Abstract
Phage therapy against bacterial pathogens has been resurrected as an alternative and supplementary anti-infective modality. Here, we observed that bacterial group motilities were impaired in Pseudomonas aeruginosa strain PA14 lysogens for some temperate siphophages; the PA14 lysogens for DMS3 and MP22 were impaired in swarming motility, whereas the PA14 lysogen for D3112 was impaired in twitching motility. The swarming and twitching motilities of PA14 were also affected in the presence of MP22 and D3112, respectively. The in vitro killing activities of D3112 and MP22 toward PA14 did not differ, and neither did their in vivo persistence in the absence of bacterial infections in mice as well as in flies. Nevertheless, administration of D3112, not MP22, significantly reduced the mortality and the bacterial burdens in murine peritonitis-sepsis and Drosophila systemic infection caused by PA14. Taken together, we suggest that a temperate phage-mediated twitching motility inhibition might be comparably effective to control the acute infections caused by P. aeruginosa.
INTRODUCTION
Pseudomonas aeruginosa causes a variety of infections, mostly in those who suffer from undesirable loss or abnormality in physicochemical or biological defense barriers against infections. Those include traumatic skin damage (burns), abnormal anatomy (some ear infections), abnormal secretion (cystic fibrosis), and compromised immunity. Several studies have revealed that clinical P. aeruginosa isolates from cystic fibrosis (CF) patients who suffer from persistent biofilm infections display a profound clonal diversity (20). Biofilm-mediated diversification may help enhance the ecological fitness of P. aeruginosa (2), even in the presence of conventional antibiotic therapy, which leads to the emergence of multiple-antibiotic-resistant bacterial clones (7, 17). Thus, physicians treating CF patients have been increasingly faced with infections caused by multiple-antibiotic-resistant isolates of P. aeruginosa. Such infections are generally treated by using two antibiotics together, which have different antibacterial mechanisms (6). The exploitation of this so-called antibiotic cocktail therapy is believed to increase the likelihood of therapeutic success through antimicrobial synergy (9, 14).
Renewed attention has been recently paid to phage therapy, which was proposed as an alternative or supplementary anti-infective modality to the conventional antibiotic therapy in controlling bacterial infections (8). Phages have been exploited to treat bacterial infections in Eastern Europe since the early 20th century (22). Phage therapy has been successful in humans for treating infectious diseases caused by various bacteria including Staphylococcus aureus, Streptococcus pneumoniae, Escherichia coli, Shigella, and Salmonella enterica, as well as P. aeruginosa (24). Furthermore, there is evidence for better therapeutic efficacy of phages than of antibiotics in that phages were successful in controlling biofilm-forming microorganisms such as P. aeruginosa, which are tolerant to the known antimicrobial drugs (13).
The prevalence of multiple-antibiotic-resistant clinical P. aeruginosa isolates has led us to explore phage therapy for managing P. aeruginosa-mediated infections, based on various natures of the virulent phages. In recent years, the therapeutic use of phages was shown to be effective in clinical ear infections caused by this bacterium (23). Moreover, the therapeutic efficacy of isolated or genetically engineered phages has been extensively investigated in various experimental models of infection by P. aeruginosa that include mouse burn wound infection (18) and gut-derived sepsis (26). We also established a nonmammalian infection model based on Drosophila systemic infection to evaluate the antibacterial efficacy of two virulent phages (12).
The growing information on the diversity and the therapeutic efficacy of phages in various infection models and the technical advances in engineering phages at the genome level (16) have prompted us to increase the repertoire of phages as leading platforms by isolating new phages and evaluating their antibacterial efficacy. Here, based on the observation that group motilities such as swarming and twitching of P. aeruginosa were affected by lysogenization of some Siphoviridae temperate phages, we evaluated their therapeutic efficacy against virulent P. aeruginosa strains.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Pseudomonas aeruginosa strain PA14 and its isogenic in-frame deletion mutants for type VI pili (pilA) and for flagella (flgK) were used as described previously (11). Bacterial cells were grown in Luria-Bertani (LB) (1% tryptone, 0.5% yeast extract, and 1% NaCl) broth with aeration or 2% Bacto agar (Difco) LB or cetrimide agar (Pseudomonas isolation agar; Difco) plates at 37°C. Overnight cultures were inoculated into the fresh LB broth with an inoculum size of 1.6 × 107 CFU/ml and grown at 37°C for 3 to 5 h with agitation to the early stationary phase (i.e., to an optical density at 600 nm [OD600] of 3.0) and used for mouse and fly experiments.
Preparation of phage strains.
