Abstract
Mersacidin, gallidermin, and nisin are lantibiotics, antimicrobial peptides containing lanthionine. They show potent antibacterial activity. All three interfere with cell wall biosynthesis by binding lipid II, but they display different levels of interaction with the cytoplasmic membrane. On one end of the spectrum, mersacidin interferes with cell wall biosynthesis by binding lipid II without integrating into bacterial membranes. On the other end of the spectrum, nisin readily integrates into membranes, where it forms large pores. It destroys the membrane potential and causes leakage of nutrients and ions. Gallidermin, in an intermediate position, also readily integrates into membranes. However, pore formation occurs only in some bacteria and depends on membrane composition. In this study, we investigated the impact of nisin, gallidermin, and mersacidin on cell wall integrity, membrane pore formation, and membrane depolarization in Bacillus subtilis. The impact of the lantibiotics on the cell envelope was correlated to the proteomic response they elicit in B. subtilis. By drawing on a proteomic response library, including other envelope-targeting antibiotics such as bacitracin, vancomycin, gramicidin S, or valinomycin, YtrE could be identified as the most reliable marker protein for interfering with membrane-bound steps of cell wall biosynthesis. NadE and PspA were identified as markers for antibiotics interacting with the cytoplasmic membrane.
INTRODUCTION
Over the last decades, bacteria have demonstrated their impressive ability to adapt to changing environmental conditions by rapidly developing and accumulating antibiotic resistances. Helped by an extensive use of antibiotics in health care and agriculture, multidrug-resistant strains, so-called superbugs, are spreading with a remarkable impact on human health. Recent statistics of the World Health Organization show that even in developed countries, infectious diseases are back among the top five causes of death, with the majority of deaths being attributed to bacterial infections (49). At the same time, approval rates of new antibiotic agents have been steadily decreasing since their apex in the 1980s (5). In view of these developments, in addition to improving hospital hygiene and to devising and implementing strategies to preserve effectiveness of available antimicrobial agents, a reinvigoration of antibiotic research and development is urgently needed to meet the superbug challenge.
One of the promising antibiotic classes for treatment of infections caused by multidrug-resistant pathogens are antimicrobial peptides, which occur naturally as components of the host defense and typically target the bacterial cell envelope. The peptide subclass of lantibiotics, which are produced by Gram-positive bacteria, is characterized by containing the nonproteinogenic amino acid lanthionine and often also other unusual amino acids. However, the group of lantibiotics is structurally and mechanistically diverse and can be further divided into class A, comprising stretched, flexible peptides, and class B, with more globular structures. The majority of peptides in both classes have been shown to inhibit cell wall biosynthesis. In contrast to class B members, some class A lantibiotics are also capable of integrating into the bacterial membrane and of pore formation, a process facilitated by binding of cell wall precursor lipid II (7, 13). This mechanistic duality depicts an interesting advantage of lantibiotics over conventional single-mechanism antibiotics. It was shown for various antibacterial compounds that inhibit targets encoded by multiple genes that resistance development is much slower than for antibiotics inhibiting a single target of a specific metabolic pathway (11, 32). As two essential complex targets, the cell wall biosynthesis machinery and the cell membrane, need to be altered to prevent cellular damage by dual-mechanism lantibiotics, resistance development by target mutation and stress adaptation is effectively deferred (10, 32). Such properties highlight antimicrobial peptides as attractive antibacterial agents.
Here, we investigate three different lantibiotics. Nisin, the first lantibiotic ever described, belongs to class A. The bactericidal effects of nisin involve two distinct mechanisms. At lower concentrations, nisin forms aggregates with lipid II, thereby preventing them from participating in cell wall biosynthesis (23, 35). After reaching a certain concentration threshold, complexes of 8 nisin and 4 lipid II molecules are formed that integrate into the bacterial membrane. They form pores large enough for ions and small amino acids to pass through (9, 23). Structurally similar gallidermin also belongs to class A and inhibits cell wall biosynthesis by lipid II binding. Potassium leakage indicating pore formation was observed in some but not all bacteria (8); however, B. subtilis was not tested. Christ et al. were able to correlate pore formation with membrane thickness and fatty acid branching, suggesting that this second mechanism of action of gallidermin is dependent on membrane composition (16). In contrast to nisin, which needs to dock to lipid II for membrane integration and pore formation, gallidermin shows very high association and low dissociation constants for binding phospholipid bilayers, indicating that it readily integrates into the membrane independently of lipid II binding. The insertion of gallidermin into the bilayer already influences membrane properties without a necessity for pore formation (16). Class B lantibiotic mersacidin is structurally and mechanistically different from nisin or gallidermin. More globular in structure, it is not able to integrate into the membrane or to form pores, so its antibacterial activity is based solely on inhibition of cell wall biosynthesis by binding to lipid II. In contrast to vancomycin, a glycopeptide which does not integrate into the membrane either, mersacidin does not seem to bind the amino acid tail of lipid II. Rather, it binds to the disaccharide headgroup of the lipid II molecule and additionally interacts with the pyrophosphate, suggesting that lipid II-bound lantibiotic molecules are localized near the outer layer of the cell membrane (6, 14).
These three lantibiotics were chosen to investigate the proteomic response of B. subtilis to cell wall biosynthesis inhibition by lipid II binding coupled with different levels of interference with membrane integrity. The physiological response of B. subtilis manifest in the proteome after antibiotic stress was previously shown to correlate with the antibacterial mechanism (2, 4, 47) and contributed to elucidating the antibacterial mechanisms of novel antibiotic compounds (12, 48). Proteomic profiles of B. subtilis treated with several cell wall biosynthesis inhibitors have been reported (4, 39, 46), but proteomic signatures indicative of specific aspects of cell wall biosynthesis inhibition have not yet been described. Utilizing lantibiotics, which range in mechanism from just binding lipid II to binding lipid II and forming large pores, and by further drawing on an extensive library of proteomic response profiles previously described (4), we identified responder proteins indicative of the different lantibiotic mechanisms. To directly correlate the proteome profiles with the influence of mersacidin, gallidermin, and nisin on B. subtilis, the impact of the lantibiotics on membrane integrity was studied in this model organism. Lantibiotic interference with cell wall biosynthesis was examined microscopically, looking at cell shape after methanol-acetic acid fixation. Potassium efflux measurements served to investigate pore formation. And finally, the influence of lantibiotics on the membrane potential was studied microscopically with the help of a green fluorescent protein (GFP)-MinD fusion protein. MinD is a cell division protein requiring an intact membrane potential for localization at the cell poles and the cell division plane (41). Based on the correlation of these physiological experiments with the proteomic response, marker proteins for different mechanisms of membrane interference could be proposed.
MATERIALS AND METHODS
Antibiotics.
All antibiotic stock solutions were prepared at 10 mg/ml. Nisin was dissolved in 0.01 M HCl, gallidermin in double-distilled water (both were purified according to Bonelli et al. [8]), and mersacidin (Sanofi-Aventis, Paris, France) in methanol. Valinomycin and gramicidin A were dissolved in dimethyl sulfoxide (DMSO), and bacitracin was dissolved in double-distilled water (all from Sigma-Aldrich, St. Louis, MO). Vancomycin (Applichem, Darmstadt, Germany) was dissolved in DMSO. Gramicidin S was synthetized and purified according to Wadhwani et al. (45) and dissolved in DMSO.
Bacterial strains and growth conditions.
