Abstract
In the INFORM-1 study, 73 patients with chronic hepatitis C virus infection received mericitabine plus danoprevir for up to 13 days. Seventy-two patients experienced a continuous decline in HCV RNA levels during treatment, and of these patients, 14 had viral loads that remained >1,000 IU/ml by day 13 and 1 met the definition for viral breakthrough. In-depth NS5B and NS3/4A population and clonal sequencing studies and mericitabine and danoprevir drug susceptibility testing were performed to assess the variability and quasispecies dynamics before and upon monotherapy or dual therapy. Sequence analysis of the viral quasispecies indicated that the mericitabine resistance mutation S282T was not present at baseline, nor was it selected (even at a low level) during treatment. Protease inhibitor resistance mutations, either as predominant or as minority species, were detected in 18 patients at baseline. No enrichment of minority protease inhibitor-resistant variants present at baseline was observed during treatment; viral population samples were fully susceptible to mericitabine and/or danoprevir, despite the presence within their quasispecies of minority variants confirmed to have reduced susceptibility to danoprevir or other protease inhibitors. It was also observed that certain NS3 amino acid substitutions affected protease inhibitor drug susceptibility in a compound-specific manner and varied with the genetic context. In summary, the slower kinetics of viral load decline observed in some patients was not due to the selection of danoprevir or mericitabine resistance during treatment. Over 2 weeks' therapy, mericitabine suppressed the selection of danoprevir resistance, results that could differ upon longer treatment periods.
INTRODUCTION
Chronic infection with hepatitis C virus (HCV) is a leading cause of liver disease worldwide (10). With the approval of the first HCV NS3/4A protease inhibitors (PIs; boceprevir and telaprevir), which are used in combination with pegylated interferon (peginterferon) and ribavirin, cure rates for treatment-naive HCV genotype 1-infected patients have improved to ∼70% (11, 15, 30). However, given the high genetic variability and population turnover of HCV, these PIs have a low barrier to the selection of PI-resistant variants, even when administered in combination with peginterferon and ribavirin (13, 28, 38). Danoprevir (RG7227) is a macrocyclic HCV protease inhibitor (3, 4). Among treatment-naive and treatment-experienced patients receiving danoprevir monotherapy for 14 days, the incidence of viral breakthrough was 27%, and the incidence of partial response was 35%. Viral breakthrough in these patients was associated with the emergence of the NS3 protease variant R155K (22).
Mericitabine (RG7128) is a nucleoside inhibitor of the HCV NS5B polymerase that acts as an RNA chain terminator and prevents elongation of RNA transcripts during replication (12, 24, 29, 37). Resistance to mericitabine in vitro is conferred by the NS5B substitution S282T (1), but no resistance-related viral breakthrough has been observed in patients treated for 2 weeks with mericitabine alone as monotherapy or for up to 24 weeks with mericitabine in combination with peginterferon-ribavirin (12, 19, 21, 29). In the phase IIb INFORM-SVR study, which investigated 12 or 24 weeks of response-guided treatment with mericitabine and ritonavir-boosted danoprevir with and without ribavirin in treatment-naive HCV genotype 1-infected patients, the S282T variant was described in one patient (6).
Viral kinetic models predict that every possible single, double, and triple mutant may exist in an HCV-infected patient before treatment and that, theoretically, resistant variants (if fit) can be rapidly selected during treatment (33). The selection and enrichment of a resistant variant depend on its baseline prevalence and fitness (20, 28). The emergence of resistance is a particular problem among patients who have not responded to prior treatment with peginterferon-ribavirin. Combining a compound with a high barrier to resistance, such as mericitabine with a PI, may delay or prevent the emergence of resistance to the PI, even in the absence of interferon.
The INFORM-1 study assessed the safety and efficacy of up to 13 days of treatment with mericitabine and danoprevir and demonstrated consistent reductions in HCV RNA (7). In this paper, we report the results of detailed resistance studies that were performed to determine the baseline prevalence of HCV variants with resistance to mericitabine, danoprevir, or other PIs (including sequence analyses of 2,937 NS3 clones from 34 samples and 1,910 NS5B clones from 21 samples). We also studied the dynamic evolution of minority variants and their impact on drug susceptibility to determine whether short-term treatment (13 days) with the combination of danoprevir and a compound with a high barrier to resistance, mericitabine, could prevent the emergence of danoprevir-resistant variants that typically emerge rapidly when danoprevir is administered alone as monotherapy.
MATERIALS AND METHODS
Patients and study design.
INFORM-1 was a phase I, randomized, double-blind, placebo-controlled, dose-escalation trial (NCT00801255) in which 87 adult patients with HCV genotype 1 infection were randomized to receive up to 13 days of oral combination therapy with danoprevir and mericitabine or placebo. Patients in cohort A were randomized to receive 3 days of monotherapy with either low-dose mericitabine at 500 mg every 12 h (q12h) (cohort A1) or low-dose danoprevir at 100 mg every 8 h (q8h) (cohort A2). Both subgroups then received the combination of mericitabine plus danoprevir for an additional 4 days. In all other cohorts (cohorts B to G), patients received concurrent treatment with mericitabine at a dosage of either 500 or 1,000 mg q12h and danoprevir at a dosage of 100 or 200 mg q8h or 600 or 900 mg q12h for 13 days. The trial was conducted in accordance with the Declaration of Helsinki. The protocol was approved by regulatory authorities and by the local research ethics committees at each study site. Written informed consent was obtained from all patients before they underwent any study-related procedures. A comprehensive description of the patient selection criteria, study design, and the primary results are published elsewhere (7).
Sample selection for viral resistance monitoring studies.
Baseline samples from all patients and samples from patients experiencing viral breakthrough, defined as an increase in HCV RNA level of at least 0.5 log10 IU/ml above the nadir, and from patients with an end-of-monotherapy (cohort A) or end-of-treatment (all other cohorts) HCV RNA level of at least 1,000 IU/ml, were analyzed for viral resistance.
Plasmid constructions.
The NS5B shuttle replicons (H77 and Con1 strains) contain restriction sites flanking the start and the end of the NS5B gene (19). The NS3 shuttle replicon based on Con1 contains restriction sites flanking the start and the end of the NS3 protease gene.
HCV RNA extraction, target gene amplification, and cloning of clinical isolates into shuttle replicon vectors for phenotypic characterization and clonal analysis.
HCV RNA extraction, reverse transcription, NS5B amplification in duplicate for each isolate, and cloning were carried out as previously described (19). Amplification of NS3/4A and/or NS3 protease sequences was carried out similarly to the NS5B amplification procedure. All NS3 protease genes (genotype 1a and 1b) were cloned into the genotype 1b Con1 shuttle replicon. Briefly, to maintain the quasispecies diversity, 96 individual colonies were pooled for each clinical isolate for drug susceptibility characterization (NS5B and NS3 protease genes). Clonal analysis was performed on each of the 96 individual colonies, and the total number of successfully sequenced clones per isolate varied from 76 to 94 (NS5B and NS3 protease).