Five temperate phage strains (D3112, DMS3, MP22, MP29, and MP42) were enriched by the plate lysate method using P. aeruginosa strain PA14, as described elsewhere (12). Phage lysate obtained from confluent phage plaques on LB plates by adding 5 ml (per plate) phage buffer (10 mM MgSO4, 10 mM Tris [pH 7.6], and 1 mM EDTA) was collected and centrifuged at 8,000 × g for 10 min at 4°C to remove the cell debris. Phage particles were precipitated by centrifugation at 10,000 × g for 30 min at 4°C from the lysate supernatant in the presence of 1 M NaCl and 10% polyethylene glycol (average molecular weight, 8,000) and then dissolved in 5 ml phage buffer. Then, the phage particles were concentrated by ultracentrifugation at 100,000 × g for 2 h at 4°C and resuspended in phage buffer. Finally, for purification of phages, the phage suspension was placed on top of a discontinuous CsCl gradient (1.45, 1.50, and 1.70 g/ml) and centrifuged at 87,000 × g for 2 h at 4°C. Phage band was collected and dialyzed.
Motility (swarming, twitching, and swimming) assays.
Motility assays were performed as described previously (5), with some modifications. For twitching motility, an overnight-grown single colony on LB agar plates was picked and stab inoculated through a thin (about 3-mm) twitching assay plate (i.e., an LB plate containing 1.5% agar) to the bottom of the petri dish. After incubation for 48 h at 30°C, a bacterial hazy zone at the interface between the agar and the surface was observed after visualization using crystal violet. For swarming motility, modified M9 medium (20 mM NH4Cl, 12 mM Na2HPO4, 8.6 mM NaCl, 1 mM MgSO4, 1 mM CaCl2, 11 mM dextrose, supplemented with 0.5% Casamino Acids [Difco]) with 0.5% Bacto agar (Difco) was used after drying under laminar flow during 60 min. Five microliters of the stationary culture suspension was spotted onto the swarming assay plates, which were then incubated at 30°C for 16 h. Swimming assay plates (i.e., LB plates containing 0.3% agar) were inoculated with a toothpick from an overnight-grown single colony on LB agar plates and incubated at 30°C for 16 h. Motility was assessed by examining the circular turbid zone formed by the bacterial cells migrating away from the point of inoculation. To test for the effect of phages on the wild-type bacterial motilities, motility assays were performed using PA14 on the appropriate motility plates containing the phages at a concentration of ∼106 PFU/ml.
Identification of D3112 integration sites.
Arbitrary PCR followed by sequencing was used to determine the phage integration site in the D3112 lysogens under the amplification conditions as described elsewhere (4), using different sets of PCR primers: the first PCR was performed using the primers Arb1MP (5′-GMT GGA TGG TTT AGC GCT CAT TC-3′), which is specific to the left end of D3112, and Arb1D (5′-GGC CAG GCC TGC AGA TGA TGN NNN NNN NNN GTA T-3′), whose PCR products were used as the template for the second PCR, using the primers Arb2MP (5′-TCC TTG CTC AAT CCT GAT CGA TAT TTT CC-3′) and Arb2D (5′-GGC CAG GCC TGC AGA TGA G-3′). The most prominent product from the second PCR was purified and sequenced using the primer Seq-MP (5′-TTA TAG GGG TTC TAG CCT ATC C-3′).
Measurement of phage titers and lytic activity in vitro.
For measurement of phage titers, 10 μl of phage samples that may contain about 103 PFU of phages was mixed with 107 CFU of the stationary-phase bacteria in 100 μl of phage buffer. After 10 min of incubation, 3 ml of top agar was added, and the mixture was plated. Plaques were visualized after 16 to 24 h of incubation at 37°C. In vitro lytic activity was measured using PA14 and PA14-derived phage lysates as described previously (12). Overnight cultures were inoculated into the fresh LB broth containing a phage strain at 2.0 × 106 PFU/ml, with an inoculum size of 1.0 × 106 CFU/ml, and then grown at 37°C. Viable bacteria were enumerated every 30 min.
Mouse experiments.
