Abstract
Persistent Pseudomonas aeruginosa infections are a major cause of morbidity and mortality in cystic fibrosis (CF) patients and are linked to the formation of a biofilm. The development of new biofilm inhibition strategies is thus a major challenge. LL-37 is the only human antimicrobial peptide derived from cathelicidin. The effects on the P. aeruginosa PAO1 strain of synthetic truncated fragments of this peptide were compared with the effects of the original peptide. Fragments of LL-37 composed of 19 residues (LL-19, LL13-31, and LL7-25) inhibited biofilm formation. The strongest antibiofilm activity was observed with the peptides LL7-37 and LL-31, which decreased the percentage of biomass formation at a very low concentration. Some peptides were also active on the bacteria within an established biofilm. LL7-31, LL-31, and LL7-37 increased the uptake of propidium iodide (PI) by sessile bacteria. The peptide LL7-37 decreased the height of the biofilm and partly disrupted it. The peptides active within the biofilm had an infrared spectrum compatible with an α-helix. LL-37, but not the peptides LL7-31 and LL7-37, showed cellular toxicity by permeabilizing the eukaryotic plasma membrane (uptake of ethidium bromide and release of lactate dehydrogenase [LDH]). None of the tested peptides affected mitochondrial activity in eukaryotic cells. In conclusion, a 25-amino-acid peptide (LL7-31) displayed both strong antimicrobial and antibiofilm activities. The peptide was even active on cells within a preformed biofilm and had reduced toxicity toward eukaryotic cells. Our results also suggest the contribution of secondary structures (α-helix) to the activity of the peptides on biofilms.
INTRODUCTION
Pseudomonas aeruginosa, a Gram-negative bacterium, is an opportunistic pathogen responsible for numerous infections. It is also the most common pathogen causing chronic respiratory infections in cystic fibrosis (CF) patients that result in significant morbidity and mortality (12, 17). Persistent P. aeruginosa infection is linked to the formation of a biofilm (11, 38), a complex community of microorganisms attached to a surface and enclosed in a self-produced extracellular matrix (6, 29).
The worldwide increase of bacterial resistance to conventional antibiotic therapy motivates the urgent search for new strategies to combat persistent infections. The cathelicidin family is a group of antimicrobial peptides (AMP) found in diverse mammalian species. They are synthesized as preproproteins with a highly conserved and inactive N-terminal domain, the cathelin domain, and a highly variable C-terminal domain with an antimicrobial activity (47). In humans, only one cathelicidin (CAP18) has been identified, from which the active C-terminal domain (LL-37) is cleaved by a serine proteinase (39). This peptide consists of 37 amino acids with two leucyl residues at its N terminus, hence its name. It is a cationic peptide with a high content of basic and hydrophobic residues, playing a key role in the innate immune system. It forms an amphipathic α-helix in contact with lipid membranes (18), dodecylphosphocholine (DPC) (31), or SDS micelles (45).
Understanding the mechanism of action of the peptide, its structural properties (31), and its interactions with bacteria is important for discovering new cellular targets for therapeutic intervention (26). Numerous structure-activity relationship studies have revealed the importance of a balance among parameters like net charge, amphipathicity and hydrophobicity in targeting cell selectivity, and antimicrobial potency (41, 42). The nonspecific membrane lysis by host defense peptides accounts for their antimicrobial activity (16, 47) and explains why they are active on both antibiotic-susceptible and -resistant strains. The in vitro antimicrobial activity of LL-37 against P. aeruginosa, including strains which are mucoid and antibiotic resistant, suggests that this peptide might lead to the development of novel antimicrobial agents (15, 43). The strategy used to develop these new drugs should take into account three major drawbacks: (i) LL-37 is a rather long peptide and expensive to synthesize, (ii) LL-37 is sensitive to proteases secreted by eukaryotic cells or prokaryotes (23), and (iii) LL-37 has some toxicity against eukaryotic cells (18). The search for an efficient, stable, and safe fragment of LL-37 was thus initiated. Wang (45) reported that the fragment KR-12 (fragment LL18-29) was toxic for bacteria but not for human cells. Removal of the first 12 N-terminal residues (LL13-37) decreased the toxicity of LL-37 toward human peripheral blood monocytes and rabbit erythrocytes without affecting its antifungal activity (46). It was also recently reported that truncated fragments of LL-37 exhibited antimicrobial and antibiofilm effects on Burkholderia thailandensis (19) and Burkholderia pseudomallei (20). While the present study was in progress, de la Fuente-Núñez et al. (9) identified, in a library of synthesized fragments, a novel 9-amino-acid peptide with antibiofilm activity superior to that of LL-37.