Bacillus subtilis 168 (trpC2) (1) was grown at 37°C under steady agitation in Belitzky minimal medium (BMM) (42). MICs were determined in a modified MIC test described previously to match the growth conditions of the proteome experiment, specifically using chemically defined medium and supplying sufficient oxygen (48). Briefly, in a test tube, 2 ml of defined medium was inoculated with 5 × 105 bacteria per ml and incubated with different lantibiotic concentrations at 37°C under steady agitation for 18 h. The MIC was defined as the lowest concentration inhibiting visible growth. In growth experiments, bacterial cultures were treated with different antibiotic concentrations in mid-exponential phase after reaching an optical density at 500 nm (OD500) of 0.35. For physiological stress experiments, including proteomic studies, antibiotic concentrations were chosen that reduced growth rates to approximately 50 to 70% compared to that of the untreated control culture.
Light microscopy.
B. subtilis 168 cultures were grown in BMM to an OD500 of 0.35 and subsequently treated with 0.75 μg/ml nisin, 6 μg/ml gallidermin, 30 μg/ml mersacidin, 10 μg/ml valinomycin, 0.025 μg/ml gramicidin A, 1.5 μg/ml vancomycin, 12 μg/ml bacitracin, or 1 μg/ml gramicidin S. After 15 min of antibiotic exposure, 200 μl of bacterial culture were immediately fixed in 1 ml of a 1:3 mixture of acetic acid and methanol. Five microliters of fixed cells were immobilized in 5 μl BMM containing 0.5% low-melting agarose at 40°C. Cells were observed with an Olympus BX51 microscope using a U-UCD8 condenser and a UPlanSApo 100XO objective. Pictures were taken using a CC12 digital color camera and cell imaging software (all components by Olympus, Hamburg, Germany).
GFP-MinD localization.
B. subtilis 1981 GFP-MinD (41) was grown overnight in BMM. Cells were then inoculated to an OD500 of 0.1 in modified BMM containing xylose instead of glucose to induce expression of the GFP-MinD fusion protein. After reaching an OD500 of 0.35, cells were exposed to antibiotics at concentrations described above. After 15 min of antibiotic stress, 5 μl of the bacterial culture was withdrawn and the nonfixed, nonimmobilized samples were imaged immediately in fluorescent mode using the described equipment with a U-LH100HGAPO burner and a U-RFL-T power supply (Olympus, Hamburg, Germany).
Potassium release.
Potassium efflux experiments were performed using a microprocessor pH meter (pH 213; Hanna Instruments, Kehl, Germany) with an MI-442 potassium electrode and MI-409F reference electrode. The electrodes were preconditioned by immersing both the potassium selective and the reference electrodes in choline buffer (300 mM choline chloride, 30 mM MES, 20 mM Tris, pH 6.5) for at least 1 h before starting calibration or measurements. Calibration was carried out prior to each determination by immersing the electrodes in fresh standard solutions containing 0.01, 0.1, or 1 mM KCl in choline buffer. B. subtilis 168 was grown in BMM and harvested at an OD500 of 1.0 to 1.5 (3,300 × g, 4°C, 3 min), washed with 50 ml cold choline buffer, and resuspended in the same buffer to an OD500 of 30. The concentrated cell suspension was kept on ice and used within 30 min. For each measurement, the cells were diluted in choline buffer (25°C) to an OD500 of about 3. Antibiotics were applied at the concentrations described above. Calculations of potassium efflux were performed according to the equations established by Orlov et al. (31). Antibiotic-induced leakage was monitored for 3 min with readings taken every 10 s and expressed relative to the total potassium release induced by nisin.
Proteome analysis.
Protein extracts for 2D-PAGE were prepared as described previously (48). In short, 5 ml of a bacterial culture in early exponential growth phase were exposed to 6 μg/ml gallidermin, 30 μg/ml mersacidin, 10 μg/ml valinomycin, or 1 μg/ml gramicidin S or left untreated as the control. When B. subtilis cultures were treated during exponential growth, nisin exhibited a very narrow window of concentrations that solicit a stress response without causing massive cell lysis. Addressing this problem, concentrations from 0.25 to 1.25 μg/ml of nisin were used in parallel for each of the replicate labeling experiments. For the proteomic analysis, only those cultures were processed that showed growth inhibition but no extensive cell lysis. Cultures treated with 0.75 μg/ml (replicates one and two) and a culture treated with 0.5 μg/ml nisin (replicate three) were chosen for proteome analysis based on the impact of nisin on the growth rate. After 10 min of antibiotic stress, proteins were pulse labeled with [35S]methionine for 5 min. Incorporation of radioactive methionine was stopped by adding 1 mg/ml chloramphenicol and an excess of cold methionine. For nonradioactive preparative gels, cells were treated with antibiotics for 30 min to allow sufficient protein accumulation for mass spectrometry. Cells were harvested by centrifugation, washed three times with Tris buffer, and disrupted by ultrasonication.
Two-dimensional (2D) gels were prepared as described previously (48). In short, 55 μg of protein for analytical and 300 μg for preparative gels were loaded onto 24-cm immobilized pH gradient (IPG) strips, pH 4 to 7 (GE Healthcare), by passive rehydration for 18 h followed by isoelectric focusing. After reduction and alkylation, proteins were separated in a second dimension by SDS-PAGE using 12.5% acrylamide gels. Images were analyzed as described by Bandow et al. (3) using Decodon Delta 2D 4.1 image analysis software (Decodon, Greifswald, Germany). Proteins found to be induced more than 2-fold in three independently performed biological experiments were defined as marker proteins.
Protein spots were excised from preparative 2D gels and transferred into 96-well microtiter plates. Tryptic in-gel digest and subsequent spotting of extracted peptides with matrix on a matrix-assisted laser desorption ionization (MALDI) target was carried out automatically with the Ettan spot handling workstation (Amersham Biosciences, Uppsala, Sweden) as described by Eymann et al. (20). MALDI-time of flight (TOF) as well as MALDI-TOF/TOF (consecutive TOF analysis) measurements were carried out on a 4800 MALDI-TOF/TOF analyzer (Applied Biosystems, Foster City, CA). For database searches, the Mascot search engine 2.1.04 (Matrix Science Ltd., London, United Kingdom) with a specific B. subtilis sequence database was used as described previously (48).
RESULTS
In order to obtain specific marker proteins that allow delineation of different mechanisms of action affecting the cell envelope, we investigated a set of lantibiotics, which inhibit bacterial cell wall biosynthesis while exhibiting different degrees of interference with the bacterial membrane. Mersacidin binds to lipid II without integrating into the cytoplasmic membrane, gallidermin binds to lipid II and can disrupt the structure of the membrane by insertion, and nisin binds to lipid II, integrates into the membrane, and leads to pore formation.