Mouse infection was carried out using female ICR mice (aged 4 weeks), following the protocol approved by the Animal Care and Use Committee at CHA University. Bacterial cells were grown to the early stationary growth phase, harvested, washed twice with phosphate-buffered saline (PBS) buffer (2.7 mM KCl, 137 mM NaCl, 10 mM Na2HPO4, and 2 mM KH2PO4, pH 7.0), and then resuspended in PBS buffer at 2 × 106 CFU/ml. To induce peritonitis, mice were infected intraperitoneally (i.p.) with 100 μl of the bacterial suspension (i.e., 2 × 105 CFU). For phage therapy, after 6 h postinfection, phage solution containing 2 × 108 PFU in PBS buffer (100 μl) was administered intramuscularly (i.m.) or intraperitoneally. For enumeration of bacterial burden from mouse tissues, infected mice were anesthetized by inhalation of ether at the designated time point. Lung, liver, and spleen samples were obtained aseptically and homogenized with a tissue homogenizer (Yamato Scientific Co., Ltd., Tokyo, Japan) in PBS buffer (1 ml). Portions of blood and homogenized tissue samples were plated onto cetrimide agar plates, which were incubated for 24 h at 37°C. For phage pharmacokinetics, phages were administered i.m. or i.p. into the uninfected mice, from which the blood, lung, and liver samples were obtained and homogenized at the designated time points (0.5, 12, 24, 36, and 48 h). The tissue homogenates were subjected to filtration, and the filtrates were used for PFU measurement by plaque assay.
Fly experiments.
The fruit fly Drosophila melanogaster strain Oregon R was grown and maintained at 25°C using the corn meal-dextrose medium, consisting of 0.93% agar, 6.24% dry yeast, 4.08% corn meal, 8.62% dextrose, 0.1% methyl paraben, and 0.45% (vol/vol) propionic acid. Infection of flies was performed by picking 4- to 5-day-old adult flies in the dorsal thorax with a 10-μm needle (Ernest F. Fullam, Inc.). The needle was dipped halfway into PBS-diluted bacterial suspension containing 107 CFU/ml from the stationary-phase cultures. Infected flies were transferred to a new medium overlaid with 100 μl phage solution containing 1010 PFU. Fly mortality was monitored for up to 48 h postinfection. Flies that died within 12 h were not included in mortality determination. Mortality studies were repeated at least three times. For phage pharmacokinetics in D. melanogaster, flies were fed with phages for 12 h and then transferred to a new medium without phage. Flies were homogenized at the designated time points (0.5, 12, 24, 36, and 48 h), and the tissue homogenates were filtered and the filtrates were used for PFU measurement by plaque assay.
Statistical analysis.
The statistical significance of the differences between the control and the treatment groups in mortality rates was determined based on Kaplan-Meier log rank statistics. The Mann-Whitney U test was used to verify the statistical significance in the numbers of viable bacteria recovered from blood and organs.
RESULTS
Identification of temperate siphophages from clinical P. aeruginosa strains.
Phages specific to P. aeruginosa can be obtained from the stationary-growth-phase culture supernatants of clinical P. aeruginosa isolates, as described previously (12). We identified two phages (named MP29 and MP42) that could form discernible plaques on the lawns of two well-studied virulent P. aeruginosa strains, PAO1 and PA14. Based on the morphology of the purified phage particles observed under a transmission electron microscope, they were found to belong to the Siphoviridae family (morphotype B of the order Caudovirales), subdivision B1, with the icosahedral head and the flexible tails of ∼150-nm length with fibers (data not shown) (1), being similar to the previously identified phages MP22, DMS3, and D3112. Both MP29 and MP42 required type IV pilus for infection (data not shown), and their genome sequences displayed synteny to one another (GenBank accession numbers: MP29, EU272036; MP42, JQ762257).
Group behavior inhibition of P. aeruginosa by temperate siphophages.
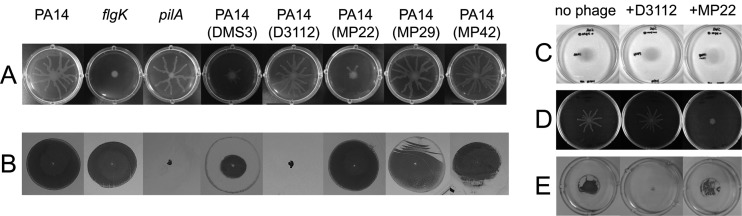
Zegans et al. (29) first observed the inhibition of swarming motility of P. aeruginosa strain PA14 by lysogenization of DMS3, which involves the CRISPR-Cas system of the host bacteria. We examined whether or not our siphophages could also have affected the group motilities (swarming and/or twitching) or the solitary motility (swimming) of P. aeruginosa when they lysogenized the PA14 strain. As shown in Fig. 1A, the PA14 lysogen for MP22 was impaired in swarming, as was the DMS3 lysogen, whereas those for D3112, MP29, and MP42 were not affected at all. This type of swarming inhibition by DMS3 was not due to the gene disruption caused by the integration of the phage genome into the host chromosome but involved the host CRISPR spacers with sequence stretches similar to those of the regions of the DMS3 genome (3). As a part of this observation, we also revealed that PAO1 lysogens for MP22 were not affected at all, although the swarming pattern by the wild-type PAO1 was different from that by PA14 (data not shown).