In this study, we screened a library of fragments of LL-37 obtained by sequentially deleting hexapeptides from the N-terminal or C-terminal domain or from both ends of the native peptide. Based on the observation that LL-37 could inhibit biofilm formation (30), the activity of these fragments was examined on the P. aeruginosa PAO1 strain, in planktonic cultures, or within a biofilm (both on the formation of a biofilm and on a pregrown biofilm). We also compared the propensity of the peptides to form an α-helix and their toxicity toward eukaryotic cells, the human umbilical vein endothelial cells (HUVEC). Our results showed that the removal of the first 6 amino acids of LL-37 decreased its toxicity toward eukaryotic cells without affecting its antimicrobial properties. In agreement with Sigurdardottir et al. (37) and Wang (45), we observed that LL13-31, the central fragment of the peptide, retained most of the antimicrobial activity of the native peptide against planktonic bacteria. Sigurdardottir et al. reported that LL14-34 had a similar even stronger antimicrobial activity than LL-37, and Wang (45) demonstrated that LL18-29 (KR-12) was the smallest fragment with antimicrobial activity against Escherichia coli. Using confocal laser scanning microscopy (CLSM), we established that the fragment LL7-31 was the smallest tested fragment with activity against bacteria within a biofilm.
MATERIALS AND METHODS
Materials.
Propidium iodide (PI), trypsin-EDTA solution, and ethidium bromide were from Life Technologies (Groningen, The Netherlands). The endothelial cell growth medium (EGM) medium was from Lonza (Verviers, Belgium). EGTA, Triton X-100, Trizma hydrochloride (TRIS), β-NADH, and sodium pyruvate were purchased from Sigma-Aldrich (St. Louis, MO).
Strains and growth conditions.
Subcultures of the P. aeruginosa reference strain (ATCC 15692 PAO1) were stored at −20°C in glycerol. The P. aeruginosa PAO1 labeled with the green fluorescent protein (GFP) was kindly provided by M. Franklin (Center for Biofilm Engineering, Bozeman, MT). Before use, bacterial colonies were spread onto tryptic soy agar (TSA) medium and incubated at 37°C for 24 h. The bacteria were plated not more than three times onto TSA medium.
Synthesis of LL-37 and of its fragments.
LL-37 and its truncated variants were synthesized by 9-fluorenylmethoxy carbonyl (Fmoc) chemistry using a MilliGen 9050 peptide synthesizer (MilliGen/Biosearch, Bedford, MA) as described previously (10). Peptides were purified by preparative reversed-phase high-pressure liquid chromatography. The purity of the peptides was at least 95%, and the authenticity of the peptides was confirmed by a Microflex LRF matrix-assisted laser desorption ionization–time of flight mass spectrometer equipped with an additional gridless reflectron (Bruker Daltonik, Bremen, Germany) as described previously (2).
Determination of the MIC and the MBC.
Susceptibility testing to the peptides was performed on planktonic cultures using the microdilution broth method according to NCCLS guidelines (25). MICs were performed in 96-well microplates (Greiner, Wemmel, Belgium), and the results were recorded after incubation at 35°C for 18 h. For minimal bactericidal concentration (MBC) determination, aliquots were removed from the wells after incubation and spread onto Mueller-Hinton agar in petri dishes and incubated overnight at 35°C. The growth results were then recorded.
Cell viability assay.
