Abstract
Daptomycin-nonsusceptible (DNS) Staphylococcus aureus is found in difficult-to-treat infections, and the optimal therapy is unknown. We investigated the activity of high-dose (HD) daptomycin plus trimethoprim-sulfamethoxazole de-escalated to HD daptomycin or trimethoprim-sulfamethoxazole against 4 clinical DNS methicillin-resistant S. aureus (MRSA) isolates in an in vitro pharmacokinetic/pharmacodynamic model of simulated endocardial vegetations (109 CFU/g). Simulated regimens included HD daptomycin at 10 mg/kg/day for 14 days, trimethoprim-sulfamethoxazole at 160/800 mg every 12 h for 14 days, HD daptomycin plus trimethoprim-sulfamethoxazole for 14 days, and the combination for 7 days de-escalated to HD daptomycin for 7 days and de-escalated to trimethoprim-sulfamethoxazole for 7 days. Differences in CFU/g (at 168 and 336 h) were evaluated by analysis of variance (ANOVA) with a Tukey's post hoc test. Daptomycin MICs were 4 μg/ml (SA H9749-1, vancomycin-intermediate Staphylococcus aureus; R6212, heteroresistant vancomycin-intermediate Staphylococcus aureus) and 2 μg/ml (R5599 and R5563). Trimethoprim-sulfamethoxazole MICs were ≤0.06/1.19 μg/ml. HD daptomycin plus trimethoprim-sulfamethoxazole displayed rapid bactericidal activity against SA H9749-1 (at 7 h) and R6212 (at 6 h) and bactericidal activity against R5599 (at 72 h) and R5563 (at 36 h). A ≥8 log10 CFU/g decrease was observed with HD daptomycin plus trimethoprim-sulfamethoxazole against all strains (at 48 to 144 h), which was maintained with de-escalation to HD daptomycin or trimethoprim-sulfamethoxazole at 336 h. The combination for 14 days and the combination for 7 days de-escalated to HD daptomycin or trimethoprim-sulfamethoxazole was significantly better than daptomycin monotherapy (P < 0.05) and trimethoprim-sulfamethoxazole monotherapy (P < 0.05) at 168 and 336 h. Combination therapy followed by de-escalation offers a novel bactericidal therapeutic alternative for high-inoculum, serious DNS MRSA infections.
INTRODUCTION
Daptomycin is a concentration-dependent cyclic lipopeptide antibiotic with activity against Gram-positive organisms that exerts its bactericidal activity by binding to and inserting into bacterial membranes, thereby leading to rapid membrane depolarization and deregulation of several cell functions, such as DNA, RNA, and protein synthesis (9, 18, 31). Daptomycin susceptibility in Staphylococcus aureus is defined as a MIC of ≤1 μg/ml, and any strains with a MIC of >1 μg/ml are referred to as daptomycin nonsusceptible (DNS) (8). The rate of DNS S. aureus in North America over the last several years is low (0.01 to 0.1%), with no trend in increasing MIC values (7, 25, 29, 30). The development of DNS in S. aureus is due to a series of incremental changes in the cell membrane and wall due to changes in gene expression or the development of genetic mutations that increase the MIC and eventually lead to DNS strains (13–16, 23, 27, 33, 37). This variability in genetic alterations leading to DNS may explain why daptomycin retains some activity against these strains. We have previously demonstrated that daptomycin at simulated doses of 6 mg/kg/day and 10 mg/kg/day in an in vitro pharmacokinetic/pharmacodynamic (PK/PD) model maintained bactericidal activity against some DNS S. aureus strains, although time to bactericidal activity was increased and regrowth often occurred (26, 28).