Fig 1.
Phage-mediated inhibition of group behaviors of PA14. (A and B) Group motilities of P. aeruginosa PA14 and its lysogens for temperate siphophages, with its isogenic mutants for pilA or flgK used as the negative controls. Photographs of swarming assay after 16 h of growth on M9 minimal media (A) and twitching assay after 48 h of growth on LB media (B) as described in Materials and Methods. (C, D, and E) Motilities of PA14 in the presence (+) or absence (no phage) of the siphophage strain (either MP22 or D3112) in the swimming (C), the swarming (D), and the twitching (E) assay plates.
To our surprise, we also found that the PA14 lysogen for D3112 displayed a defect in twitching motility, which was not observed in the other siphophage lysogens tested (Fig. 1B). We were able to exclude the possibility of lysogeny-mediated inactivation of a gene critical for type IV pilus biogenesis or twitching motility by enumerating the five independent integration sites using arbitrary PCR: four are within the potential coding regions of PA14_15000, PA14_38310, PA14_60020, and PA14_69060, and one is at the intergenic region between PA14_61640 (phr) and PA14_61650 (pagL). This result was consistent with the previous findings that D3112 randomly integrates into the host genome (21, 25). Two mutants for the aforementioned genes (PA14_69060 and PA14_60020) were readily available from the PA14 NR set (15), whose twitching motilities were not affected at all (data not shown). To further determine whether the impaired twitching was a common consequence of D3112 lysogenization, we analyzed additional D3112-lysogenized strains as well. One hundred D3112-lysogenized bacteria tested were incapable of twitching motility (data not shown). This result appears to differ from the recent observation from DMS3, whose lysogenization in the PA14 background inhibits swarming motility only by interacting with the host CRISPR region (29). Taken together, these data suggest that D3112 lysogeny alone might be sufficient to cause the loss of twitching motility.
To assess the capability of group behavior inhibition of the wild-type bacteria by the phages that are exogenously administered, motility assays were performed using the wild-type bacteria on the plates amended with phage lysates (Fig. 1C to E). MP22, but not D3112, could inhibit the swarming motility of PA14, whereas the twitching motility was impaired upon the addition of D3112, but not MP22. Neither of the two phages could affect the swimming motility at the same concentration as in the swarming and twitching motility assays. Although we did not determine the lysogenization status of the bacteria in the motility plates, these data could substantiate the suggestion that the motility impairments were not due to the stochastic events such as lysogeny-dependent host gene disruption but could be attributed to the phage-derived mechanism of twitching inhibition. Furthermore, the finding that the twitching motility of the PAO1 lysogen for D3112 was clearly impaired led us to the conclusion that the twitching inhibition by D3112 did not involve the host-dependent CRISPR-Cas system (data not shown) and thus was independent of the background of host bacteria.
Lytic activity of temperate siphophages toward P. aeruginosa strains.
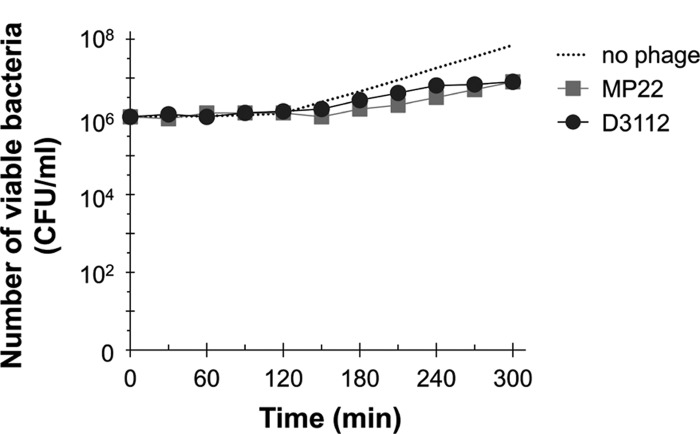
To address how efficiently these siphophages kill virulent P. aeruginosa strains, we first determined the lytic activity of MP22 and D3112 based on the multistep bacterial killing curve. PA14 cells (106 CFU) were infected with one of the aforementioned phages at 2 × 106 PFU (i.e., at the multiplicity of infection [MOI] of 2), and the viability was measured (Fig. 2). The number of viable cells that had been infected with either MP22 or D3112 was gradually increasing, but significantly less than that of the uninfected bacteria. This is in contrast to the lytic activity of a virulent myophage, MPK1, which displays a steep decrease in the bacterial viability only after 30 min (12; data not shown). The number of viable bacteria did not significantly decrease, and the growth of the resistant bacteria very rapidly resumed (within 5 h of incubation), with a growth rate comparable to that of the uninfected bacteria (data not shown), suggesting that these temperate phages may be capable of rapid lysogenization upon the host bacteria.