The permeability of the bacterial membrane after exposure to the different fragments of LL-37 was assessed using PI. Briefly, an overnight growth culture of P. aeruginosa PAO1 in modified BM2 medium [0.2 g/liter MgSO4 · 2H2O, 0.0005 g/liter FeSO4 · 7H2O, 1.25 g/liter Na2HPO4, 0.5 g/liter KH2PO4, 0.1 g/liter (NH4)2SO4, 0.05 g/liter glucose] was washed three times and resuspended in modified BM2 medium supplemented with 0.5% (wt/vol) Casamino Acids to a final optical density at 600 nm (OD600) of 0.100. Serial dilutions of the antimicrobial peptides (final concentrations, 0 to 10 μM; final volume, 100 μl) were added to the wells of a black-walled microplate. One hundred microliters of the bacterial suspension containing PI (final concentration, 10 μM) was added to the wells. The emitted fluorescence was measured at excitation and emission wavelengths of 544 and 620 nm, respectively, using a BioTek multidetection microplate reader, during 1 h.
An enumeration of the viable cells was performed after exposure of the bacteria to various concentrations of LL-37 at 37°C for 1 h. Aliquots were diluted 102- to 106-fold. One milliliter of each dilution was plated in duplicate on a TSA plate, and the number of colonies was counted after incubation at 32°C for 48 h. The logarithmic decrease in CFU from treated biofilms was calculated using the following formula: logarithmic decrease = log(CFUc) − log(CFUx), where c corresponds to bacteria incubated in the absence of the peptide and x corresponds to bacteria incubated in the presence of the peptide.
Assay of the formation of a biofilm on an inert support.
This assay was performed as described previously (30). Briefly, an overnight growth culture in BM2 medium, centrifuged and washed twice in BM2 medium supplemented with 0.5% (wt/vol) Casamino Acids, was adjusted to an OD600 of 1.00 ± 0.05. The bacterial suspension and the peptides were added to the wells of a polystyrene microplate (Greiner, Wemmel, Belgium) and incubated at 37°C for 18 h. The formation of the biofilm was evaluated using the crystal violet staining technique (40). The medium was removed, and the wells were washed three times and dried for 20 min to remove nonadherent cells. The wells were filled with a 1% (wt/vol) crystal violet solution for 20 min before being washed three times with distilled water. The wells were read at 590 nm after solubilization of the dye with 33% acetic acid. An enumeration of the viable cells was performed after exposure for 18 h to 10 μM LL-37, LL7-37, LL13-37, and LL-31 as described previously.
Biofilm assay in a microtiter plate using CLSM.
One milliliter of an overnight growth culture of the GFP-tagged P. aeruginosa PAO1 strain in tryptic soy broth (TSB) supplemented with 150 μg/ml carbenicillin was added to modified BM2 medium supplemented with 0.5% (wt/vol) Casamino Acids to reach an OD600 of 0.350. Two hundred fifty microliters of the bacterial suspension was added to the wells of a polystyrene 96-well microtiter plate with a μClear base (Greiner Bio-One, France). The plate was incubated at 37°C on an orbital shaker (30 rpm). The medium was changed after 1 h to remove nonattached bacteria. After 24 h of incubation, the bacteria were rinsed and treated with the peptides (0, 20, 50, or 100 μM) in the presence of 10 μM PI. After a 25-min exposure to the antimicrobial agents, the bacteria were rinsed and examined by CLSM with a Leica SP5 (Leica Microsystems Inc., Buffalo Grove, IL) using a 63× objective with a 1.2 numerical aperture. Excitation was performed using two lasers at 488 and 561 nm, and the fluorescence emission was collected from 500 to 550 nm and from 570 to 700 nm. Images were scanned at 600 Hz and analyzed with the Imaris software (Bitplane, Zurich, Switzerland).
Study of a biofilm in a CDC biofilm reactor.