The development of DNS in S. aureus has occurred mainly in patients with complicated infections, such as osteomyelitis, septic arthritis, or endocarditis (6, 32). Optimal therapy for these types of infections requires agents with bactericidal activity to ensure a rapid reduction of the bacterial burden and the highest probability of clinical cure. Unfortunately, clinicians commonly turn to bacteriostatic agents such as linezolid, tigecycline, trimethoprim-sulfamethoxazole, clindamycin, and tetracyclines when they encounter a DNS S. aureus infection. Clinical data on successful treatment of infections caused by DNS S. aureus is limited to a few case reports (32). The optimal therapy for the treatment of complicated infections caused by DNS S. aureus in which a bactericidal regimen is desired is currently unknown. Utilizing the activity daptomycin maintains against DNS strains, we have previously demonstrated a novel combination of daptomycin at 6 mg/kg/day, and trimethoprim-sulfamethoxazole was rapidly bactericidal against two DNS S. aureus strains, including one that was also a heteroresistant vancomycin-intermediate Staphylococcus aureus (hVISA) strain, in a 3-day in vitro PK/PD model of simulated endocardial vegetations (SEV) (35). Since our first study utilized only two strains, a relatively short duration of treatment, and the standard dose of daptomycin (6 mg/kg/day), further data are needed to characterize the potential benefit of this novel antibiotic combination. As daptomycin therapy is often salvage therapy after vancomycin and the potential exists under these circumstances to influence daptomycin's activity, it would be prudent to utilize high-dose daptomycin (10 mg/kg) and therefore examine this dose in combination with trimethoprim-sulfamethoxazole over a prolonged period of time in the in vitro PK/PD SEV model. Additionally, since the types of infections caused by DNS S. aureus strains often require therapy lasting several weeks, the optimal length of combination therapy would need to be evaluated. The objective of the study was to examine the combination of high-dose daptomycin with trimethoprim-sulfamethoxazole for 14 days or de-escalated to daptomycin or trimethoprim-sulfamethoxazole at day 7 against 4 clinical DNS methicillin-resistant Staphylococcus aureus (MRSA) strains in a 14-day in vitro PK/PD model with SEV.
MATERIALS AND METHODS
Bacterial strains.
A total of four clinical DNS MRSA isolates were evaluated: SA H9749-1 (VISA isolate recovered from a patient with osteomyelitis at St. Joseph's Hospital Center, Syracuse, NY), R6212 (hVISA isolate from Detroit Medical Center), R5599 (MRSA blood isolate from Albany Medical Center), and R5563 (MRSA blood isolate).
Antimicrobials.
Daptomycin was provided by Cubist Pharmaceuticals. Trimethoprim-sulfamethoxazole was commercially purchased from Sigma-Aldrich Co. (St. Louis, MO). Stock solutions of each antimicrobial were prepared daily per Clinical and Laboratory Standards Institute recommendations (8).
Media.
Mueller-Hinton II broth (Difco, Detroit, MI) with 25 mg/liter calcium and 12.5 mg/liter magnesium (MHBII) was used for MIC testing and in vitro PK/PD models, except for those with daptomycin. Due to the dependency of daptomycin on calcium for antimicrobial activity, Mueller-Hinton II broth was supplemented (SMHBII) to 50 mg/liter calcium for MIC testing and to 75 mg/liter calcium (equivalent to 50 mg/liter of calcium in the presence of albumin) for in vitro PK/PD models (19). Colony counts were determined using tryptic soy agar (TSA; Difco, Detroit, MI) plates. The development of resistance to daptomycin and trimethoprim-sulfamethoxazole was evaluated using either MHBII and agar (Bacto; Difco, Detroit, MI) supplemented with 50 mg/liter of calcium (due to the dependency of daptomycin on calcium for antimicrobial activity) or brain heart infusion agar (BHIA; Difco, Detroit, MI) plates containing the respective antimicrobial at a concentration of three times the MIC.
Susceptibility testing.
MICs of study antimicrobial agents were determined by broth microdilution at 106 CFU/ml according to Clinical and Laboratory Standards Institute guidelines (8). All were incubated at 37°C for 18 to 24 h.
SEVs.
Organism stocks were prepared by inoculating three TSA plates for overnight growth at 37°C. Organisms were swabbed from the growth plates into 5-ml test tubes containing SMHBII, resulting in a concentration of approximately 1010 CFU/ml. Simulated endocardial vegetations (SEVs) were prepared in 1.5 ml siliconized Eppendorf tubes by mixing 0.05 ml of organism suspension (final inoculum, 109 CFU/g), 0.5 ml of human cryoprecipitated antihemophilic factor from volunteer donors (American Red Cross, Detroit, MI), and 0.025 ml of platelet suspension (platelets mixed with normal saline; 250,000 to 500,000 platelets per clot; American Red Cross, Detroit, MI). Bovine thrombin (0.05 ml; 5,000 U/ml; GenTrac, Inc., Middleton, WI) was added to each tube after insertion of a sterile monofilament line into the mixture. The resultant simulated vegetations were then removed from the Eppendorf tubes with a sterile disposable plastic needle (Becton, Dickinson, Sparks, MD) and introduced into the model. This methodology results in SEVs consisting of approximately 3 to 3.5 g/dl of albumin and 6.8 to 7.4 g/dl of total protein (1).