Fig 2.
Lytic activities of the temperate phages MP22 and D3112 toward PA14 in vitro. The PA14 culture suspensions (106 CFU) were mixed with the phage lysates of 2 × 106 PFU of MP22 (■) or D3112 (●) in LB medium and then incubated further. The number of viable bacteria (CFU) was determined at the appropriate dilutions to count 102 CFU.
Pharmacokinetics of temperate siphophages in mice and flies.
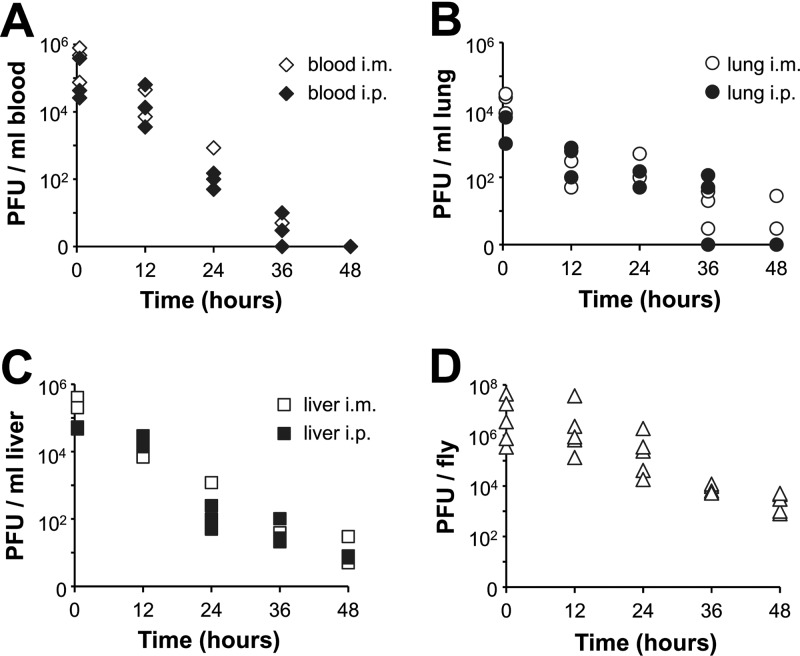
Prior to assessment of antibacterial efficacy, the pharmacokinetics was examined to address the in vivo stability of the siphophages, depending on the routes by which phages were delivered in mice. Phages (2 × 107 PFU) were injected via the i.p. or i.m. route into uninfected mice. Three mice from each group receiving phage samples i.p. or i.m. were euthanized at 0.5, 12, 24, 36, and 48 h postinfection. The number of phages was determined from blood (Fig. 3A), lung (Fig. 3B), and liver (Fig. 3C). In each tissue examined, a consistent pattern of the relative PFU levels after administration of D3112 by the different routes was observed: the i.m. route yielded higher levels than the i.p. route, which is similar to what has been observed in the myophage MPK1 and in a podophage, MPK6 (12). Under these conditions, D3112 displayed intermediate persistence between MPK1 and MPK6. Interestingly, D3112 was hardly recovered from lung in both administration routes, whereas MPK6 was hardly recovered from blood (12), suggesting that the phage virion structure may be associated with the preference of host tissue infiltration. In parallel experiments, the pharmacokinetics of MP22 did not differ from those of D3112 (data not shown).
Fig 3.
Pharmacokinetics of D3112 in mice and flies. (A, B, and C) Pharmacokinetics of D3112 in mice. Phage samples (2 × 107 PFU in PBS) of D3112 were administered intramuscularly (i.m., open symbols) or intraperitoneally (i.p., solid symbols) into uninfected mice. Groups of mice (n = 3) were sacrificed at 0.5, 12, 24, 36, and 48 h postinfection, blood (A), lung (B), and liver (C) were extracted, and their homogenates were used to determine the PFU per unit volume (ml) as described in Materials and Methods. The PFU/ml are shown on a log scale. (D) Pharmacokinetics of D3112 in flies. Phage samples (5 × 109 PFU in PBS) of D3112 were overlaid on the surface of the fly media for 12 h before flies were transferred to new media without phage. Groups of flies (n = 5) were collected at 0.5, 12, 24, 36, and 48 h, and their homogenates were removed to determine the PFU per fly, which are shown on a log scale.