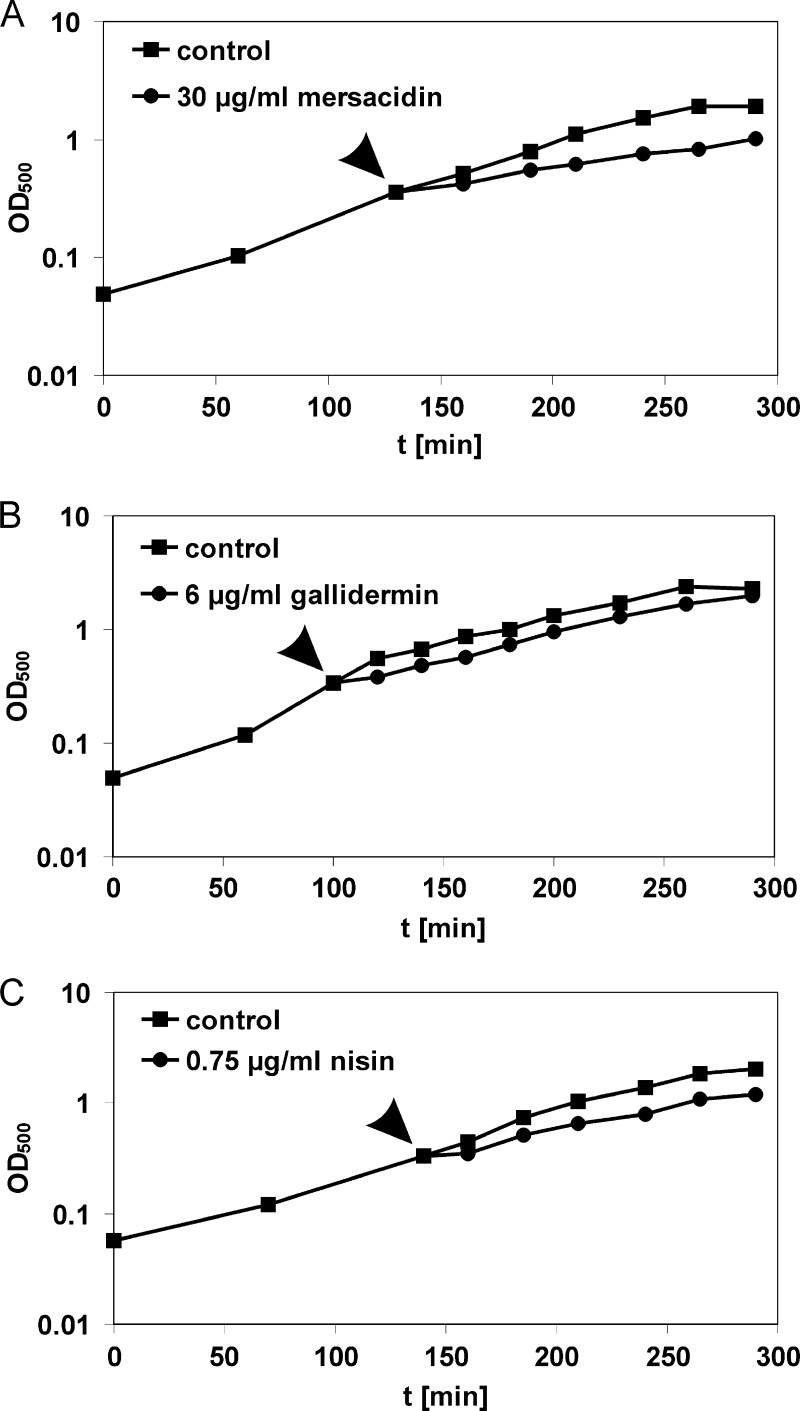
To find appropriate conditions for proteome analyses and the characterization of the effects of lantibiotics on the cell envelope, MICs of the lantibiotics were determined under conditions resembling those of the proteome experiment, using the chemically defined medium and incubation under steady agitation. The MICs were 30 μg/ml for mersacidin, 6 μg/ml for gallidermin, and 5 μg/ml for nisin. Based on these MICs, growth experiments were performed to identify antibiotic concentrations that reduce growth rates without causing massive cell lysis or completely stopping bacterial metabolism. This is essential, as proteomic experiments require a stress level high enough to induce a proteomic response but at the same time allowing protein biosynthesis to continue. While for mersacidin and gallidermin the MICs were optimal stressor concentrations, nisin caused massive cell lysis when applied at concentrations near the MIC during exponential growth. However, cells surviving the acute nisin shock resumed growth after some time (data not shown), explaining why the MIC is significantly higher than the acutely tolerated concentration. We determined the optimal stressor concentration for nisin to be 0.75 μg/ml (Fig. 1).
Fig 1.
Effects of lantibiotic treatment on B. subtilis growth. B. subtilis was grown in defined medium to an OD500 of 0.35. Cultures were split and either treated with lantibiotics or left untreated. (A) Mersacidin, 30 μg/ml; (B) gallidermin, 6 μg/ml; and (C) nisin, 0.75 μg/ml. The time points of antibiotic addition are marked by arrowheads.
Characterization of the impact of lantibiotics on the B. subtilis cell wall.
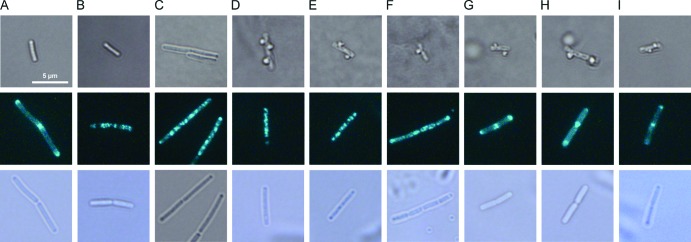
All tested lantibiotics were expected to influence cell wall integrity, as they prevent incorporation of cell wall precursors by binding lipid II. Their impact on cell wall integrity was investigated by microscopic examination of the cell shape after acetic acid-methanol fixation (Fig. 2, top) using a method described by Schneider et al., who investigated cell wall biosynthesis inhibitors vancomycin and plectasin (36). Upon inhibition of cell wall biosynthesis, small holes occur in the peptidoglycane layer, where new cell wall material is no longer incorporated. When the perforated cells are then fixed in a 1:3 mixture of acetic acid and methanol, the cytoplasmic membrane extrudes through holes in the cell wall, appearing as bubbles on the cell surface. To ensure that this phenomenon occurs specifically upon impairment of cell wall integrity and does not occur after treatment with agents impairing membrane integrity or simply as a result of sample handling, we used untreated, valinomycin-treated, and gramicidin A-treated B. subtilis cultures as negative controls. Valinomycin is a cyclic peptide carrier ionophore that selectively transports potassium ions across the bacterial membrane, while peptide channel ionophore gramicidin A transports monovalent cations (18). Neither the untreated controls (Fig. 2A) nor the valinomycin or gramicidin A-treated cultures (Fig. 2B or C, respectively) displayed any membrane extrusions. As expected, all lantibiotics led to membrane extrusions (Fig. 2E, F, G), demonstrating their influence on cell wall biosynthesis in B. subtilis. Further, we tested the cyclic peptide antibiotic gramicidin S (Fig. 2D), which served as a positive control for depolarization experiments. Gramicidin S had been suggested to affect membrane-bound processes indirectly by causing delocalization of membrane-associated enzymes (25, 26, 44, 55). Indeed, after gramicidin S treatment, cells also showed membrane extrusions, corroborating an effect of gramicidin S on cell wall biosynthesis.
Fig 2.
Influence of mersacidin, gallidermin, and nisin on B. subtilis cell wall integrity and membrane potential. (Top) Light microscopy images of B. subtilis fixed with acetic acid-methanol and immobilized with low-melting agarose show the influence of lantibiotics and comparator compounds on cell wall integrity. (Center) Fluorescence microscopy images of nonfixed cells show GFP-MinD localization after treatment with envelope-targeting agents. Cultures of B. subtilis 168 or B. subtilis 1981 GFP-MinD were grown until early exponential phase, divided into aliquots, and stressed with antibiotics for 15 min: (A) untreated control; (B) valinomycin (10 μg/ml); (C) gramicidin A (0.025 μg/ml); (D) gramicidin S (1 μg/ml); (E) nisin (0.75 μg/ml); (F) gallidermin (6 μg/ml); (G) mersacidin (30 μg/ml); (H) bacitracin (12 μg/ml); (I) vancomycin (1.5 μg/ml). (Bottom) Corresponding light microscopy pictures of the B. subtilis GFP-MinD cells shown above. Note that B. subtilis GFP-MinD mutants typically grow longer than wild-type B. subtilis due to MinD overexpression.
Lantibiotics' influence on B. subtilis membrane potential.