GFP-tagged P. aeruginosa PAO1 biofilms were grown in CDC biofilm reactors (Biosurface Technologies Inc., Bozeman, MT). This method is routinely used to study the formation of biofilms by P. aeruginosa grown with high shear and continuous flow (13). It consists of growing biofilms on glass coupons held in rods immersed in a glass vessel. A continuous flow provides nutrient medium at a constant flow rate, and a shear is generated by a magnetic stirring. Briefly, 1 ml of an overnight planktonic bacterial culture in TSB (300 mg/liter) at 37°C was added to 350 ml TSB (300 mg/liter) and poured in the reactor. The reactor was incubated for 24 h at room temperature with constant stirring at 120 rpm. The reactor was then connected to a nutrient carboy containing TSB (100 mg/liter) to allow a continuous flow at a 12-ml/min rate for 24 h. Biofilms grown on the glass coupons were removed from the reactor, rinsed, and exposed ex situ to different concentrations of the fragment LL7-37 in the presence of 60 μM PI for 25 min. After contact, the biofilms were rinsed, mounted on the stage of the confocal microscope, and observed using a 63× water immersion objective with a 0.9 numerical aperture. Excitation was performed using two lasers at 488 and 561 nm, and the fluorescence emission was collected from 500 to 550 nm and from 570 to 700 nm. Images were scanned at 600 Hz and analyzed with the Imaris software (Bitplane, Zurich, Switzerland).
Determination of the secondary structures of the peptides using ATR-FTIR.
The peptides went through two cycles of dissolution in 10 mM HCl, followed by lyophilization to eliminate traces of trifluoroacetic acid. The peptides were then dissolved in 1 mM HEPES (pH 7.3) to a 1 mM final concentration. One microliter of the peptide solution was spread on a diamond internal reflection element and dried under a stream of nitrogen. In our experimental conditions, the peptide sample on the attenuated total reflection (ATR) element was never completely dehydrated since it was equilibrated with water vapor. The spectra were obtained on a Bruker IFS55 Fourier transform infrared (FTIR) spectrophotometer (Bruker, Ettlingen, Germany) equipped with a mercury-cadmium-telluride (MCT) detector (broadband, 120,000 to 420 cm−1, liquid nitrogen cooled) at a resolution of 2 cm−1 and were subtracted for residual water vapor spectral contribution to the signal. Similar experiments were performed with deuterated peptides. Briefly, after lyophilization of the peptides, they were dissolved in a D2O solution for 1 h. One microliter of the deuterated peptide solution was deposited on the diamond, and the spectra were obtained after a rapid flushing with nitrogen gas saturated with D2O as described previously (44). The exchange was monitored by the decrease of the amide II peak area around 1,550 cm−1 which is sensitive to hydrogen/deuterium (H/D) exchange compared with the amide I peak area (1,600 to 1,700 cm−1 region) taken as a reference.
Measurement of the effect of the peptide LL-37 and some of its truncated fragments on the integrity of HUVEC.
The permeabilization of the plasma membrane of HUVEC was estimated as described previously (24). Briefly, HUVEC were cultured in EGM medium at 37°C in a humidified atmosphere of 5% CO2. The cells were detached using 10% trypsin-EDTA and plated overnight in a black-walled 96-well microtiter plate. The medium was removed, and ethidium bromide was added to the wells at a 25 μM final concentration. The plates were incubated at 37°C in a BioTek multidetection microplate reader. Measurement of the fluorescence was performed at excitation and emission wavelengths of 540 and 590 nm, respectively. Ten minutes after the start of the assay, the cells were exposed to various concentrations of the peptides and the emitted fluorescence was measured every 65 s. The incubation in the presence of the peptides was performed for 60 min. The maximal uptake of ethidium bromide was measured after exposure of the cells to 0.5% (wt/vol) Triton X-100.
The toxicity of the peptide LL-37 and some of its truncated fragments on HUVEC was also estimated by measuring the release of lactate dehydrogenase (LDH). After incubation of the cells for 12 h, the medium was replaced and various concentrations of the peptides were added to each well. The plate was incubated at 37°C for 10 min. At the end of the incubation, the plates were centrifuged at 1,200 rpm for 10 min and 30 μl of the supernatant was transferred to a new clear-walled 96-well microtiter plate. An NADH solution (56 mM TRIS, 5.6 mM EGTA, and 170 μM β-NADH) was then added to the wells of the second plate. Just before the start of the assay, pyruvate was added (final concentration, 1.22 mM). The activity of LDH was assayed at 30°C for 20 min. The absorbance was measured at 340 nm. Blank samples were obtained using the supernatants of untreated cells. Total cellular LDH activity was measured after cytolysis of HUVEC by sonication.