Since we established the fly infection model to assess the antibacterial efficacy of phages toward P. aeruginosa, we further determined the pharmacokinetics of D3112, which was administered to flies by feeding. Uninfected flies were fed with D3112 at 5 × 109 PFU for 12 h and transferred to a new medium without phage feeding. Figure 3D shows the results of the pharmacokinetics of phage feeding in the fly model. D3112 phages are more stable than MPK6 in fly bodies as well and did not completely disappear within 48 h.
Therapeutic efficacy of temperate siphophages in mice and flies.
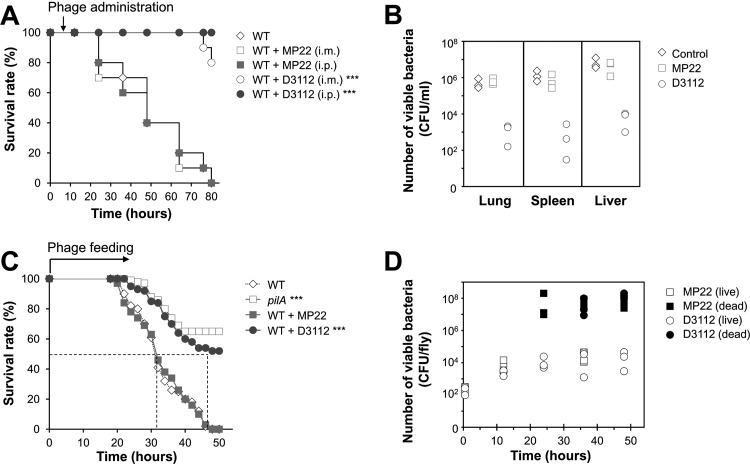
To determine the antibacterial efficacy of phages in vivo, we tested for the protective ability of D3112 and MP22 in mice and flies that had been infected with P. aeruginosa. D3112 and MP22 were injected either i.m. or i.p. at 2 × 107 PFU of P. aeruginosa strain PA14 in 100 μl PBS after 6 h postinfection. D3112 phages significantly protected the infected mice compared to the untreated mice, whereas MP22 phages showed no protective effect (Fig. 4A). The i.p. administration displayed slightly better efficacy than did the i.m. administration for D3112, most likely due to the infection model we used, as observed previously (12).
Fig 4.
Protective effect of D3112 in mice and flies. (A) Mortality of PA14 (2 × 105 CFU)-infected mice (n = 20) with either MP22 (□ and ■) or D3112 (○ and ●) at 2 × 108 PFU per 100 μl, which was administered by the i.m. route (open symbols) or by the i.p. route (solid symbols) at 6 h postinfection. One hundred percent of PA14-infected mice with MP22 treatment or without phage treatment (♢) died within 84 h. Statistical significance, based on the log rank test, was set at P values of <0.001 (***). (B) Bacterial burdens in lung, spleen, and liver of mice treated with or without i.p. administration of either MP22 or D3112 (2 × 108 PFU in 100 μl). The numbers of viable bacteria (CFU) were determined from the organs of live mice at 24 h postinfection. The CFU per unit volume (ml) were determined as described in Materials and Methods and are shown on a log scale. Symbols: ♢, no phage treatment; □, MP22; ○, D3112. (C) Mortality of PA14 (either wild type [WT] or its isogenic pilA mutant)-infected flies (n = 33) fed with phages. Infected flies were transferred to a new medium overlaid with nothing (♢ and □) or phage samples (5 × 109 PFU) of either MP22 (■) or D3112 (●). The dotted line represents the time required to reach 50% mortality. Statistical significance, based on the log rank test, was set at P values of <0.001 (***). (D) Bacterial burdens in fly homogenates fed with phages were measured as the number of bacteria (CFU) from the homogenates of an individual live (open symbols) or dead (solid symbols) fly at 0.5 h, 12 h, 24 h, or 48 h postinfection. The CFU per fly are shown on a log scale. Symbols: □ and ■, MP22 feeding; ○ and ●, D3112 feeding.
We also investigated whether the administration of phages could inhibit the bacterial proliferation by measuring the bacterial loads in mouse organs such as spleen, lung, and liver from live animals at 24 h postinfection (Fig. 4B). Treatment with D3112, but not MP22, could significantly reduce the bacterial loads by about 2 to 4 log in lung, spleen, and liver from live mice at 24 h postinfection.
