immun. 79:674–687 [DOI] [PMC free article] [PubMed] [Google Scholar]