Population and clonal sequencing of target gene from clinical isolates.
Population and clonal sequencing spanning the NS5B and/or NS3 protease-coding regions was performed using primers covering both DNA strands. Sequencing reactions were performed using an ABI 3730xl DNA analyzer, and chromatograms were analyzed using Sequencher and VNTI software (19).
Compounds.
β-d-2′-Deoxy-2′-fluoro-2′-C-methyl cytidine (parent compound of mericitabine) was obtained from Pharmasset, Inc. (Princeton, NJ). Danoprevir was obtained from Intermune, Inc. (Brisbane, CA). Telaprevir was obtained from Acme Bioscience, Inc. (Menlo Park, CA).
Determinations of 50% effective concentrations (EC50s).
Four million cured Huh7 cells were transfected with 10 μg of in vitro-transcribed replicon RNA from genotype 1b or genotype 1a reference strains or from clinical isolate-derived transient replicons as previously described (19).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study have been deposited in GenBank with accession numbers JX437195 to JX437368.
RESULTS
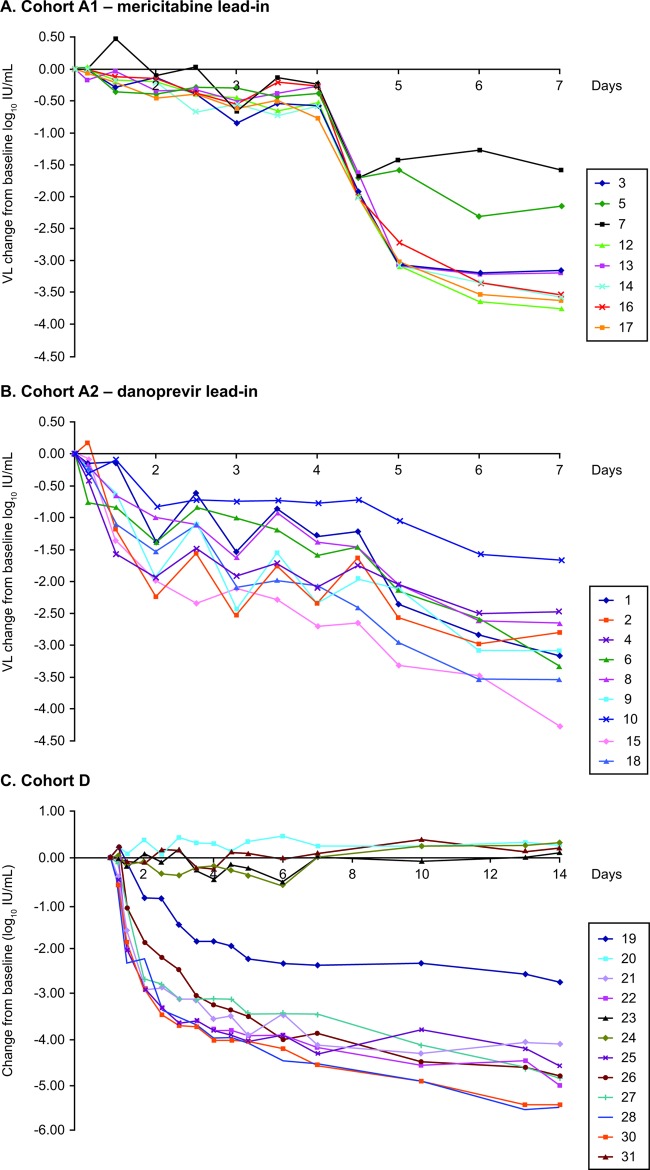
HCV load kinetics were assessed throughout the trial. Among patients in cohort A, initial slower declines in viral load were observed in patients who received low-dose mericitabine lead-in monotherapy in cohort A1 than in patients who received low-dose danoprevir lead-in monotherapy in cohort A2. After the addition of the second drug at day 4, HCV RNA levels declined similarly in both subgroups and were generally similar at the end of dosing (day 7). The median decrease in serum HCV RNA level at the end of treatment was 3.4 log10 IU/ml in cohort A1 and 3.1 log10 IU/ml in cohort A2.
Among the patients in cohorts B to G who received 13 days of mericitabine-danoprevir combination therapy, the median decrease in serum HCV RNA at the end of treatment ranged from 3.7 to 5.2 log10 IU/ml, with no significant differences between treatment-naive and treatment-experienced patients (7).
Of the 73 patients who received active drug in all cohorts, 72 (98.6%) experienced a continuous decline in HCV RNA levels during treatment. Only one patient, in cohort C, who received treatment with mericitabine at 500 mg q12h and danoprevir at 200 mg q8h, met the definition for virologic breakthrough. HCV RNA levels in this individual decreased by 4.5 log10 IU/ml and then increased by 1.45 log10 IU/ml between the nadir and the end of dosing (day 13). Virologic breakthrough in this individual was not associated with resistance to mericitabine or danoprevir (7).
Fourteen patients had HCV RNA levels that remained above 1,000 IU/ml at the end of treatment (10 in cohort A, 4 in cohorts B to G). We wanted to determine whether the slower decline in HCV RNA in these patients was due to treatment with low drug doses, the presence of drug-resistant variants at baseline, and/or the selection of resistant variants during treatment.
Inhibition of PI-resistant virus present at baseline after short-term combination therapy.
No mericitabine resistance mutation was detected in any patient by population sequencing of baseline samples. However, a number of mutations previously described to affect susceptibility to PIs were detected in 15 of 73 patients who received mericitabine plus danoprevir and 3 of 14 patients who received placebo. These included Q80K (in 8% of all patients), T54S (7%), V55A (2%), V36M (1%), Q80R (1%), R155K (1%), and D168E (1%) (3, 17, 18, 25, 26, 31, 36, 41).
Despite the presence of preexisting PI resistance mutations, HCV RNA decreased continuously in the 15 patients who received mericitabine plus danoprevir treatment and was <1,000 IU/ml in 12 of 15 patients at the end of treatment. In one patient (patient 19; Fig. 1), the danoprevir resistance mutation D168E was present in 91% of clones at baseline and was correlated with a 37-fold reduction in danoprevir susceptibility (at the population level) (7). Baseline samples (at the population level) from this patient were fully susceptible to mericitabine (0.4-fold change versus control).
Fig 1.
(A) Viral load (VL) kinetics of patients (boxed numbers) in cohort A who received mericitabine alone on days 1 to 3 and then mericitabine plus danoprevir on days 4 to 7; (B) viral load kinetics of patients in cohort A who received danoprevir alone on days 1 to 3 and then mericitabine plus danoprevir on days 4 to 7; (C) viral load kinetics of patients in cohort D. Includes a patient with a D168E resistance mutation (patient 19) who received mericitabine and danoprevir for 13 days. Other patients from the same cohort are presented as a comparison (patients 20, 23, 24, and 31 received placebo).