In vitro PK/PD model.
A two-compartment in vitro model consisting of a 250-ml glass apparatus with ports where the SEVs were suspended was utilized for all simulations. The apparatus was prefilled with media, and antibiotics were administered as boluses over a 336-h time period into the central compartment via an injection port. The model apparatus was placed in a 37°C water bath throughout the procedure, and a magnetic stir bar was placed in the media for thorough mixing of the drug in the model. Fresh medium was continuously supplied and removed from the compartment along with the drug via a peristaltic pump (Masterflex; Cole-Parmer Instrument Company, Chicago, IL) set to simulate the half-lives of the antibiotics. Simulated antibiotic regimens were daptomycin at 10 mg/kg every 24 h (estimated total peak and half-life [T1/2], 141.1 mg/liter and 8 h, respectively) for 14 days and trimethoprim-sulfamethoxazole at 160/800 mg every 12 h (2.4/100 mg/liter, 10/10 h) for 14 days (4, 20). Combination simulated antibiotic regimens included daptomycin plus trimethoprim-sulfamethoxazole for 14 days, daptomycin plus trimethoprim-sulfamethoxazole for 7 days followed by daptomycin for 7 days, and daptomycin plus trimethoprim-sulfamethoxazole for 7 days followed by trimethoprim-sulfamethoxazole for 7 days. The elimination rate for the combination models was set for the antibiotic with the shorter half-life, and the antibiotic with the longer half-life was supplemented into the second chamber (5). All simulated drug regimens were performed in duplicate to ensure reproducibility.
Pharmacodynamic analysis.
Two simulated endocardial vegetations were removed from each model at 0, 4, 8, 24, 32, 48, 72, 96, 120, 144, 168, 176, 192, 200, 216, 240, 264, 288, 312, and 336 h. The SEVs were homogenized and diluted in cold saline to be plated onto TSA plates. To minimize antibiotic carryover, all samples were diluted at least 10-fold before plating. If the drug concentration at the anticipated dilution was within 1 tube dilution of the MIC or higher, then vacuum filtration (100-μl samples washed through a 0.45-μm filter with normal saline) was used to avoid antibiotic carryover. Plates were then incubated at 37°C for 24 h, at which time a colony count was performed. These methods have a lower limit of detection of 1 log10 CFU/g. The total reduction in log10 CFU/g over 336 h was determined by plotting time-kill curves based on the number of remaining organisms over the 336-h time period. Bactericidal activity (99.9% kill) was defined as a ≥3-log10 CFU/g reduction in colony count from the initial inoculum. Bacteriostatic activity was defined as a <3-log10 CFU/g reduction in colony count from the initial inoculum, while inactive was defined as no observed reductions from the initial inocula. The time to achieve a 99.9% bacterial load reduction was determined by linear regression (if r2 ≥ 0.95) or by visual inspection.
Pharmacokinetic analysis.
Pharmacokinetic samples were obtained, through the injection port of each model (duplicate samples) at 0, 4, 8, 24, 32, 48, 72, 96, 120, 144, 168, 176, 192, 200, 216, 240, 264, 288, 312, and 336 h for verification of target antibiotic concentrations. All samples were stored at −70°C until they were ready for analysis. Daptomycin concentrations were determined by a previously validated bioassay (Micrococcus luteus ATCC 9341; antibiotic assay medium 1; standards of 200, 100, and 50 mg/liter) (2). Blank 1/4-in disks were placed on a preswabbed plate and spotted with 10 μl of the standard or sample. For trimethoprim, we utilized a bioassay (S. aureus R1867; Mueller-Hinton Agar; standards of 40, 20, and 10 mg/liter) (35). Briefly, 1/4-in holes were punched in the preswabbed agar plate and filled with 50 μl of the standards or samples. This assay has a lower limit of detection of 10 mg/liter and a between-day percent coefficient of variation of 7.15. A separate trimethoprim model was run at 10 times the targeted concentration to utilize this assay due to detection limits. For sulfamethoxazole, we utilized a bioassay (S. aureus R1867; Mueller-Hinton agar; standards of 125, 75, and 50 mg/liter) (35). Briefly, 1/4-in holes were punched in the preswabbed agar plate and filled with 50 μl of the standard or sample, which contained 6 mg/liter of trimethoprim. This assay had a between-day coefficient of variation of 9.0%. A separate model was run for sulfamethoxazole to evaluate pharmacokinetic parameters. Each standard was tested in duplicate. Plates were incubated for 18 to 24 h at 37°C, at which time the zone sizes were measured using a protocol reader (Microbiology International, Frederick, MD). The half-lives, areas under the concentration-time curve from 0 to 24 h (AUC0-24 h), AUC24 h/MIC, and peak concentrations of the antibiotics were determined by the trapezoidal method utilizing PK Analyst software (version 1.10; MicroMath Scientific Software, Salt Lake City, UT).