The antibacterial efficacy of D3112 phage was further evaluated, based on their protective effects from PA14-induced mortality in the fly infection model. We administered the D3112 phage by overlaying 5 × 109 PFU on top of fly media with PA14-infected flies. As shown in Fig. 4C, D3112, but not MP22, protected the infected flies, in contrast to the unfed flies, which was in agreement with the reduction of bacterial loads in the live flies (Fig. 4D). These results suggest that D3112 phage are highly effective to control P. aeruginosa-induced peritonitis by inhibiting bacterial twitching motility in vivo, rather than by killing the bacteria, since MP22 did not show any protective effect at all with the similar lytic activity in vitro (Fig. 2).
DISCUSSION
Phages have successfully treated experimental bacterial infections caused by P. aeruginosa in model animals, which include gut-derived sepsis, peritonitis-sepsis, and burn wound infections in mice (12, 18, 26). Skin grafts on guinea pigs were protected from P. aeruginosa infections by topical administration of phages (23). Recently, we reported that phages could also enhance the survival of Drosophila melanogaster flies that had been subjected to systemic infections by P. aeruginosa (12). Phages have been used for many years in Eastern Europe to treat a variety of bacterial infections, but valuable information on controlled evaluation of their efficacy as well as potential adverse effects, which could include toxicity, allergy, and effects on other bacteria, is lacking. Since phages are composed largely of protein and DNA, few toxic or allergic effects might be expected. No such adverse effects were noted when phages were administered intravenously to 200 people as a test of immune function (27). Our experimental infection settings also indicate that harmful effects are minimal in that the animals were not affected at all when phages were administered in amounts of >1010 PFU.
Almost all of the isolated P. aeruginosa phages are members of the viral order Caudovirales including the three major families, Siphoviridae (siphophages), Podoviridae (podophages), and Myoviridae (myophages), which are distinguished primarily by their virion morphology. It is intuitively understood that the structural characteristics of the phages, which are largely associated with their virion morphology, might also be related to their stability in vitro as well as in vivo. Although our studies on the pharmacokinetics of Caudovirales phages in animal tissues are not comprehensive, we observed that myophages are the most stable whereas the podophages are the least stable in both mice and flies (12). The most apparent difference between them is the tail structure. Further investigation needs to be done to investigate whether the tail structure of the phages might affect the overall stability of the phage particles in host tissues. By this approach, the phage tails can be engineered to modulate the pharmacokinetic properties of the phages, as was done to engineer the tail structure of the R-type pyocins to specifically kill E. coli cells (28).
Large repertoires of native as well as genetically engineered phages have been intensively exploited to treat bacterial infections, but all of the phages used were virulent organisms whose antibacterial efficacy was attributed to the killing activity toward the target bacteria. This study, however, is the first report indicating that temperate phages could also be exploited to control bacterial infections, based on their ability to modulate the bacterial group behaviors upon infection and/or lysogenization, rather than their killing activity. Since group behaviors such as social motilities, biofilm formation, and quorum sensing are crucial in the virulence and/or survival mechanisms of P. aeruginosa (10), the possibility of therapy exploiting the temperate phages that affect those properties can be a new platform that needs to be further developed. The major obstacle in using temperate phages for antibacterial therapy may be that they can integrate themselves relatively randomly into the host chromosome, potentially leading to unexpected consequences regarding virulence traits of the bacteria (19). However, there has been no direct evidence for that, and we may be unable to predict whether the dynamic and complicated interactions between the temperate phages and the bacteria within the infected hosts are either beneficial or detrimental to each party, unless some phage-specified virulence determinants are transmitted during the phage-bacteria interactions as in other bacteria. Thus, a careful and comprehensive evaluation of the antibacterial efficacy of temperate phages in addition to virulent phages or the mixture of both kinds needs to be performed, under concerns regarding the potential side effects derived from unpredictable mutations and gene transfer (for temperate phages) and cytolysis-derived endotoxin generation (for virulent phages), which can be facilitated by the extensive use of nonmammalian live animal infections as in this study.
ACKNOWLEDGMENTS
This work was supported by grants from the National Research Foundation (NRF) of South Korea (2011-0015953) and from CHA University.