No on-treatment selection of mericitabine or danoprevir resistance after 3 days monotherapy or up to 13 days dual therapy.
Ten of the 17 patients in cohort A (3 who received the mericitabine lead-in in cohort A1 and 7 who received the danoprevir lead-in in cohort A2) had HCV RNA levels of >1,000 IU/ml at the end of treatment (day 7), thus allowing PCR amplification and further analyses. Nine of these 10 patients were infected with genotype 1a.
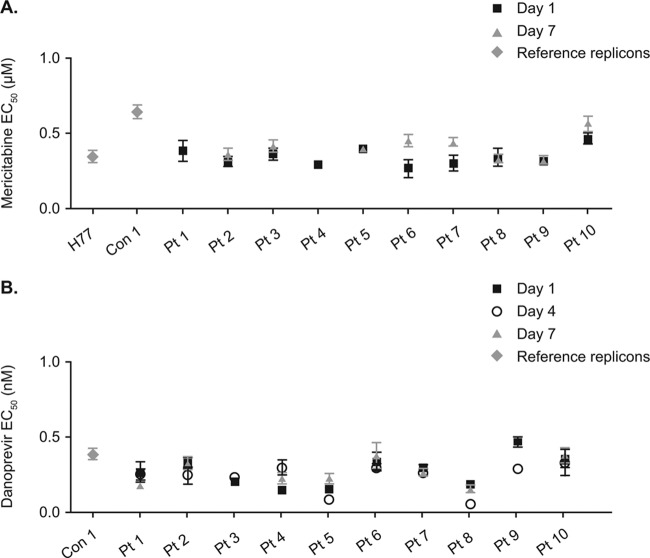
Sequencing and phenotypic analyses of the NS3/4A and NS5B regions were performed at baseline and at treatment days 4 and 7. No mericitabine or danoprevir resistance mutations were detected by population sequencing at the end of monotherapy (day 4) or at the end of dual therapy (day 7) in any of these patients (Table 1), which is in contrast to results in patients treated for 14 days with danoprevir monotherapy, among whom mutant variants were detected in approximately 50% of patients (22). Samples from all 10 patients were fully susceptible to mericitabine and danoprevir at baseline, with no change in susceptibility (at the population level) in on-treatment samples, which suggests that the amino acid substitutions observed in some samples did not alter the susceptibility to either compound (Fig. 2A and B). No danoprevir or mericitabine resistance mutations were observed at baseline or at the end of monotherapy in any of the seven patients (five from cohort A1 and two from cohort A2) whose viral loads were >1,000 IU/ml by day 4 but <1,000 IU/ml by day 7 (Table 1).
Table 1.
Population sequence analysis of samples obtained at day 4 and 7 of treatment from 10 patients in cohort Aa
| Population and patient (HCV genotype) | Treatment regimen | Day | Amino acid change(s) from baseline (day 1) sample |
|
|---|---|---|---|---|
| NS5B | NS3 protease | |||
| Viral load > 1,000 IU/ml on day 7 | ||||
| 1 (1a) | Danoprevir days 1–7 + mericitabine days 4–7 | 4 | K77K/R, K100K/R, H374H/Q, M423I/M, C/Y451C, K/R523K, I/M539I | Q9P/Q |
| 7 | NA | Q9P/Q, N174N/S | ||
| 2 (1a) | Danoprevir days 1–7 + mericitabine days 4–7 | 4 | N/S81N, C/S110C, F/Y162F, T377A, R510K/R, A/P540A | I71I/M, A/M/T/V147A, I/V170I |
| 7 | C/S110C, Q124Q/R, F/Y162F | A/T40T, A/M/T/V147A, I/V170I | ||
| 3 (1a) | Mericitabine days 1–7 + danoprevir days 4–7 | 4 | I184I/V, F572F/I | None |
| 7 | R48R/G, K/R401R, A/T580A | None | ||
| 4 (1a) | Danoprevir days 1–7 + mericitabine days 4–7 | 4 | NAb | None |
| 7 | NA | None | ||
| 5 (1a) | Mericitabine days 1–7 + danoprevir days 4–7 | 4 | S/T5S, S62N/S, K/R254K, S/T473T, K/R510K | None |
| 7 | S/T5S, V11I/V, M187M/V, A327A/V, S/T473T, K/R510K, K517K/R | None | ||
| 6 (1a) | Danoprevir days 1–7 + mericitabine days 4–7 | 4 | V71A/V, R98K/R, I520I/V, K531K/R | None |
| 7 | D/G/N/S62N/S, S431G/S | I/V18V | ||
| 7 (1a) | Mericitabine days 1–7 + danoprevir days 4–7 | 4 | None | N174N/S |
| 7 | A185A/V, K510K/R | None | ||
| 8 (1a) | Danoprevir days 1–7 + mericitabine days 4–7 | 4 | S180R/S, Y276F, A/T580A | I/V114I |
| 7 | Q124Q/R, I/L462L, T520I/T, G558E/G, A/T580A | A87A/T, I/V114I | ||
| 9 (1a) | Danoprevir days 1–7 + mericitabine days 4–7 | 4 | N/S117S, D/E/K/N124N, Q206R, I/V405I, D444G | Nonec |
| 7 | V116V/L, D/E/K/N124N, S326S/G, A334A/T, I/V405I | NA | ||
| 10 (1b) | Danoprevir days 1–7 + mericitabine days 4–7 | 4 | None | None |
| 7 | None | None | ||
| Viral load < 1,000 IU/ml on day 7 (and >1,000 IU/ml on day 4) | ||||
| 12 (1a) | Mericitabine days 1–7 + danoprevir days 4–7 | 4 | S543G/S | NA |
| 13 (1a) | Mericitabine days 1–7 + danoprevir days 4–7 | 4 | A112A/S, N/S120N, A/G198G, F526F/L | NA |
| 14 (1b) | Mericitabine days 1–7 + danoprevir days 4–7 | 4 | None | V48V/I |
| 15 (1a) | Danoprevir days 1–7 + mericitabine days 4–7 | 4 | NA | None |
| 16 (1a) | Mericitabine days 1–7 + danoprevir days 4–7 | 4 | I/V262I, F/I/S/T267T | NA |
| 17 (1b) | Mericitabine days 1–7 + danoprevir days 4–7 | 4 | None | None |
| 18 (1a) | Danoprevir days 1–7 + mericitabine days 4–7 | 4 | E/K69E, K/M72K, N130S, G549S | T95T/A |
The patients included those with end-of-treatment serum HCV RNA levels of ≥1,000 IU/ml or with HCV RNA levels of ≥1,000 IU/ml at the end of monotherapy.
NA, sequence not available.
Telaprevir resistance mutation V36M was present at baseline and day 4.
Fig 2.