In addition to cell wall biosynthesis inhibition, gallidermin and nisin are expected to influence membrane integrity due to integration into the membrane and pore formation, respectively. Therefore, we investigated the influence of lantibiotics on the membrane potential monitoring microscopically the localization of a GFP-MinD fusion protein (Fig. 2, central and bottom panels). MinD is part of the cell division regulation machinery of B. subtilis. As long as the membrane potential remains intact, MinD is attached to the membrane and localizes at the cell poles and in the cell division plain. It was shown that MinD localization is altered by membrane-depolarizing agents like valinomycin or proton ionophore carbonyl cyanide m-chlorophenylhydrazone (CCCP), due to the interruption of electrostatic interactions between MinD and the membrane surface (41). This phenomenon was employed here to investigate depolarizing properties of antibiotic agents. As described by Strahl and Hamoen (41), valinomycin was used as a positive control, as it causes rapid depolarization (Fig. 2B). Disruption of the membrane potential results in delocalization of GFP-MinD, which appears in fluorescing spots irregularly distributed throughout the cells. Nisin and gallidermin (Fig. 2E and F, respectively) exhibited the same GFP-MinD distribution pattern as valinomycin, indicating the collapse of the membrane potential (Fig. 2). In contrast, mersacidin did not disturb the polar and septal localization of GFP-MinD (Fig. 2G). These findings suggest that nisin and gallidermin but not mersacidin cause membrane depolarization in B. subtilis. Delocalization of GFP-MinD was also caused by the membrane-integrating antimicrobial peptide gramicidin S (Fig. 2D), which is neither an ionophore nor a pore former (25, 44).
Lantibiotics' ability to form pores in the B. subtilis membrane.
In order to investigate the ability of mersacidin, gallidermin, and nisin to form pores in B. subtilis, we measured the lantibiotic-induced potassium release using a potassium-sensitive electrode (Fig. 3). Consistent with the literature, pore-forming lantibiotic nisin led to strong, immediate potassium release, whereas the potassium ionophore valinomycin (positive control) led to release of the same amount of potassium, following a slower kinetic characteristic for carrier ionophores. In contrast, as described previously, mersacidin did not cause an increase in extracellular potassium levels (6, 9, 14, 23). Gallidermin, which had not yet been tested regarding its ability to form pores in B. subtilis, caused only a minor potassium release of up to 20% compared to nisin, suggesting that it does not effectively form pores in B. subtilis under the tested conditions.
Fig 3.
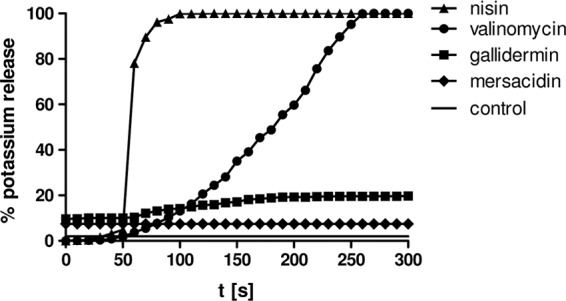
Potassium release from B. subtilis induced by mersacidin, gallidermin, nisin, and valinomycin. Potassium efflux from living cells was monitored with a potassium-sensitive electrode. Ion leakage is expressed relative to the total amount of potassium released after addition of the pore-forming lantibiotic nisin (100% value). Antibiotics were added at 50 s and applied at the same concentrations as for proteome analysis: 0.75 μg/ml nisin, 6 μg/ml gallidermin, 30 μg/ml mersacidin, and 10 μg/ml valinomycin. The symbol-free solid line represents the baseline without antibiotics.
Proteomic response to lantibiotic treatment.
Comparative proteome analyses were carried out to investigate whether or not the proteomic response would reveal marker proteins for the lantibiotic mechanism of action of cell wall biosynthesis inhibition by lipid II binding and how far it could provide insights into additional membrane-impairing properties of lantibiotics. To this end, B. subtilis was grown to early exponential phase and stressed with the appropriate concentrations of lantibiotics. [35S]-l-methionine pulse labeling was used to label selectively those proteins newly synthesized under antibiotic stress. This procedure allows monitoring of the acute bacterial stress response with exquisite sensitivity. The soluble intracellular protein fraction was then separated by 2D-PAGE. Representative proteome expression profiles of mersacidin, gallidermin, and nisin are shown in Fig. 4. Proteins induced at least 2-fold are referred to as marker proteins. They mirror the physiological stress conditions the cells are facing and proved valuable for studying antibiotic mechanisms of action.
Fig 4.
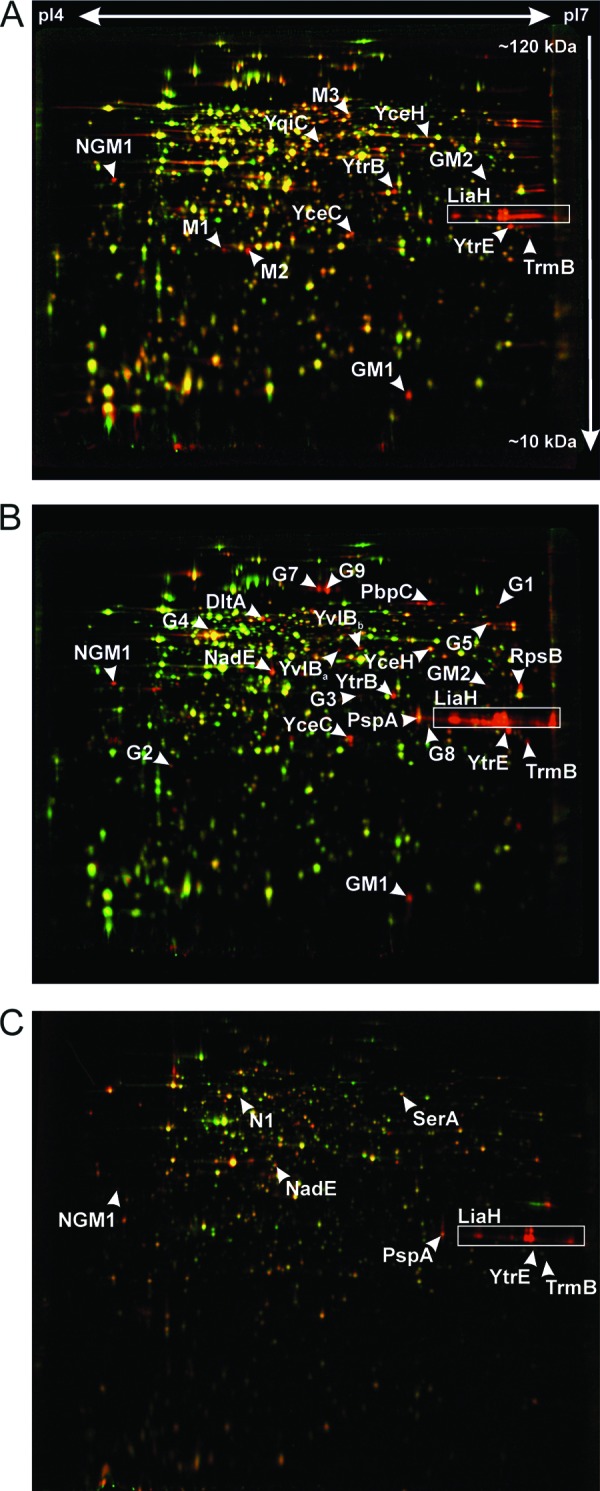
Differential proteome analysis of B. subtilis 168 in response to mersacidin (A), gallidermin (B), and nisin (C). 2D gel-based protein synthesis profiles of the controls false colored in green were overlaid with those of the antibiotic-treated samples false colored in red. In the overlays, downregulated proteins appear green, upregulated proteins appear red, and proteins expressed at equal rates appear yellow. Unidentified proteins are labeled as follows: M, induced by mersacidin; G, induced by gallidermin; N, induced by nisin; NGM, induced by nisin, gallidermin, and mersacidin.