Resistance.
Development of resistance was evaluated at 168 and 336 h. One hundred-μl samples from each time point were plated on BHIA or SMHBII agar plates containing three times the drug MIC to assess the development of resistance. Plates were examined for growth after 48 h of incubation at 37°C.
Statistical analysis.
Changes in CFU/g at 168 and 336 h were compared by two-way analysis of variance with Tukey's post hoc test. A P value of ≤0.05 was considered significant. All statistical analyses were performed using SPSS statistical software (Release 20.0; SPSS, Inc., Chicago, IL).
RESULTS
MIC values for the evaluated isolates are displayed in Table 1. All isolates are DNS and trimethoprim-sulfamethoxazole susceptible. Isolate SA H9749-1 is a VISA strain (vancomycin MIC of 8 to 16 μg/ml via broth microdilution), and isolate R6212 was an hVISA strain (ratio with Mu3 of 0.96 by modified population analysis). Observed pharmacokinetic parameters (total peak and T1/2) were 145.8 to 146.1 mg/liter (targeted at 141.1 mg/liter) and 6.9 to 8.2 h (targeted at 8 h) for daptomycin, 23.6 to 25.1 mg/liter (targeted at 24 [2.4 mg/liter × 10]) and 9.3 to 10.1 h (targeted at 10 h) for trimethoprim, and 98 to 104.8 mg/liter (targeted at 100 mg/liter) and 9.6 to 10.2 h (targeted at 10 h) for sulfamethoxazole.
Table 1.
MICs for evaluated isolatesa
| Isolate | MICs (μg/ml) |
||
|---|---|---|---|
| DAP | SXT | VAN | |
| SA H9749-1 | 4 | 0.03/0.6 | 8–16 |
| R6212 | 4 | 0.06/1.19 | 2 |
| R5599 | 2 | 0.06/1.19 | 1 |
| R5563 | 2 | 0.03/0.6 | 2 |
DAP, daptomycin; SXT, trimethoprim-sulfamethoxazole; VAN, vancomycin.
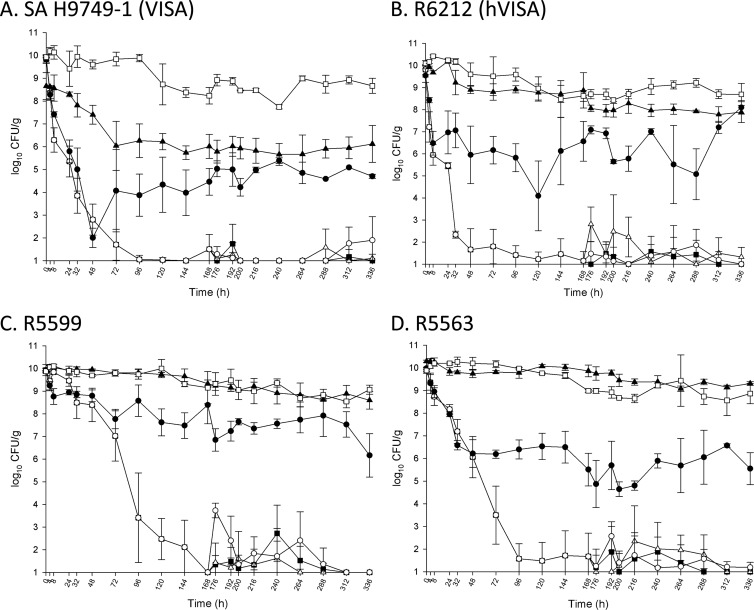
The quantitative changes in the log10 CFU/g for the tested regimens against the four tested strains are displayed in Fig. 1A to D and Table 2. Against all four tested strains, the novel combination of daptomycin plus trimethoprim-sulfamethoxazole for the first 7 days produced a greater than 8 log10 CFU/g reduction by 168 h (7 days), and this decrease was sustained to 336 h (14 days) for the combination regimen and de-escalation to daptomycin at 10 mg/kg or trimethoprim-sulfamethoxazole. The combination of daptomycin plus trimethoprim-sulfamethoxazole produced rapid and sustained bactericidal activity against the VISA strain SA H9749-1 (7 h) and the hVISA strain R6212 (6 h). Time to sustained bactericidal activity with the combination of daptomycin plus trimethoprim-sulfamethoxazole was longer for MRSA R5599 (72 h) and MRSA R5563 (36 h). Trimethoprim-sulfamethoxazole was not bactericidal against any tested strains. Daptomycin alone displayed sustained bactericidal activity against two of the tested strains at 14 h (VISA strain SA H9749-1) and 32 h (MRSA strain R5563). Daptomycin plus trimethoprim-sulfamethoxazole, the combination de-escalated to daptomycin, and the combination de-escalated to trimethoprim-sulfamethoxazole were significantly better than daptomycin (P < 0.05), trimethoprim-sulfamethoxazole (P < 0.05), and the growth control (P < 0.05) at 168 and 336 h. No organisms with MIC values of 3× the initial MIC or greater were recovered for either drug at 168 or 336 h.