(A) Phenotypic characterization of susceptibility to mericitabine in samples obtained at baseline and during treatment from 10 patients in cohort A with an end-of-treatment serum HCV RNA level of ≥1,000 IU/ml; (B) phenotypic characterization of susceptibility to danoprevir in samples obtained at baseline and during treatment from 10 patients in cohort A with an end-of-treatment serum HCV RNA level of ≥1,000 IU/ml.
Four patients (three infected with genotype 1a and one infected with genotype 1b) in cohorts B to G who were treated for 13 days had HCV RNA levels of >1,000 IU/ml at the end of treatment. These included patient 40, in whom HCV RNA levels decreased by 2.9 log10 IU/ml from baseline to 14,400 IU/ml at the end of treatment, and patients 46, 51, and 59, who experienced 2.5-, 3.5-, and 3.0-log10-IU/ml reductions in viral load to 2,000, 1,410, and 1,290 IU/ml, respectively, by the end of treatment (Table 1). Population and clonal sequencing at baseline did not detect any known resistance mutation in NS5B or NS3/4A. Amplification of day 14 samples from patients 46, 51, and 59 was not successful. Baseline and end-of-treatment samples from patient 40 were susceptible to both mericitabine (EC50s, 0.44 ± 0.04 μM and 0.32 ± 0.02 μM, respectively) and danoprevir (EC50s, 0.37 ± 0.06 nM and 0.83 ± 0.08 nM, respectively).
Quasispecies composition and dynamics after 3 days of monotherapy followed by 4 days of dual therapy.
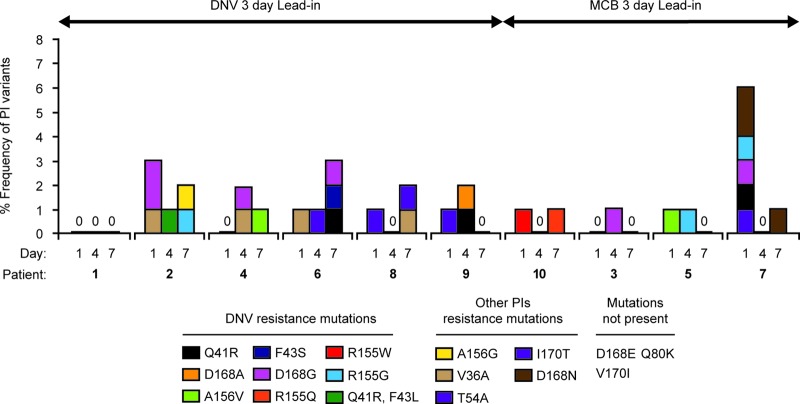
Clonal analysis (76 to 94 clones per sample) was performed on the 10 subjects in cohort A with HCV RNA levels of >1,000 IU/ml at the end of treatment (day 7) to assess their quasispecies composition and dynamics at baseline, day 4 (end of monotherapy), and day 7 (end of combination therapy) (Fig. 3).
Fig 3.
Presence and viral dynamics of low-frequency PI-resistant variants in cohort A patients with an end-of-treatment serum HCV RNA level of ≥1,000 IU/ml. DNV, danoprevir; MCB, mericitabine.
Baseline samples from two of the three patients in cohort A1 that had an HCV RNA level of >1,000 IU/ml at day 7 contained at frequencies of 1 to 2% minority variants that confer resistance to danoprevir or to another PI but not to mericitabine. The observed PI-resistant variants were not enriched during mericitabine monotherapy or mericitabine-danoprevir dual therapy (Fig. 3).
Baseline samples from five of the seven patients in cohort A2 that had an HCV RNA level of >1,000 IU/ml at day 7 contained at low frequencies (1 to 2%) minority variants that conferred resistance to danoprevir or to another PI but not to mericitabine. These variants were not enriched during danoprevir monotherapy or mericitabine-danoprevir dual therapy. In the two patients with no PI-resistant variants at baseline, low-frequency PI resistance variants were observed at day 4 and/or day 7 (Fig. 3).
No mericitabine-resistant variants were selected during monotherapy. Moreover, the dynamics of the NS3 variants detected within patients' quasispecies at baseline were similar after danoprevir or mericitabine monotherapy, suggesting that they arose as the result of random mutagenesis rather than being driven by drug-associated selective pressure.
Drug susceptibility studies of variants containing PI-resistant amino acid substitutions.
Susceptibility to danoprevir and telaprevir was assessed in 41 replicon clones isolated from the quasispecies of patients included in this resistance analysis. Eighteen of 41 clones represented consensus sequences, and the sequences of the remaining 23 clones differed from the consensus sequence by having amino acid substitutions that either were known to confer PI resistance or occurred at residues that are associated with PI resistance (and for which the effect on PI susceptibility has not yet been described) (Table 2). Most clones that contained PI resistance mutations had a lower replication capacity than the corresponding consensus clone.
Table 2.