Footnotes
Published ahead of print 20 August 2012
REFERENCES
- 1. Ackermann HW. 2001. Frequency of morphological phage descriptions in the year 2000. Arch. Virol. 146:843–857 [DOI] [PubMed] [Google Scholar]
- 2. Boles BR, Thoendel M, Singh PK. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. U. S. A. 101:16630–16635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cady KC, O'Toole GA. 2011. Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J. Bacteriol. 193:3433–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caetano-Anolles G. 1993. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 3:85–94 [DOI] [PubMed] [Google Scholar]
- 5. Chung I-Y, Choi KB, Heo Y-J, Cho Y-H. 2008. Effect of PEL exopolysaccharide on the wspF mutant phenotypes in Pseudomonas aeruginosa PA14. J. Microbiol. Biotechnol. 18:1227–1234 [PubMed] [Google Scholar]
- 6. Conway SP, Brownlee KG, Denton M, Peckham DG. 2003. Antibiotic treatment of multidrug-resistant organisms in cystic fibrosis. Am. J. Respir. Med. 2:321–332 [DOI] [PubMed] [Google Scholar]
- 7. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 8. Curtin JJ, Donlan RM. 2006. Using bacteriophages to reduce formation of catheter-associated biofilms by Staphylococcus epidermidis. Antimicrob. Agents Chemother. 50:1268–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. El Solh AA, Alhajhusain A. 2009. Update on the treatment of Pseudomonas aeruginosa pneumonia. J. Antimicrob. Chemother. 64:229–238 [DOI] [PubMed] [Google Scholar]
- 10. Hausner M, Wuertz S. 1999. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 65:3710–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heo Y-J, Chung I-Y, Choi KB, Lau GW, Cho Y-H. 2007. Genome sequence comparison and superinfection between two related Pseudomonas aeruginosa phages, D3112 and MP22. Microbiology 153:2885–2895 [DOI] [PubMed] [Google Scholar]
- 12. Heo Y-J, et al. 2009. Antibacterial efficacy of phages against Pseudomonas aeruginosa infections in mice and Drosophila melanogaster. Antimicrob. Agents Chemother. 53:2469–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hunt SM, Werner EM, Huang B, Hamilton MA, Stewart PS. 2004. Hypothesis for the role of nutrient starvation in biofilm detachment. Appl. Environ. Microbiol. 70:7418–7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kutateladze M, Adamia R. 2010. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol. 28:591–595 [DOI] [PubMed] [Google Scholar]
- 15. Liberati NT, et al. 2006. An ordered, non-redundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103:2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu TK, Koeris MS. 2011. The next generation of bacteriophage therapy. Curr. Opin. Microbiol. 14:524–531 [DOI] [PubMed] [Google Scholar]
- 17. Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051–1060 [DOI] [PubMed] [Google Scholar]
- 18. McVay CS, Velasquez M, Fralick JA. 2007. Phage therapy of Pseudomonas aeruginosa infection in a mouse burn wound model. Antimicrob. Agents Chemother. 51:1934–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mizuuchi K, Craigie R. 1986. Mechanism of bacteriophage mu transposition. Annu. Rev. Genet. 20:385–429 [DOI] [PubMed] [Google Scholar]
- 20. Nguyen D, Singh PK. 2006. Evolving stealth: genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc. Natl. Acad. Sci. U. S. A. 103:8305–8306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rehmat S, Shapiro JA. 1983. Insertion and replication of the Pseudomonas aeruginosa mutator phage D3112. Mol. Gen. Genet. 192:416–423 [DOI] [PubMed] [Google Scholar]
- 22. Slopek S, Weber-Dabrowska B, Dabrowski M, Kucharewicz-Krukowska A. 1987. Results of bacteriophage treatment of suppurative bacterial infections in the years 1981–1986. Arch. Immunol. Ther. Exp. (Warsz.) 35:569–583 [PubMed] [Google Scholar]
- 23. Soothill J, Hawkins C, Anggard E, Harper D. 2004. Therapeutic use of bacteriophages. Lancet Infect. Dis. 4:544–545 [DOI] [PubMed] [Google Scholar]
- 24. Sulakvelidze A, Alavidze Z, Morris JG., Jr 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang PW, Chu L, Guttman DS. 2004. Complete sequence and evolutionary genomic analysis of the Pseudomonas aeruginosa transposable bacteriophage D3112. J. Bacteriol. 186:400–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Watanabe R, et al. 2007. Efficacy of bacteriophage therapy against gut-derived sepsis caused by Pseudomonas aeruginosa in mice. Antimicrob. Agents Chemother. 51:446–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wedgwood RJ, Ochs HD, Davis SD. 1975. The recognition and classification of immunodeficiency diseases with bacteriophage phiChi 174. Birth Defects Orig. Artic. Ser. 11:331–338 [PubMed] [Google Scholar]
- 28. Williams SR, Gebhart D, Martin DW, Scholl D. 2008. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl. Environ. Microbiol. 74:3868–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zegans ME, et al. 2009. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J. Bacteriol. 191:210–219 [DOI] [PMC free article] [PubMed] [Google Scholar]