Fig 1.
Activity of tested regimens in the in vitro pharmacokinetic/pharmacodynamic model of simulated endocardial vegetations. Closed squares, daptomycin plus trimethoprim-sulfamethoxazole (DSXT) for 14 days; open triangle, DSXT for 7 days followed by SXT for 7 days; open circles, DSXT for 7 days followed by daptomycin at 10 mg/kg/day (D10) for 7 days; closed circles, D10 for 14 days; closed triangles, SXT for 14 days; open squares, growth control.
Table 2.
Activity of tested regimens in the in vitro pharmacokinetic/pharmacodynamic modela
| Isolate | Time (h) to 99.9% kill |
Change in log10 CFU/g at: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 168 h |
336 h |
||||||||||||||
| DAP | SXT | DSXT | −DAP | −SXT | DAP | SXT | DSXT | −DAP | −SXT | DAP | SXT | DSXT | −DAP | −SXT | |
| SA H | 14 | NA | 7 | 7 | 7 | 5.35 | 2.65 | 8.34 | 8.34 | 8.34 | 5.11 | 2.54 | 8.84 | 7.94 | 8.75 |
| R6212 | NA | NA | 6 | 6 | 6 | 2.98 | 0.67 | 8.33 | 8.33 | 8.33 | 1.44 | 1.65 | 8.81 | 8.81 | 8.49 |
| R5599 | 336 | NA | 72 | 72 | 72 | 1.48 | 0.58 | 8.93 | 8.93 | 8.93 | 3.7 | 1.31 | 8.93 | 8.93 | 8.93 |
| R5563 | 32 | NA | 36 | 36 | 36 | 4.47 | 0.42 | 8.21 | 8.21 | 8.21 | 4.43 | 0.98 | 8.88 | 8.69 | 8.88 |
Time to 99.9% kill refers to the time to bactericidal activity sustained at 336 h. DAP, daptomycin at 10 mg/kg/day for 14 days; SXT, trimethoprim-sulfamethoxazole for 14 days; DSXT, daptomycin plus trimethoprim-sulfamethoxazole for 14 days; −DAP, daptomycin plus trimethoprim-sulfamethoxazole for 7 days followed by daptomycin for 7 days; −SXT, daptomycin plus trimethoprim-sulfamethoxazole for 7 days followed by trimethoprim-sulfamethoxazole; SA H, SA H9749-1; NA, not applicable.
DISCUSSION
As stated previously, the optimal therapy for DNS MRSA infections is complicated, as the majority are deep-seated, high-inoculum infections requiring long-term therapy, and evidence is limited (6, 21). Optimal therapy would be cost-effective, have minimal side effects, and display bactericidal activity, particularly for endocarditis, in which failure due to bacteriostatic antibiotics is well documented (12, 24). Daptomycin nonsusceptibility in S. aureus is a complicated phenotype that can arise from multiple genetic alterations and is variable among strains. The mechanism of DNS in S. aureus is due to the additive effects of many potential changes found in some, but not all, strains. One common mechanism is increased cell surface charge, thus repulsing the positively charged daptomycin-calcium complex. This process may occur via translocation of positively charged phospholipids to the outer side of the cytoplasmic membrane, lysinylation of membrane phosphatidylglycerol, and/or increased d-alanylation of cell wall teichoic acids (10, 11, 14, 17, 23, 36). Increased cell wall thickness, alterations in membrane proteins, and changes in membrane fluidity are also often associated with DNS S. aureus (10, 16, 22). Since DNS in S. aureus is due to the cumulative affect of many of these changes and is not due to one change conferring high-level nonsusceptibility, it is rational that daptomycin retains activity against some of these strains. Previous results from our laboratory have shown that daptomycin maintains variable levels of activity against DNS S. aureus (26, 28). The results of the current study were consistent with daptomycin demonstrating delayed bactericidal activity against VISA SA H9749-1 (14 h) and MRSA R5563 (32 h) and a lack of sustained bactericidal activity against the other two strains. The enhancement of this remaining activity in combination with trimethoprim-sulfamethoxazole was examined in this study as a potential combination therapy for DNS MRSA strains.