Comparative susceptibility to danoprevir and telaprevir of HCV-containing NS3 mutations in baseline samples from patients in cohorts A to D
| Strain or site(s) of polymorphism and patient sample (HCV genotype), day | Clone no. | Sequence/difference from consensus (amino acid) | Frequency in viral populationa | Replication | Danoprevir |
Telaprevir |
||
|---|---|---|---|---|---|---|---|---|
| EC50 ± SEM (nM) | Fold shiftb | EC50 ± SEM (μM) | Fold shiftb | |||||
| Con1 | 1 | 0.19 ± 0.020 (16)c | 0.13 ± 0.025 (15) | |||||
| Residue 170 | ||||||||
| 1 (1a), day 1 | A04 | Consensus | 58.2 | 0.017 ± 0.008 (2) | 0.11 ± 0.013 (2) | 1 | 0.41 ± 0.041 (2) | 1 |
| A10 | V170I | 9.9 | 0.031 ± 0.012 (2) | 0.12 ± 0.008 (2) | 1.10 | 0.21 ± 0.023 (2) | 0.51 | |
| 1 (1a), day 7 | A02 | Consensus | 32.9 | 0.097 ± 0.022 (4) | 0.25 ± 0.033 (2) | 1 | 0.10 ± 0.025(2) | 1 |
| A06 | V170I | 14.5 | 0.046 ± 0.003 (2) | 0.12 ± 0.029 (3) | 0.48 | 0.053 ± 0.004 (2) | 0.53 | |
| 2 (1a), day 1 | A04 | Consensus | 33.7 | 0.24 ± 0.040 (2) | 0.17 ± 0.041 (2) | 1 | 0.21 ± 0.019 (2) | 1 |
| A02 | I170V | 3.6 | 0.061 ± 0.023 (2) | 0.10 ± 0.006 (2) | 0.58 | 0.27 ± 0.022 (2) | 1.28 | |
| 5 (1a), day 1 | A02 | Consensus | 76.2 | 0.028 ± 0.003 (4) | 0.080 ± 0.020 (3) | 1 | 0.022 ± 0.001 (2) | 1 |
| D03 | I170V | 2.5 | 0.022 ± 0.004 (4) | 0.11 ± 0.016 (2) | 1.37 | 0.039 ± 0.010 (2) | 1.77 | |
| 7 (1a), day 1 | A03 | Consensus | 56.6 | 0.038 ± 0.009 (4) | 0.12 ± 0.034 (3) | 1 | 0.060 ± 0.015 (2) | 1 |
| C11 | I170T | 1.3 | 0.006 ± 0.001 (6) | 0.32 ± 0.027 (5) | 2.66 | 0.35 ± 0.030 (3) | 5.83 | |
| Residue 155 | ||||||||
| 2 (1a), day 7 | D06 | R155G | 1.1 | No replication | ||||
| 7 (1a), day 1 | F04 | R155G | 1.3 | No replication | ||||
| 10 (1b), day 1 | A02 | Consensus | 28.0 | 0.22 ± 0.030 (2) | 0.11 ± 0.010 (4) | 1 | 0.096 ± 0.011 (2) | 1 |
| A12 | R155W | 1.2 | 0.001 ± 0.00 (4) | 3.06 ± 0.21 (2)d | 27.81 | 0.20 ± 0.033 (2)d | 2.08 | |
| 10 (1b), day 7 | A01 | Consensus | 68.0 | 0.090 ± 0.024 (3) | 0.12 ± 0.012 (2) | 0.14 ± 0.004 (2) | ||
| B04 | R155Q | 1.0 | 0.001 ± 0.00 (2)e | |||||
| 20 (1a), day 1 | A05f | Consensus (155K) | 62.2 | 0.020 ± 0.001 (3) | >100 (2) | >833 | 0.78 ± 0.11 (2) | 14.44 |
| E07g | K155R | 1.1 | 0.038 ± 0.006 (3) | 0.12 ± 0.017 (3) | 1 | 0.054 ± 0.006 (2) | 1 | |
| 20 (1a), day 4 | A02f | Consensus (155K) | 79.3 | 0.005 ± 0.001 (3) | >100 (2) | 0.90 ± 0.088 (2) | ||
| A08 | L106P, R155K | 1.1 | No replication | |||||
| 20 (1a), day 10 | A01g | K155R | 29.9 | 0.080 ± 0.022 (3) | 0.11 ± 0.029 (3) | 1 | 0.043 ± 0.005 (3) | 1 |
| A05f | Consensus (155K) | 47.1 | 0.013 ± 0.002 (3) | >100 (2) | >909 | 0.63 ± 0.084 (3) | 14.65 | |
| 20 (1a), day 14 | A01g | Consensus (155R) | 48.2 | 0.063 ± 0.025 (3) | 0.11 ± 0.033 (3) | 1 | 0.051 ± 0.005 (3) | 1 |
| A04f | R155K | 36.4 | 0.019 ± 0.002 (2) | >100 (2) | >909 | 0.72 ± 0.11 (3) | 14.11 | |
| Residues D168E and Q80K | ||||||||
| 19 (1a), day 1 | A02h | Consensus (168E) | 64.8 | 0.38 ± 0.141 (2) | 9.58 ± 1.69 (2) | 53.22 | 0.041 ± 0.005 (2) | 1.36 |
| C06 | Q80K, E168D, V170I | 1.1 | 0.38 ± 0.100 (2) | 1.81 ± 0.12 (2) | 10.05 | 0.068 ± 0.009 (2) | 1.21 | |
| H11 | E168D, V170I | 1.1 | 0.22 ± 0.082 (2) | 0.18 ± 0.052 (2) | 1 | 0.056 ± 0.011 (2) | 1 | |
| 19 (1a), day 3 | A01h | Consensus (168E) | 87.8 | 0.30 ± 0.13 (2) | 18.76 ± 1.671 (2) | 0.12 ± 0.011 (2) | ||
| C07 | F43Y, A156V | 1.2 | No replication | |||||
| Residues known to confer resistance to danoprevir in vitro | ||||||||
| 2 (1a), day 7 | A01 | Consensus | 71.9 | 0.026 ± 0.004 (4) | 0.14 ± 0.022 (5) | 1 | 0.23 ± 0.037 (2) | |
| H12 | A156G | 1.1 | 0.009 ± 0.004 (2) | 0.17 ± 0.026 (2) | 1.21 | 0.34 ± 0.093 (3) | 1.47 | |
| 5 (1a), day 1 | A02 | Consensus | 76.2 | 0.028 ± 0.003 (4) | 0.080 ± 0.020 (3) | 1 | 0.022 ± 0.001 (2) | |
| H07 | A156V | 1.2 | 0.008 ± 0.004 (4) | 10.5 ± 2.96 (3) | 131.25 | >30 (2) | >1,363 | |
| 6 (1a), day 7 | A02 | Consensus | 51.7 | 0.023 ± 0.007 (2) | 0.12 ± 0.011 (2) | 1 | 0.082 ± 0.012 (2) | 1 |
| B12 | Q41R | 1.1 | 0.014 ± 0.006 (2) | 11.14 ± 0.38 (2) | 92.83 | 1.04 ± 0.022 (2) | 12.70 | |
| 9 (1a), day 4 | A04 | Consensus (V36 M) | 69.7 | 0.035 ± 0.017 (2) | 0.17 ± 0.021 (2) | 1 | 0.67 ± 0.015 (2) | 1 |
| C08 | Q41R, V36 M | 1.1 | 0.005 ± 0.002 (2) | 8.48 ± 1.31 (2) | 49.90 | 7.46 ± 1.442 (2) | 11.13 | |
| 11i (1b), day 15 | A08 | Consensus | 57.9 | 0.020 ± 0.008 (2) | 0.17 ± 0.023 (2) | 0.027 ± 0.008 (2) | ||
| C10 | F43S | 1.1 | No replication | |||||
| 19 (1a), day 4 | A07 | Consensus | 36.8 | 0.16 ± 0.076 (2) | 0.13 ± 0.019 (2) | 1 | 0.16 ± 0.031 (2) | 1 |
| D09 | Q41R, F43L | 1.1 | 0.004 ± 0.001 (2) | 8.28 ± 1.38 (2) | 63.70 | 0.71 ± 0.12 (2) | 4.75 | |
| Residue 36, 2 (1a), day 1 | A04 | Consensus | 33.7 | 0.24 ± 0.040 (2) | 0.17 ± 0.041 (2) | 1 | 0.21 ± 0.019 (2) | 1 |
| E02 | V36A | 1.1 | 0.068 ± 0.003 (2) | 0.32 ± 0.047 (2) | 1.88 | 0.52 ± 0.094 (2) | 2.47 | |
Percentage out of total number of sequenced clones within each sample. The total number of sequenced clones varied from 76 to 94 per sample.