Twenty-five (25) marker protein spots were induced by treatment with gallidermin, 13 by mersacidin, and 8 by nisin (Table 1). Using MALDI-TOF/TOF, we were able to identify 13, 7, and 6 protein spots, respectively. The remaining protein spots were clearly visible on autoradiographs reflecting protein synthesis after antibiotic treatment but did not accumulate to any appreciable extent. Although these unidentified protein spots do not allow rational insights into the antibiotic mechanism, they still serve as antibiotic-specific markers, as they are reproducibly induced after treatment.
Table 1.
Responder proteins induced by nisin, gallidermin, and mersacidin
| Responder protein | Induction factora |
Protein name | Function | Regulator | |||
|---|---|---|---|---|---|---|---|
| Nisin (25% inhibition) | Nisin (50% inhibition) | Gallidermin | Mersacidin | ||||
| LiaH | 6.6 | 19 | 86 | 92 | Modulator of liaIHGFSR operon expression | ||
| TrmB | 3.4 | 2.8 | 9.1 | 8.6 | tRNA [guanin-N(7)]methyltransferase | ||
| YtrE | 1.6 | 2.1 | 21 | 14 | Similar to ABC transporter | ||
| NGM1 | 1.6 | 2.5 | 13 | 8.5 | Not identified | ||
| PspA | 1.8 | 5.5 | 6.4 | Phage shock protein A | Protection against cell envelope stress | SigW | |
| NadE | 1.6 | 2.3 | 6.7 | NAD synthetase | Energy limitation-mediated stress response | SigB | |
| YceC | 13 | 4.2 | Similar to tellurium resistance protein | Stress response | SigW/SigB | ||
| YceH | 5.8 | 2.4 | Similar to toxic anion resistance protein | Stress response | SigW/SigB | ||
| YtrB | 14 | 9.8 | Similar to ABC transporter | Putative ABC transporter ATPase subunit | YtrA | ||
| GM1 | 7.9 | 8.3 | Not identified | ||||
| GM2 | 8.6 | 6.7 | Not identified | ||||
| SerA | 1.2 | 2.6 | d-3-Phosphoglycerate dehydrogenase | Serine biosynthesis/oxidative stress/NADH regeneration | |||
| N1 | 1 | 13 | Not identified | ||||
| DltA | 4.2 | d-Alanyl:d-alanine carrier protein ligase | Modification of lipoteichoic acids | SigX | |||
| YvlBa | 3.6 | Unknown | Stress response | SigW | |||
| YvlBb | 7 | Unknown | Stress response | ||||
| PbpC | 27 | Penicillin binding protein 3 | Cell wall biosynthesis | ||||
| RpsB | 4.1 | Ribosomal protein S2 | Translation | ||||
| G1 | 7.2 | Not identified | |||||
| G2 | 3 | Not identified | |||||
| G3 | 3.9 | Not identified | |||||
| G4 | 2.4 | Not identified | |||||
| G5 | 3.3 | Not identified | |||||
| G6 | 47 | Not identified | |||||
| G7 | 5.8 | Not identified | |||||
| G8 | 2.8 | Not identified | |||||
| G9 | 11 | Not identified | |||||
| YqiG | 5 | NADH-dependent flavin oxidoreductase | Electron transport | ||||
| M1 | 7.3 | Not identified | |||||
| M2 | 7.2 | Not identified | |||||
| M3 | 3.2 | Not identified | |||||
Bold numbers indicate reliable marker proteins, more than 2-fold induced in three independent biological replicates.
DISCUSSION
In this study, we set out to characterize the stress response of B. subtilis to three different lantibiotics with gradually different mechanisms of action regarding interactions with the cytoplasmic membrane but each inhibiting cell wall biosynthesis by binding to precursor lipid II. It was previously described that mersacidin acts solely on cell wall biosynthesis, whereas gallidermin is able to integrate into the membrane, although pores are formed only in some bacterial species and not others (8, 16). Nisin was shown to integrate into the membrane upon lipid II binding, forming pores of 2 to 2.5 nm in diameter (50), if concentrations exceed a certain threshold (8, 38). In preparation of the proteomic study, we tested nisin, gallidermin, and mersacidin for their effects on the B. subtilis cell envelope under growth conditions matching those of the proteomic profiling analysis. As expected, all lantibiotics significantly affected cell wall integrity as shown microscopically (Fig. 2). Protein localization studies using GFP-labeled MinD showed protein delocalization after treatment with nisin and gallidermin, indicating disruption of the membrane potential. Mersacidin tested negative for MinD-GFP delocalization. Significant potassium release demonstrating pore formation was observed only for nisin but not gallidermin or mersacidin (Fig. 2 and 3). Taken together, these results suggest that in B. subtilis, both nisin and gallidermin effectively depolarize the membrane, with only nisin forming pores. Our results indicate that the GFP-MinD delocalization observed for gallidermin may be due to membrane depolarization caused by the lantibiotic integrating into the membrane rather than to pore formation. As Christ et al. demonstrated, gallidermin leads to effective pore formation in lipid bilayers containing mainly acyl chains up to 15 C atoms in length. Limited pore formation is observed in membranes composed predominantly of 17 C lipid tails (16). Consistently, Bonelli et al. reported pore formation for Staphylococcus simulans and Micrococcus flavus (both 15 C) but not for Lactococcus lactis (17 C) (8). Additionally to the membrane thickness, a large amount of branched-chain fatty acids in the membrane seems to negatively affect pore formation by gallidermin (16). The B. subtilis membrane consists mostly of 15 C lipid chains but at the same time possesses about 80% branched-chain fatty acids (17). Therefore, it is likely that gallidermin is not able to form pores in this organism due to membrane density rather than thickness. However, integration of gallidermin into the membrane might affect the physicochemical properties of the membrane, resulting in depolarization and consequently MinD-GFP delocalization. Similar effects have already been described for cationic antimicrobial peptide gramicidin S, a depolarizing agent. At sublytical concentrations, gramicidin S seems to disturb the membrane structure leading to dysfunction of membrane protein complexes. It uncouples electron transport by inhibiting cytochrome bd quinol oxidase and consequently disrupts the electrochemical gradient (25, 30, 44, 55). Gramicidin S also affects cell wall integrity (Fig. 2), potentially also due to disturbing the membrane-bound cell wall biosynthesis machinery by integrating into the membrane. There are also reports that gramicidin S increases membrane permeability for potassium ions (26), which might be the effect rather than the cause of membrane depolarization (30).
Based on their common mechanism of inhibiting cell wall biosynthesis by binding lipid II, the three lantibiotics were ideally suited for comparative proteomic studies aimed at describing proteomic signatures for cell wall biosynthesis inhibition. At the same time, this collection of lantibiotics in combination with the existing library of proteomic response profiles could be used to correlate the differences in the proteome according to the differences in membrane interaction. Many of the identified marker proteins are known to be upregulated in response to cell envelope stress (Table 1). YvlB, for instance, belongs to the SigW regulon induced by cell envelope stress. Taking into account the lantibiotic proteome profiles as well as previously published reference patterns of an extensive antibiotic proteome response library (4), we delineated three groups of marker proteins (Table 2, Fig. 5): those indicative of cell wall biosynthesis inhibition, those indicating membrane stress, and those induced in response to general envelope stress.
Table 2.
Marker proteins for antibiotics targeting membrane integrity and/or cell wall biosynthesis correlated with influence on cell envelope integrityc
| Compound | Mode of action | Cell wall integrity | MinD delocalization | Potassium leakage | NadE | PspA | YceC | YceH | TrmB | LiaH | YtrE | YtrB | NGM1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Valinomycinb | Carrier ionophore | − | + | + | x | x | x | x | |||||
| Gramicidin Aa | Channel ionophore | − | + | + | x | x | x | ||||||
| Gramicidin Sb | Membrane integration | + | + | − | x | x | x | x | x | x | |||
| Nisin | Pore formation and cell wall biosynthesis | + | + | + | x | x | x | x | x | x | |||
| Gallidermin | Membrane integration and cell wall biosynthesis | + | + | − | x | x | x | x | x | x | x | x | x |
| Mersacidin | Cell wall biosynthesis | + | − | − | x | x | x | x | x | x | x | ||
| Bacitracina | Cell wall biosynthesis | + | − | − | x | x | x | x | x | ||||
| Vancomycina | Cell wall biosynthesis | + | − | − | x | x | x |
Previously published (4).