In this study, we evaluated the activity of daptomycin plus trimethoprim-sulfamethoxazole against four clinical DNS MRSA strains, including one VISA and one hVISA strain, under high-inoculum conditions (>9.5 log10 CFU/g). The combination was able to produce a substantial reduction of greater than 8 log10 CFU/g against all four strains tested. Additionally, for 168 to 336 h (days 7 to 14), the combination, de-escalation to daptomycin, and de-escalation to trimethoprim-sulfamethoxazole were able to sustain this decrease in colony counts and keep viable colony counts close to detection limits. Based on these in vitro data, the combination of daptomycin plus trimethoprim-sulfamethoxazole provides a possible treatment option for high-inoculum infections with DNS MRSA, including those in which bactericidal therapy would be optimal. The high susceptibility of S. aureus to trimethoprim-sulfamethoxazole of greater than 98% among clinical isolates in the United States supports the potential wide use of this combination (30). Additionally, these in vitro data support the de-escalation to either daptomycin or trimethoprim-sulfamethoxazole. This ability to de-escalate while maintaining regimen activity provides clinicians with the option to stop one agent if necessary due to cost (daptomycin), side effects (increased serum creatinine with trimethoprim-sulfamethoxazole or increased creatine phosphokinase with daptomycin), or regimen complexity. Interestingly, time to bactericidal activity was shorter for the VISA (SA H9749-1) and hVISA (R6212) strains than for the two tested MRSA strains R5599 and R5563. While the mechanism for this increased activity is unknown, it is possible the mechanism of synergy for daptomycin plus trimethoprim-sulfamethoxazole is more potent against these isolates.
Preliminary clinical data have been favorable regarding this combination to treat osteomyelitis caused by S. aureus with reduced susceptibility to both vancomycin and daptomycin (34). The first patient received high-dose daptomycin and trimethoprim-sulfamethoxazole as salvage therapy for vertebral osteomyelitis with SA H9749-1 (vancomycin MIC of 8 to 16 and daptomycin MIC of 4 μg/ml) and achieved a clinical cure. The second patient received daptomycin plus trimethoprim-sulfamethoxazole as salvage therapy for chronic vertebral osteomyelitis with concurrent prolonged bacteremia from SA F31774 (vancomycin MIC of 4 and daptomycin MIC of 4 μg/ml). This combination cleared the bacteremia in 48 h but was stopped after 7 days due to an increase in serum creatinine. The patient's regimen was then changed to daptomycin plus rifampin. Unfortunately, the patient subsequently deteriorated for a week on this combination before dying.
While this study was strengthened by its use of four DNS MRSA strains with various levels of susceptibility to vancomycin and its 14-day study duration, it still has some limitations. While the treatment duration in this in vitro model of simulated endocardial vegetations was 14 days, this is still shorter than the 4 to 6 weeks typically required to treat infections such as staphylococcal endocarditis (3). Based on data for the second patient reported above, more than 1 week of combination therapy may be required in vivo before de-escalation. In conclusion, the combination of daptomycin plus trimethoprim-sulfamethoxazole produced sustained bactericidal activity against four DNS MRSA strains, including one VISA strain and one hVISA strain, and this inoculum decrease was maintained when de-escalating to daptomycin or trimethoprim-sulfamethoxazole. Further clinical research examining this combination and the optimal time to de-escalate is warranted.
ACKNOWLEDGMENTS
We are grateful to St. Joseph's Hospital Center, Syracuse, NY, for SA H9749-1 and to Albany Medical Center for R5599.
This work was funded by an investigator-initiated grant from Cubist Pharmaceuticals.
M.J.R. has received grant support, consulted for, or provided lectures for Astellas, Cubist, Forest, Clinical Therapeutics, and Rib-X and is supported in part by grant R21AI092055 for the NIAID.