Fold shift was calculated using the variant that did not contain any resistance mutation.
Values in parentheses are numbers of patients.
EC50 determination is approximate, as inhibition curves did not reach 100%.
Level of replication too low for EC50 determination.
Identical clones within a patient.
Identical clones within a patient.
Identical clones within a patient.
The patient that experienced viral breakthrough while on dual therapy (8).
Amino acid changes at residue V36 (V36A or V36M) did not confer resistance to danoprevir (1.7- and 0.9-fold increases in EC50s versus the EC50 for the Con1 reference control). V36M confers low-level resistance to telaprevir (5-fold increase in EC50 versus that for the Con1 reference strain) (42). The mutation Q41R conferred resistance to both danoprevir (50- to 100-fold increases in EC50s versus the EC50 for the consensus clone from the same sample) and telaprevir (<12-fold increases).
Samples with changes at residue 43 were evaluated for their susceptibility to danoprevir. These included F43Y/A156V, which was observed in 1.2% of clones from patient 19 on day 3; Q41R/F43L, which was observed in 1.1% of clones from patient 2 on day 4; and F43S, which was observed in 1% of clones from patient 84, who experienced viral breakthrough on study day 15. The Q41R/F43L variant was the only one that replicated in vitro and subsequently showed an increase in EC50 similar to that observed for the mutant with a single mutation, Q41R, suggesting that F43L does not affect susceptibility to danoprevir.
Effects of substitutions at amino acid Q80 on danoprevir susceptibility depended on the genetic context. Q80K conferred low-level resistance to danoprevir (10-fold) but not to telaprevir in a clone from patient 19 (C06, genotype 1a) (Table 2). Three other genotype 1a isolates that contained predominantly Q80K (from patients 42 and 52) or a Q80K/Q mixture (from patient 71) were evaluated (as a pool of 96 clones) and showed no significant reduction in susceptibility to danoprevir (EC50s, 0.28 ± 0.05 nM [2-fold increase compared to a Con1 EC50 of 0.14 ± 0.002 nM], 0.29 ± 0. 016 nM [2-fold increase], and 0.09 ± 0.001 nM [0.64-fold increase], respectively) (Table 2).
Substitutions at residue 155 were evaluated in different isolates and therefore different genetic contexts. R155K-containing clones showed high-level resistance (>800-fold) to danoprevir, medium-level resistance (∼15-fold) to telaprevir, and also a reduced replication capacity. One R155K-containing clone that had an additional mutation at position 106 (L to P), R155G-containing variants from two genotype 1a-infected patients, and several R155Q-containing clones could not be evaluated because of low/no replication in vitro. Reduced susceptibility to danoprevir (27-fold) but not to telaprevir was observed in a variant containing R155W.
The A156G substitution did not confer resistance to either danoprevir or telaprevir; in contrast, A156V conferred high-level resistance to danoprevir and to telaprevir (>100-fold and >1,000-fold increases in EC50s, respectively) (Table 2).
Amino acid change I/V170I was observed in the on-treatment population sequence of one patient in cohort A and was found at a low prevalence in the quasispecies of four baseline samples (Table 1 and 2). This substitution had no significant effect on susceptibility to danoprevir (2.6-fold decrease versus the consensus clone; Table 2). The substitution I170T was observed in one genotype 1a genetic context and conferred low-level resistance (6-fold) to telaprevir but not to danoprevir.
DISCUSSION
Interferon-free therapy could become a new option for the treatment of chronic hepatitis C virus infection. Recent clinical studies in small cohorts of HCV-infected patients have shown that high sustained virological response rates can be achieved using combinations of two direct-acting antiviral agents (DAAs) (2, 8, 23). The success of interferon-free regimens hinges on two major elements that enable a durable response: profound viral suppression and a high barrier to resistance. Resistance to danoprevir in patients receiving danoprevir monotherapy is conferred by the mutation NS3 R155K (22). Low-level resistance to mericitabine is conferred through the NS5B mutation S282T (1).
The INFORM-1 study showed that an interferon-free regimen of two DAAs (mericitabine and danoprevir) could produce a rapid and continual decline in HCV RNA through 13 days of treatment with no resistance-related viral breakthrough, as assessed by population and clonal sequence and drug susceptibility analysis (6). In contrast, danoprevir monotherapy was associated with viral load breakthrough and resistance selection by day 7 in five of eight patients (22). This suggests that mericitabine effectively suppressed the emergence of danoprevir-resistant virus during up to 13 days of treatment, even in regimens that contained suboptimal doses of either mericitabine, danoprevir, or both drugs. It is acknowledged that these results could differ with longer treatment durations if clearance of the danoprevir-resistant virus by mericitabine is not maintained.
Viral breakthrough due to selection of drug-resistant HCV has been detected within as little as 3 days of the start of treatment with telaprevir monotherapy (5, 13, 32–35), and resistant HCV variants have emerged within 8 days of the initiation of treatment with telaprevir, peginterferon alfa-2a (40KD), and ribavirin triple-combination therapy (16). For this reason, the absence of protease-resistant variants after 13 days of combination therapy in this trial suggests that mericitabine has the potential to prevent or delay the emergence of PI-resistant variants.
The high viral replication rate and the nucleotide misincorporation rate of the HCV NS5B polymerase result in the formation of a viral quasispecies, resulting in a situation in which many viral sequence variants, including drug-resistant variants, can preexist in untreated patients. DAA-resistant virus was detected in untreated patients by population sequencing of NS3- and NS5B-coding sequences. The most frequent mutations were associated with resistance to nonnucleoside polymerase inhibitors (data not shown). Less frequent were mutations associated with resistance to PIs, while the mutation S282T, which confers in vitro resistance to mericitabine, PSI-7977, and IDX-184, was not detected in any sample, similar to previous reports (14, 19, 39, 40).
This study provides a comprehensive clonal assessment of the prevalence of minority variants affecting PI susceptibility (a total of 2,937 clonal variants were analyzed) and the dynamics of such variants within the quasispecies upon treatment.
The NS5B S282T variant was not detected by population or clonal analysis in any samples collected at baseline or during treatment, consistent with previous studies where patients received mericitabine alone for 2 weeks or in combination with peginterferon alfa-2a (40KD) plus ribavirin for up to 24 weeks (12, 19, 21, 29). Position 282 of the NS5B polymerase is located near the active binding site and is highly conserved across all HCV genotypes. If the serine at this position is required for polymerase function, this would explain the low in vitro replicative capacity of S282T-containing mutant replicons (1, 27) and could explain the high barrier to resistance observed in vivo.