2D gel images not shown.
+, envelope functionality as measured in cell wall integrity assay, MinD delocalization assay, or potassium release assay was impaired; −, control-like envelope functionality; x, marker protein upregulated at least twofold in response to antibiotic treatment.
Fig 5.
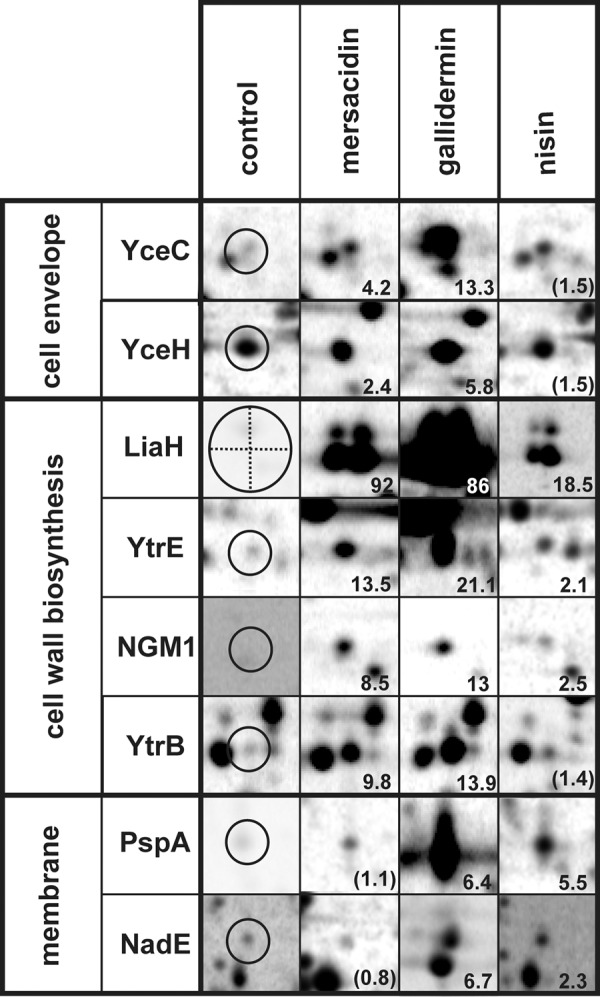
Details of autoradiographs of 2D gels depicting the specific marker proteins for cell wall biosynthesis, membrane stress, and general cell envelope stress. Induction factors are displayed in the lower right corner as the average over three independent biological replicates.
Markers for interference with cell wall biosynthesis by lipid II binding.
Four proteins, namely, LiaH, YtrE, and TrmB, as well as unidentified protein NGM1, were induced by all tested lantibiotics. LiaH belongs to the LiaRS regulon. It forms multimeric ringlike structures consisting of 36 identical subunits that protect cell envelope integrity probably by binding to the membrane upon cell wall stress (51). LiaRS has been shown before to be induced by various cell wall biosynthesis inhibitors, such as bacitracin, nisin, ramoplanin, and to a lesser extent vancomycin, measuring liaI-lacZ reporter gene activation (29). Further, the LiaRS system was induced after severe secretion stress, suggesting that protein accumulation in the cell envelope may also trigger LiaH induction (24). Besides the three lantibiotics investigated here, two inhibitors targeting membrane-bound steps of cell wall precursor synthesis were previously analyzed using proteomic profiling. Vancomycin prevents transglycosylation by binding to the amino acid side chain of lipid II, while bacitracin prevents recycling of the bactoprenol carrier that transports cell wall precursor molecules through the membrane (15). LiaH was upregulated in response to all lantibiotics, as well as bacitracin and gramicidin S, but not vancomycin. A possible explanation might be that the lipid II binding site of vancomycin is the amino acid chain of lipid II, which is remote from the membrane (15), while for all other compounds, the lipid II binding sites are in close proximity to the membrane.
YtrE and YtrB, which are upregulated by all investigated cell wall biosynthesis inhibitors with the exception of nisin, are encoded in the ytrABCDEF operon and form two cytoplasmic ATPase subunits of a putative ABC transporter (52). Earlier studies by Yoshida et al. demonstrated impaired acetoin import in ytr-deficient knockout mutants (53). However, it should be noted that the YtrF substrate recognition subunit shares homology with both antimicrobial peptide efflux systems and the FtsX permease involved in cell division. Delayed spore formation was observed in the absence of FtsX (21) as well as in ytr-deficient mutants (53), possibly indicating similar functions of both proteins. Although it is yet unclear how YtrE/YtrB induction might be connected to the antibiotic mechanism, they serve as specific markers for cell wall biosynthesis inhibition. YtrE is the only marker protein shared by all lantibiotics, vancomycin, and bacitracin. It is not induced by other antibiotics tested and is, therefore, the most reliable marker for inhibition of a membrane-bound step of cell wall biosynthesis.
TrmB is a tRNA-modifying [guanine-N(7)-]-methyltransferase involved in tRNA maturation (54). What role it might play in the response to lantibiotic treatment remains unclear. Like LiaH, unidentified NGM1 is induced by all lantibiotics and bacitracin but not vancomycin, demonstrating a different cell response to vancomycin.
Likewise, the response pattern induced by the lipopeptide friulimicin B differs from that of cell wall biosynthesis-targeting antibiotics that bind proximal to the membrane. In contrast to the lipid II-binding lantibiotics tested here, friulimicin B binds to bactoprenol monophosphate, thus inhibiting carrier recycling. As a result, cell wall biosynthesis and biosynthesis of wall teichoic acids and carbohydrate-containing capsular material are impaired, and cell envelope modification reactions such as glycosylations are inhibited. All these pathways depend on bactoprenol-mediated building block shuffling to the outside (37). As expected for an antibiotic interfering with cell wall biosynthesis, friulimicin B leads to membrane extrusions in the methanol-acetic acid assay. It does not depolarize the bacterial membrane in the GFP-MinD assay (data not shown) and does not cause potassium leakage (37). Friulimicin B shares a high mode of action similarity with bacitracin, which binds to bactroprenol pyrophosphate. However, while bacitracin induces all cell wall biosynthesis marker proteins, friulimicin B induces only the unidentified marker proteins NGM1 and GM1 but not LiaH or YtrE (data not shown). In a previous study, Wecke et al. further showed induction of YceC and YceH (46), which in the present study were upregulated in response to several of the cell envelope-targeting antibiotics but were not correlated strictly with either interference with the membrane or with cell wall biosynthesis.
A clear distinction based on the proteome response can be made between inhibition of membrane-bound steps of cell wall biosynthesis and inhibition of either intracellular or extracellular steps. This could be shown using d-cycloserine or methicillin, respectively. d-Cycloserine, which inhibits alanine racemase, and d-alanine:d-alanine ligase, two intracellular enzymes of the pathway (28, 43), elicit a completely different proteomic response with no marker proteins overlapping with any of the other investigated cell wall biosynthesis inhibitors (4). Methicillin inhibits penicillin-binding protein 2 (PBP-2), a membrane-anchored transpeptidase and transglycosylase (33); however, the methicillin binding site is not located near the membrane anchor, and a direct interaction of methicillin with the cytoplasmic membrane has not been reported. For this compound in B. subtilis, no acute stress response at all was observed on the proteome level (4), suggesting that B. subtilis cells do either not sense the consequences of methicillin action and/or are unable to react to them. Based on these results, differences in the proteomic response to cell wall biosynthesis inhibition relating to the localization of the steps inhibited and potentially even the proximity of the antibiotic binding sites to the bacterial membrane can be observed.