Footnotes
Published ahead of print 20 August 2012
REFERENCES
- 1. Akins RL, Rybak MJ. 2001. Bactericidal activities of two daptomycin regimens against clinical strains of glycopeptide intermediate-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus faecium, and methicillin-resistant Staphylococcus aureus isolates in an in vitro pharmacodynamic model with simulated endocardial vegetations. Antimicrob. Agents Chemother. 45:454–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akins RL, Rybak MJ. 2000. In vitro activities of daptomycin, arbekacin, vancomycin, and gentamicin alone and/or in combination against glycopeptide intermediate-resistant Staphylococcus aureus in an infection model. Antimicrob. Agents Chemother. 44:1925–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baddour LM, et al. 2005. Infective endocarditis: diagnosis, antimicrobial therapy, and management of complications: a statement for healthcare professionals from the Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease, Council on Cardiovascular Disease in the Young, and the Councils on Clinical Cardiology, Stroke, and Cardiovascular Surgery and Anesthesia, American Heart Association: endorsed by the Infectious Diseases Society of America. Circulation 111:e394–e434 doi:10.1161/CIRCULATIONAHA.105.165564 [DOI] [PubMed] [Google Scholar]
- 4. Benvenuto M, Benziger DP, Yankelev S, Vigliani G. 2006. Pharmacokinetics and tolerability of daptomycin at doses up to 12 milligrams per kilogram of body weight once daily in healthy volunteers. Antimicrob. Agents Chemother. 50:3245–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blaser J. 1985. In-vitro model for simultaneous simulation of the serum kinetics of two drugs with different half-lives. J. Antimicrob. Chemother. 15(Suppl A):125–130 [DOI] [PubMed] [Google Scholar]
- 6. Boucher HW, Sakoulas G. 2007. Perspectives on daptomycin resistance, with emphasis on resistance in Staphylococcus aureus. Clin. Infect. Dis. 45:601–608 [DOI] [PubMed] [Google Scholar]
- 7. Castanheira M, Jones RN, Sader HS. 2008. Update of the in vitro activity of daptomycin tested against 6710 Gram-positive cocci isolated in North America (2006). Diagn. Microbiol. Infect. Dis. 61:235–239 [DOI] [PubMed] [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2011. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 9th ed Approved standard M7-A9. CLSI, Wayne, PA [Google Scholar]
- 9. Cotroneo N, Harris R, Perlmutter N, Beveridge T, Silverman JA. 2008. Daptomycin exerts bactericidal activity without lysis of Staphylococcus aureus. Antimicrob. Agents Chemother. 52:2223–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cui L, et al. 2010. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 54:5222–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ernst CM, et al. 2009. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 5:e1000660 doi:10.1371/journal.ppat.1000660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Finberg RW, et al. 2004. The importance of bactericidal drugs: future directions in infectious disease. Clin. Infect. Dis. 39:1314–1320 [DOI] [PubMed] [Google Scholar]
- 13. Friedman L, Alder JD, Silverman JA. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:2137–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jones T, et al. 2008. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 52:269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Julian K, et al. 2007. Characterization of a daptomycin-nonsusceptible vancomycin-intermediate Staphylococcus aureus strain in a patient with endocarditis. Antimicrob. Agents Chemother. 51:3445–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaatz GW, Lundstrom TS, Seo SM. 2006. Mechanisms of daptomycin resistance in Staphylococcus aureus. Int. J. Antimicrob. Agents 28:280–287 [DOI] [PubMed] [Google Scholar]
- 17. Kilelee E, Pokorny A, Yeaman MR, Bayer AS. 2010. Lysyl-phosphatidylglycerol attenuates membrane perturbation rather than surface association of the cationic antimicrobial peptide 6W-RP-1 in a model membrane system: implications for daptomycin resistance. Antimicrob. Agents Chemother. 54:4476–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Laganas V, Alder J, Silverman JA. 2003. In vitro bactericidal activities of daptomycin against Staphylococcus aureus and Enterococcus faecalis are not mediated by inhibition of lipoteichoic acid biosynthesis. Antimicrob. Agents Chemother. 47:2682–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lamp KC, Rybak MJ, Bailey EM, Kaatz GW. 1992. In vitro pharmacodynamic effects of concentration, pH, and growth phase on serum bactericidal activities of daptomycin and vancomycin. Antimicrob. Agents Chemother. 36:2709–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. LaPlante KL, Leonard SN, Andes DR, Craig WA, Rybak MJ. 2008. Activities of clindamycin, daptomycin, doxycycline, linezolid, trimethoprim-sulfamethoxazole, and vancomycin against community-associated methicillin-resistant Staphylococcus aureus with inducible clindamycin resistance in murine thigh infection and in vitro pharmacodynamic models. Antimicrob. Agents Chemother. 52:2156–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu C, et al. 2011. Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin. Infect. Dis. 52:e18–e55 doi:10.1093/cid/ciq146 [DOI] [PubMed] [Google Scholar]