While it has been shown that DAA-resistant variants may exist at low frequencies within the quasispecies of untreated patients, their clinical relevance has not yet been determined. Variants containing danoprevir resistance mutations were detected as minority species at baseline in 4 out of 12 studied patients (10 in cohort A, 1 in cohort B, 1 in cohort E). These variants were not enriched during treatment, suggesting that the slower kinetics of plasma HCV RNA reduction in patients in cohort A were not associated with the presence and/or selection of virus resistant to danoprevir and/or mericitabine. The dynamics of the minority PI-resistant variants at baseline and throughout treatment were similar after danoprevir or mericitabine monotherapy. The low frequency of resistant variants was, therefore, likely due not to drug pressure but, rather, to random mutagenesis caused by the error-prone nature of the NS5B polymerase. In addition, relative fitness and immune pressure may determine the relative proportion of minority variants within a given patient's quasispecies (9). As an example of intrinsic quasispecies dynamics, the NS3 R155K substitution was predominant at baseline (98% of clones) in a patient who received placebo (patient 20) and then reverted to wild type by day 13 (55% wild type), likely because of the lower fitness of this variant compared with wild type. To address the potential relevance of minority variants on susceptibility to PIs, extensive phenotypic studies were performed on individual variants that either contained amino acid substitutions known to confer resistance to PIs or had variations that occurred at residues known to be associated with resistance to PIs but that have not yet been described. While certain amino acid changes, such as NS3 Q41R, affected susceptibility to both linear (i.e., telaprevir) and macrocyclic (i.e., danoprevir) PIs, other changes affected susceptibility to specific drugs (V36M, R155W, I170T, or Q80K). Importantly, all patients achieved a continuous viral load decline, suggesting that minority species with reduced drug susceptibility could be effectively suppressed by the combination of mericitabine and danoprevir.
In summary, the present study showed that minority variants resistant to PIs exist within quasispecies of treatment-naive patients as either predominant or minority species. New NS3 amino acid substitutions affecting susceptibility to telaprevir (V36M, I170T) or danoprevir (R155W, Q80K), or both drugs (Q41R), were observed in individual minority variant clones and were characterized for the first time. Importantly, this study shows that the genetic context can affect susceptibility to PIs, highlighting the importance of phenotypic studies in resistance monitoring. Minority PI-resistant variants that are present at baseline can be effectively suppressed by the inclusion of the nucleoside analog mericitabine in a combination regimen and are unlikely to be clinically relevant. This study shows that HCV RNA levels of >1,000 IU/ml at day 13 in 14 patients in the INFORM-1 study were not due to the selection of resistance to mericitabine or danoprevir. This suggests that the combination of a PI with a nucleoside polymerase inhibitor that has a high barrier to resistance, such as mericitabine, has the potential to reduce or prevent the emergence of clinically significant resistance to PIs even when PI-resistant variants are present at baseline.
The primary limitation of this study is the short duration of combination therapy, which likely explains why not all patients achieved HCV RNA levels below 1,000 IU/ml at the end of dual-combination therapy. Longer-duration studies are needed to confirm that mericitabine can prevent the emergence of resistance and to determine the durability of resistance suppression when mericitabine is combined with a PI or other DAA. These encouraging results support further studies of this dual-combination interferon-free regimen.
ACKNOWLEDGMENTS
We thank Jennifer Wong, Amritha Seshaadri, and Aren Ewing for excellent technical assistance and Volker Lohmann for providing the Huh7 Lunet cells. We acknowledge Saundra Clausen and Nixy Zutshi for the maintenance of the replicon and Huh7 cells, Sharon Jiang for DNA sequencing, and Rohit Kulkarni for biostatistical support. We also thank Lewyn Li for his advice on sequence analysis.
Support for third-party writing assistance for the manuscript was provided by F. Hoffmann-La Roche Ltd.
Footnotes
Published ahead of print 6 August 2012
REFERENCES
- 1. Ali S, et al. 2008. Selected replicon variants with low-level in vitro resistance to the hepatitis C virus NS5B polymerase inhibitor PSI-6130 lack cross-resistance with R1479. Antimicrob. Agents Chemother. 52:4356–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chayama K, et al. 2012. Dual therapy with the nonstructural protein 5A inhibitor, daclatasvir, and the nonstructural protein 3 protease inhibitor, asunaprevir, in hepatitis C virus genotype 1b-infected null responders. Hepatology 55:742–748 [DOI] [PubMed] [Google Scholar]
- 3. Forestier N, et al. 2011. Treatment of chronic hepatitis C patients with the NS3/4A protease inhibitor danoprevir (ITMN-191/RG7227) leads to robust reductions in viral RNA: a phase 1b multiple ascending dose study. J. Hepatol. 54:1130–1136 [DOI] [PubMed] [Google Scholar]
- 4. Forestier N, et al. 2011. Antiviral activity of danoprevir (ITMN-191/RG7227) in combination with pegylated interferon alpha-2a and ribavirin in patients with hepatitis C. J. Infect. Dis. 204:601–608 [DOI] [PubMed] [Google Scholar]
- 5. Forestier N, et al. 2007. Antiviral activity of telaprevir (VX-950) and peginterferon alfa-2a in patients with hepatitis C. Hepatology 46:640–648 [DOI] [PubMed] [Google Scholar]
- 6. Gane EJ, et al. 2012. Interferon-free treatment with a combination of mericitabine and danoprevir/r with or without ribavirin in treatment-naive HCV genotype-1 infected patients. J. Hepatol. 56(Suppl S2):S555–S556 [Google Scholar]
- 7. Gane EJ, et al. 2010. Oral combination therapy with a nucleoside polymerase inhibitor (RG7128) and danoprevir for chronic hepatitis C genotype 1 infection (INFORM-1): a randomised, double-blind, placebo-controlled, dose-escalation trial. Lancet 376:1467–1475 [DOI] [PubMed] [Google Scholar]
- 8. Gane EJ, et al. 2011. Once daily PSI-7977 plus RBV: pegylated interferon-alfa not required for complete rapid viral response in treatment-naive patients HCV GT2 or GT3, abstr 34. 54(Suppl):377A [Google Scholar]
- 9. Gaudieri S, et al. 2009. Hepatitis C virus drug resistance and immune-driven adaptations: relevance to new antiviral therapy. Hepatology 49:1069–1082 [DOI] [PubMed] [Google Scholar]
- 10. Ghany MG, Nelson DR, Strader DB, Thomas DL, Seeff LB. 2011. An update on treatment of genotype 1 chronic hepatitis C virus infection: 2011 practice guideline by the American Association for the Study of Liver Diseases. Hepatology 54:1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jacobson IM, et al. 2011. Telaprevir for previously untreated chronic hepatitis C virus infection. N. Engl. J. Med. 364:2405–2416 [DOI] [PubMed] [Google Scholar]