Marker proteins for interference with membrane integrity.
PspA and NadE are both induced by nisin and gallidermin as well as gramicidin A, gramicidin S, and valinomycin. Among all agents tested so far, only compounds acting on the bacterial membrane led to induction of these two proteins, supporting that they are specific marker proteins for membrane stress.
Interestingly, upregulation of PspA and NadE was observed only at higher nisin concentrations, leading to approximately 50% growth inhibition, while the cell wall biosynthesis-specific markers LiaH and TrmB were upregulated already under moderate stress conditions, causing approximately 25% growth inhibition (Table 1). This is in line with a two-staged mechanism of action of nisin, which is characterized by lipid II complexation at lower concentrations and pore formation above a threshold concentration (9, 23). PspA, like LiaH, forms homomultimeric ringlike structures (22). Although Standar et al. showed that those 36-mers are able to form large scaffolds (40), more recent studies demonstrated localization of PspA at midcell and at the cell poles by use of a GFP fusion (19). It was shown that PspA is able to bind to membrane phospholipids and prevent proton leakage. It is thought to counteract external membrane damage and to stabilize the membrane during membrane transport (27). Structural homology between PspA and LiaH suggests similar functions in stabilizing the cell envelope by binding to the membrane surface. Our results indicate that PspA is responsive to membrane damage, while LiaH is induced by lipid II-mediated cell wall stress. However, the different triggers inducing both stress proteins remain to be identified (51).
NAD synthase NadE, which catalyzes the final step in NAD synthesis, is regulated on the transcriptional level by the housekeeping sigma factor SigA and by the alternative sigma factor SigB coordinating the general stress response in B. subtilis in response to environmental stress and energy limitation. As a consequence of antibiotic-related impairment of membrane integrity evidenced by the disturbed membrane potential, energy limitation is a possible explanation for NadE upregulation.
Differentiation between pore formation by nisin and membrane integration by gallidermin could not be securely established based on specific marker proteins. This may be due to rapid killing of cells once nisin concentrations reach the necessary threshold for pore formation, hardly leaving a chance for a coordinated stress response. For this reason, pore formation might be difficult to distinguish from other forms of membrane stress by proteome analysis alone. However, monitoring concentration-dependent killing kinetics already allows differentiation between compounds that integrate into the membrane and those that form pores. Rapid bacteriolytic pore formers leave only a very narrow concentration window for proteomic profiling experiments and should induce the membrane stress-specific marker proteins PspA and NadE at the appropriate antibiotic concentrations.
Marker proteins for envelope stress.
Putative tellurium resistance protein YceC, induced by gallidermin and mersacidin, was also induced by several of the envelope-targeting compounds in our proteomic response library, namely, valinomycin, gramicidin S, bacitracin, and vancomycin, and can therefore serve as a general marker for envelope stress. Putative anion resistance protein YceH was also upregulated in response to gallidermin, mersacidin, valinomycin, gramicidin S, and gramicidin A. YceC and YceH are under the dual control of SigW and SigB, but to our knowledge their functions are yet unknown.
In conclusion, using lantibiotics and a number of other antibiotics with different targets related to the cellular envelope, we described marker proteins correlating with cell wall biosynthesis inhibition, membrane stress, and general cell envelope stress. The established marker proteins can now be used to characterize new antibiotic agents, particularly lantibiotics, with regard to their antibacterial mechanisms.
ACKNOWLEDGMENTS
Mersacidin was provided by Sanofi-Aventis (formerly Hoechst AG). We gratefully acknowledge Jia Yii Airina Lee from Pennsylvania State University as well as Christoph H. R. Senges (Ruhr University Bochum) for assistance with the bubble and MinD-GFP assays. We are also grateful for excellent support by the technical staff at Rubion, Ruhr University Bochum, and to Michaele Josten, University of Bonn, for purifying nisin.
This work was financially supported by “Innovative Antibiotics from NRW,” a grant from the state of North Rhine-Westphalia (NRW), Germany, and the European Union, European Regional Development Fund, “Investing in Your Future,” to J.E.B. and H.G.S.
Footnotes
Published ahead of print 27 August 2012
REFERENCES
- 1. Anagnostopoulos C, Spizizen J. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bandow JE. 2005. Proteomic approaches to antibiotic drug discovery. Curr. Protoc. Microbiol. 1:1F.2. [DOI] [PubMed] [Google Scholar]
- 3. Bandow JE, et al. 2008. Improved image analysis workflow for 2D gels enables large-scale 2D gel-based proteomics studies—COPD biomarker discovery study. Proteomics 8:3030–3041 [DOI] [PubMed] [Google Scholar]
- 4. Bandow JE, Brötz H, Leichert LI, Labischinski H, Hecker M. 2003. Proteomic approach to understanding antibiotic action. Antimicrob. Agents Chemother. 47:948–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bandow JE, Metzler-Nolte N. 2009. New ways of killing the beast: prospects for inorganic-organic hybrid nanomaterials as antibacterial agents. Chembiochem 10:2847–2850 [DOI] [PubMed] [Google Scholar]
- 6. Barrett MS, Wenzel RP, Jones RN. 1992. In vitro activity of mersacidin (M87-1551), an investigational peptide antibiotic tested against Gram-positive bloodstream isolates. Diagn. Microbiol. Infect. Dis. 15:641–644 [DOI] [PubMed] [Google Scholar]
- 7. Bierbaum G, Sahl HG. 2009. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 10:2–18 [DOI] [PubMed] [Google Scholar]
- 8. Bonelli RR, Schneider T, Sahl HG, Wiedemann I. 2006. Insights into in vivo activities of lantibiotics from gallidermin and epidermin mode-of-action studies. Antimicrob. Agents Chemother. 50:1449–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Breukink E, et al. 2003. Lipid II is an intrinsic component of the pore induced by nisin in bacterial membranes. J. Biol. Chem. 278:19898–19903 [DOI] [PubMed] [Google Scholar]
- 10. Brötz-Österhelt H, Sass P. 2010. Postgenomic strategies in antibacterial drug discovery. Future Microbiol. 5:1553–1579 [DOI] [PubMed] [Google Scholar]
- 11. Brötz-Österhelt H, Brunner NA. 2008. How many modes of action should an antibiotic have? Curr. Opin. Pharmacol. 8:564–573 [DOI] [PubMed] [Google Scholar]
- 12. Brötz-Oesterhelt H, et al. 2005. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 11:1082–1087 [DOI] [PubMed] [Google Scholar]
- 13. Brötz H, Sahl HG. 2000. New insights into the mechanism of action of lantibiotics—diverse biological effects by binding to the same molecular target. J. Antimicrob. Chemother. 46:1–6 [DOI] [PubMed] [Google Scholar]
- 14. Brötz H, Bierbaum G, Leopold K, Reynolds PE, Sahl HG. 1998. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob. Agents Chemother. 42:154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bugg TDH, Braddick D, Dowson CG, Roper DI. 2011. Bacterial cell wall assembly: still an attractive antibacterial target. Trends Biotechnol. 29:167–173 [DOI] [PubMed] [Google Scholar]