- 22. Mishra NN, et al. 2011. Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides. Antimicrob. Agents Chemother. 55:526–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mishra NN, et al. 2009. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:2312–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pankey GA, Sabath LD. 2004. Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of Gram-positive bacterial infections. Clin. Infect. Dis. 38:864–870 [DOI] [PubMed] [Google Scholar]
- 25. Pfaller MA, Sader HS, Jones RN. 2007. Evaluation of the in vitro activity of daptomycin against 19615 clinical isolates of Gram-positive cocci collected in North American hospitals (2002–2005). Diagn. Microbiol. Infect. Dis. 57:459–465 [DOI] [PubMed] [Google Scholar]
- 26. Rose WE, Leonard SN, Rybak MJ. 2008. Evaluation of daptomycin pharmacodynamics and resistance at various dosage regimens against Staphylococcus aureus isolates with reduced susceptibilities to daptomycin in an in vitro pharmacodynamic model with simulated endocardial vegetations. Antimicrob. Agents Chemother. 52:3061–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rose WE, et al. 2008. Daptomycin activity against Staphylococcus aureus following vancomycin exposure in an in vitro pharmacodynamic model with simulated endocardial vegetations. Antimicrob. Agents Chemother. 52:831–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rose WE, Rybak MJ, Kaatz GW. 2007. Evaluation of daptomycin treatment of Staphylococcus aureus bacterial endocarditis: an in vitro and in vivo simulation using historical and current dosing strategies. J. Antimicrob. Chemother. 60:334–340 [DOI] [PubMed] [Google Scholar]
- 29. Sader HS, et al. 2009. Evaluation of vancomycin and daptomycin potency trends (MIC creep) against methicillin-resistant Staphylococcus aureus isolates collected in nine U.S. medical centers from 2002 to 2006. Antimicrob. Agents Chemother. 53:4127–4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sader HS, Jones RN. 2009. Antimicrobial susceptibility of Gram-positive bacteria isolated from US medical centers: results of the Daptomycin Surveillance Program (2007–2008). Diagn. Microbiol. Infect. Dis. 65:158–162 [DOI] [PubMed] [Google Scholar]
- 31. Safdar N, Andes D, Craig WA. 2004. In vivo pharmacodynamic activity of daptomycin. Antimicrob. Agents Chemother. 48:63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sharma M, Riederer K, Chase P, Khatib R. 2008. High rate of decreasing daptomycin susceptibility during the treatment of persistent Staphylococcus aureus bacteremia. Eur. J. Clin. Microbiol. Infect. Dis. 27:433–437 [DOI] [PubMed] [Google Scholar]
- 33. Silverman JA, Oliver N, Andrew T, Li T. 2001. Resistance studies with daptomycin. Antimicrob. Agents Chemother. 45:1799–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Steed M, Avery L, Woodruff A, Rybak M. September 17, 2011. Evaluation of daptomycin (DAP) plus trimethoprim-sulfamethoxazole (TMP/SMX) or rifampin activity in an in vitro pharmacokinetic/pharmacodynamic (PK/PD) model against VISA and daptomycin non-susceptible Staphylococcus aureus (SA) isolates from osteomyelitis cases, abstr. A2-019. Abstr. 51st Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC [Google Scholar]
- 35. Steed ME, Vidaillac C, Rybak MJ. 2010. Novel daptomycin combinations against daptomycin-nonsusceptible methicillin-resistant Staphylococcus aureus in an in vitro model of simulated endocardial vegetations. Antimicrob. Agents Chemother. 54:5187–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang SJ, et al. 2009. Enhanced expression of dltABCD is associated with the development of daptomycin nonsusceptibility in a clinical endocarditis isolate of Staphylococcus aureus. J. Infect. Dis. 200:1916–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang SJ, et al. 2009. Regulation of mprF in daptomycin-nonsusceptible Staphylococcus aureus strains. Antimicrob. Agents Chemother. 53:2636–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]