- 12. Jensen DM, et al. 2010. High rates of early viral response, promising safety profile and lack of resistance-related breakthrough in HCV GT 1/4 patients treated with RG7128 plus PegIFN alfa-2a (40KD)/RBV: planned week 12 interim analysis from the PROPEL study. Hepatology 52(Suppl 1):360A20578152 [Google Scholar]
- 13. Kieffer TL, et al. 2007. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology 46:631–639 [DOI] [PubMed] [Google Scholar]
- 14. Kuntzen T, et al. 2008. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naïve patients. Hepatology 48:1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kwo P, et al. 2010. Efficacy of boceprevir, an NS3 protease inhibitor, in combination with peginterferon alfa-2b and ribavirin in treatment-naive patients with genotype 1 hepatitis C infection (SPRINT-1): an open-label, randomised, multicentre phase 2 trial. Lancet 376:705–716 [DOI] [PubMed] [Google Scholar]
- 16. Lawitz E, Rodriguez-Torres M, Muir AJ, Kieffer TLML KA, McHutchison JG. 2008. Antiviral effects and safety of telaprevir, peginterferon alfa-2a, and ribavirin for 28 days in hepatitis C patients. J. Hepatol. 49:163–169 [DOI] [PubMed] [Google Scholar]
- 17. Lenz O, et al. 2011. Treatment outcome and resistance analysis in HCV genotype 1 patients previously exposed to TMC435 monotherapy and re-treated with TMC435 in combination with PEG/IFN alpha-2a/ribavirin. J. Hepatol. 54(Suppl 1):S482–S483 [Google Scholar]
- 18. Lenz O, et al. 2010. In vitro resistance profile of the hepatitis C virus NS3/4A protease inhibitor TMC435. Antimicrob. Agents Chemother. 54:1878–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Le Pogam S, et al. 2010. RG7128 alone or in combination with pegylated interferon-alpha2a and ribavirin prevents hepatitis C virus (HCV) replication and selection of resistant variants in HCV-infected patients. J. Infect. Dis. 202:1510–1519 [DOI] [PubMed] [Google Scholar]
- 20. Le Pogam S, et al. 2008. Existence of hepatitis C virus NS5B variants naturally resistant to non-nucleoside, but not to nucleoside, polymerase inhibitors among untreated patients. J. Antimicrob. Chemother. 61:1205–1216 [DOI] [PubMed] [Google Scholar]
- 21. Le Pogam S, et al. 2010. No evidence of drug resistance or baseline S282T resistance mutation among GT1 and GT4 HCV infected patients on nucleoside polymerase inhibitor RG7128 and Peg-IFN-/RBV combination treatment for up to 12 weeks: interim analysis from the PROPEL study. Hepatology 52(Suppl 1):701–702A [Google Scholar]
- 22. Lim SR, et al. 2012. Virologic escape during danoprevir (ITMN-191/RG7227) monotherapy is hepatitis C virus subtype dependent and associated with R155K substitution. Antimicrob. Agents Chemother. 56:271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lok AS, et al. 2012. Preliminary study of two antiviral agents for hepatitis C genotype 1. N. Engl. J. Med. 366:216–224 [DOI] [PubMed] [Google Scholar]
- 24. Ma H, et al. 2007. Characterization of the metabolic activation of hepatitis C virus nucleoside inhibitor beta-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine (PSI-6130) and identification of a novel active 5′-triphosphate species. J. Biol. Chem. 282:29812–29820 [DOI] [PubMed] [Google Scholar]
- 25. McCown M, Rajyaguru S, Kular S, Cammack N, Najera I. 2009. Suppression of in vitro resistance development after treatment of the HCV replicon with NS3/4a inhibitor ITMN-191 (R7227) in combination with nucleoside inhibitor R7128 or R1626. J. Hepatol. 50(Suppl 1):S349 [Google Scholar]
- 26. Merck & Co. Inc 2011. Victrelis (boceprevir) prescribing information. Merck & Co. Inc., Whitehouse Station, NJ: http://www.accessdata.fda.gov/drugsatfda_docs/label/2011/202258lbl.pdf [Google Scholar]
- 27. Migliaccio G, et al. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164–49170 [DOI] [PubMed] [Google Scholar]
- 28. Pawlotsky JM. 2011. Treatment failure and resistance with direct-acting antiviral drugs against hepatitis C virus. Hepatology 53:1742–1751 [DOI] [PubMed] [Google Scholar]
- 29. Pockros P, et al. 2011. First SVR data with the nucleoside analogue polymerase inhibitor mericitabine (RG7128) combined with peginterferon/ribavirin in treatment-naive HCV G1/4 patients: interim analysis from the JUMP-C trial. J. Hepatol. 54:S538 [Google Scholar]
- 30. Poordad F, et al. 2011. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 364:1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reesink H, et al. 2009. Antiviral activity and safety of TMC435 combined with pegylated interferon and ribavirin in hepatitis C patients with genotype 1 who had previous exposure to TMC435. Hepatology 50(Suppl):1024A [Google Scholar]
- 32. Reesink HW, et al. 2006. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase Ib, placebo-controlled, randomized study. Gastroenterology 131:997–1002 [DOI] [PubMed] [Google Scholar]
- 33. Rong L, Dahari H, Ribeiro RM, Perelson AS. 2010. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci. Transl. Med. 2:30ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sarrazin C, et al. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767–1777 [DOI] [PubMed] [Google Scholar]
- 35. Sarrazin C, et al. 2007. SCH 503034, a novel hepatitis C virus protease inhibitor, plus pegylated interferon alpha-2b for genotype 1 nonresponders. Gastroenterology 132:1270–1278 [DOI] [PubMed] [Google Scholar]
- 36. Seiwert S, et al. Sequence variation of NS3/4A in HCV replicons exposed to ITMN-191 concentrations encompassing those likely to be achieved following clinical dosing. J. Hepatol. 46(Suppl 1):S244–S245 [Google Scholar]
- 37. Stuyver LJ, et al. 2006. Inhibition of hepatitis C replicon RNA synthesis by beta-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine: a specific inhibitor of hepatitis C virus replication. Antivir. Chem. Chemother. 17:79–87 [DOI] [PubMed] [Google Scholar]
- 38. Susser S, et al. 2009. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology 50:1709–1718 [DOI] [PubMed] [Google Scholar]
- 39. Trevino A, et al. 2011. Natural polymorphisms associated with resistance to new antivirals against HCV in newly diagnosed HIV-HCV-coinfected patients. Antivir. Ther. 16:413–416 [DOI] [PubMed] [Google Scholar]
- 40. Uzgiris A, et al. High genetic barrier to HCV resistance presented by PSI-6130, abstr 83. Global Antiviral J. 3(Suppl 2):89 [Google Scholar]
- 41. Vertex Pharmaceuticals Ltd 2011. Incivek (telaprevir) prescribing information. Vertex Pharmaceuticals Ltd., Cambridge, MA: http://www.Natap.org/2011/HCV/INCIVEKUSPI.pdf [Google Scholar]
- 42. Zhou Y, et al. 2008. Phenotypic characterization of resistant Val36 variants of hepatitis C virus NS3-4A serine protease. Antimicrob. Agents Chemother. 52:110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]