- 16. Christ K, et al. 2008. Membrane lipids determine the antibiotic activity of the lantibiotic gallidermin. J. Membr. Biol. 226:9–16 [DOI] [PubMed] [Google Scholar]
- 17. Clejan S, Krulwich TA, Mondrus KR, Seto-Young D. 1986. Membrane lipid composition of obligately and fakultatively alkalophilic strains of Bacillus spp. J. Bacteriol. 168:334–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Duax WL, et al. 1996. Molecular structure and mechanisms of action of cyclic and linear ion transport antibiotics. Biopolymers 40:141–155 [DOI] [PubMed] [Google Scholar]
- 19. Engl C, et al. 2009. In vivo localizations of membrane stress controllers PspA and PspG in Escherichia coli. Mol. Microbiol. 73:382–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eymann C, et al. 2004. A comprehensive proteome map of growing Bacillus subtilis cells. Proteomics 4:2849–2876 [DOI] [PubMed] [Google Scholar]
- 21. Garti-Levi S, Hazan R, Kain J, Fujita M, Ben-Yehuda S. 2008. The FtsEX ABC transporter directs cellular differentiation in Bacillus subtilis. Mol. Microbiol. 69:1018–1028 [DOI] [PubMed] [Google Scholar]
- 22. Hankamer BD, Elderkin SL, Buck M, Nield J. 2004. Organization of the AAA+ adaptor protein PspA is an oligomeric ring. J. Biol. Chem. 279:8862–8866 [DOI] [PubMed] [Google Scholar]
- 23. Hasper HE, de Kruijff B, Breukink E. 2004. Assembly and stability of nisin-lipid II pores. Biochemistry 43:11567–11575 [DOI] [PubMed] [Google Scholar]
- 24. Hyyryläinen HL, Sarvay M, Kontinen VP. 2005. Transcriptome analysis of the secretion stress response of Bacillus subtilis. Appl. Microbiol. Biotechnol. 67:389–396 [DOI] [PubMed] [Google Scholar]
- 25. Kaprel'iants AS, et al. 1977. Membranes of bacteria and mechanism of action of the antibiotic gramicidin S. Biokhimiia. 42:329–337 [PubMed] [Google Scholar]
- 26. Katsu T, et al. 1989. Mechanism of membrane damage induced by the amphiphilic peptides gramicidin S and melittin. Biochim. Biophys. Acta 983:135–141 [DOI] [PubMed] [Google Scholar]
- 27. Kobayashi R, Suzuki T, Yoshida M. 2007. Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol. Microbiol. 66:100–109 [DOI] [PubMed] [Google Scholar]
- 28. Lambert MP, Neuhaus FC. 1972. Mechanism of d-cycloserine action: alanine racemase from Escherichia coli W. J. Bacteriol. 109:1156–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mascher T, Zimmer SL, Smith TA, Helmann JD. 2004. Antibiotic-inducible promotor regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48:2888–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mogi T, Ui H, Shiomi K, Omura S, Kita K. 2008. Gramicidin S identified as a potent inhibitor for cytochrome bd-type quinol oxidase. FEBS Lett. 582:2299–2302 [DOI] [PubMed] [Google Scholar]
- 31. Orlov DS, Nguyen T, Lehrer RI. 2002. Potassium release, a useful tool for studying antimicrobial peptides. J. Microbiol. Methods 49:325–328 [DOI] [PubMed] [Google Scholar]
- 32. Peschel A, Sahl HG. 2006. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4:529–536 [DOI] [PubMed] [Google Scholar]
- 33. Pinho MG, de Lencastre H, Tomasz A. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. U. S. A. 98:10886–10891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rabilloud T, Strub JM, Luche S, van Dorsselaer A, Lunardi J. 2001. A comparison between SyproRuby and ruthenium II Tris (bathophenanthroline disulfonate) as fluorescent stains for protein detection in gels. Proteomics 1:699–704 [DOI] [PubMed] [Google Scholar]
- 35. Reisinger P, Seidel H, Tschesche H, Hammes WP. 1980. The effect of nisin on murein synthesis. Arch. Microbiol. 127:187–193 [DOI] [PubMed] [Google Scholar]
- 36. Schneider T, et al. 2010. Plectasin, a fungal defensin, targets the bacterial cell wall precursor lipid II. Science 328:1168–1172 [DOI] [PubMed] [Google Scholar]
- 37. Schneider T, et al. 2009. The lipopeptide antibiotic friulimicin B inhibits cell wall biosynthesis through complex formation with bactoprenol phosphate. Antimicrob. Agents Chemother. 53:1610–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schneider T, Sahl HG. 2010. An oldie but a goodie—cell wall biosynthesis as antibiotic target pathway. Int. J. Med. Microbiol. 300:161–169 [DOI] [PubMed] [Google Scholar]
- 39. Singh VK, Jayaswal RK, Wilkinson BJ. 2001. Cell wall-active antibiotic induced proteins of Staphylococcus aureus identified using a proteomic approach. FEMS Microbiol. Lett. 199:79–84 [DOI] [PubMed] [Google Scholar]
- 40. Standar K, et al. 2008. PspA forms large scaffolds in Escherichia coli. FEBS Lett. 582:3585–3589 [DOI] [PubMed] [Google Scholar]
- 41. Strahl H, Hamoen LW. 2010. Membrane potential is important for bacterial cell division. Proc. Natl. Acad. Sci. U. S. A. 27:12281–12286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stülke J, Hanschke R, Hecker M. 1993. Temporal activation of betaglucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 139:2041–2045 [DOI] [PubMed] [Google Scholar]
- 43. Vicario PP, Green BG, Katzen HM. 1987. A single assay for simultaneously testing effectors of alanine racemase and/or d-alanine:d-alanine ligase. J. Antibiot. (Tokyo) 40:209–216 [DOI] [PubMed] [Google Scholar]
- 44. Vostroknutova GN, Bulgakova VG, Udalova TP, Sepetov NF, Sibel'dina LA. 1981. Localization of gramicidin S on the cytoplasmic membrane of Bacillus brevis and its effect on the activity of membrane enzymes. Biokhimiia 46:657–666 [PubMed] [Google Scholar]
- 45. Wadhwani P, Afonin S, Ieronimo M, Buerck J, Ulrich AS. 2006. Optimized protocol for synthesis of cyclic gramicidin S: starting amino acid is key to high yield. J. Org. Chem. 71:55–61 [DOI] [PubMed] [Google Scholar]
- 46. Wecke T, et al. 2009. Daptomycin versus friulimicn B: in-depth profiling of Bacillus subtilis cell envelope stress responses. Antimicrob. Agents Chemother. 53:1619–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wenzel M, Bandow JE. 2011. Proteomic signatures in antibiotic research. Proteomics 11:3256–3268 [DOI] [PubMed] [Google Scholar]
- 48. Wenzel M, et al. 2011. Proteomic signature of fatty acid biosynthesis inhibition available for in vivo mechanism-of-action studies. Antimicrob. Agents Chemother. 55:2590–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. WHO 2012. The evolving threat of antimicrobial resistance—options for action. World Health Organization, Geneva, Switzerland: http://whqlibdoc.who.int/publications/2012/9789241503181_eng.pdf [Google Scholar]
- 50. Wiedemann I, Benz R, Sahl HG. 2004. Lipid II-mediated pore formation by the peptide antibiotic nisin—a black lipid membrane study. J. Bacteriol. 186:3259–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wolf D, et al. 2010. In-depth profiling of LiaR response in Bacillus subtilis. J. Bacteriol. 192:4680–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xiao Z, Xu P. 2007. Acetoin metabolism in bacteria. Crit. Rev. Microbiol. 33:127–140 [DOI] [PubMed] [Google Scholar]
- 53. Yoshida KI, Fujita Y, Ehrlich SD. 2000. An operon for a putative ATP-binding cassette transport system involved in acetoin utilization of Bacillus subtilis. J. Bacteriol. 182:5454–5461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zegers I, et al. 2006. Crystal structure of Bacillus subtilis TrmB, the tRNA (m7G46) methyltransferase. Nucleic Acids Res. 34:1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang L, Dhillon P, Yan H, Farmer S, Hancock REW. 2000. Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3317–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]