MTT test on HUVEC.
The toxicity of the peptides on the mitochondrial activity of eukaryotic cells was estimated by measuring the ability of the cells to reduce the yellow 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl tetrazolium bromide (MTT) to purple formazan as described previously (35). HUVEC were grown overnight in a 96-well microtiter plate at 37°C in a CO2 atmosphere. The next day, the incubation medium was discarded and replaced by fresh medium containing various concentrations of 8 different LL-37 fragments. The cells were incubated at 37°C for 20 min, and the medium was removed before the addition of 100 μl MTT solution (1 mg/ml medium) to the wells. After the incubation of the cells with MTT for 3.5 h, formazan crystals developed in respiring and early apoptotic cells. These crystals were solubilized with dimethyl sulfoxide, and the absorbance was measured at 540 nm with the microplate reader.
Statistical analysis.
Results were analyzed using a one-way analysis of variance (ANOVA), followed by Bonferroni's multiple-comparison posttest performed for each dose by using GraphPad 4.03 (GraphPad Software, San Diego, CA).
RESULTS
Characteristics of the peptides used in the study.
Planktonic cultures of PAO1 were tested for their sensitivity to the peptides according to NCCLS guidelines (25). As shown in Table 1, LL-37 was the most active peptide (MIC, 25 μM). PAO1 was equally sensitive to the fragments LL-31 and LL7-37 (MIC, 50 μM). PAO1 was slightly less sensitive to LL7-31 and LL13-31 (MIC, 100 μM). All other fragments had a MIC of >100 μM. These results suggested the contribution of the central part of the peptide (LL13-31) to the inhibition activity by the peptide of bacterial growth. Bactericidal activity required concentrations of >100 μM for all tested peptides.
Table 1.
Sequences and characteristics of LL-37 and some of its truncated fragmentsa
| Peptide | Alternative name | Amino acid sequence | MW | MIC (μM) | MBC (μM) | Agadir |
|---|---|---|---|---|---|---|
| LL-37 | LL-37 | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES | 4,494 | 25 | >100 | 5.10 |
| LL-31 | LL-31 | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNL | 3,824 | 50 | >100 | 3.72 |
| LL-25 | LL-25 | LLGDFFRKSKEKIGKEFKRIVQRIK | 3,065 | >100 | >100 | 1.19 |
| LL-19 | LL-19 | LLGDFFRKSKEKIGKEFKR | 2,327 | >100 | >100 | 0.92 |
| LL-13 | LL-13 | LLGDFFRKSKEKI | 1,581 | >100 | >100 | 0.84 |
| LL7-31 | RK-25 | RKSKEKIGKEFKRIVQRIKDFLRNL | 3,131 | 100 | >100 | 4.06 |
| LL13-31 | IG-19 | IGKEFKRIVQRIKDFLRNL | 2,374 | 100 | >100 | 4.33 |
| LL7-25 | RK-19 | RKSKEKIGKEFKRIVQRIK | 2,372 | >100 | >100 | 0.73 |
| LL13-25 | IG-13 | IGKEFKRIVQRIK | 1,615 | >100 | >100 | 0.71 |
| LL7-37 | RK-31 | RKSKEKIGKEFKRIVQRIKDFLRNLVPRTES | 3,801 | 50 | >100 | 5.68 |
| LL13-37 | IG-25 | IGKEFKRIVQRIKDFLRNLVPRTES | 3,044 | >100 | >100 | 5.87 |
| LL19-37 | RI-19 | RIVQRIKDFLRNLVPRTES | 2,341 | >100 | >100 | 2.08 |
| LL25-37 | KD-13 | KDFLRNLVPRTES | 1,575 | >100 | >100 | 0.09 |
The MIC and the MBC have been determined as described in Materials and Methods. The Agadir index has been calculated using the helix/coil transition program AGADIR (22).
The Agadir index is a measure of the propensity of the peptides to form an α-helix (22). This index varied widely (from less than 1 to more than 5) among the various fragments. Removal of 12 amino acids from the C-terminal domain (LL-25) but not from the N-terminal domain (LL13-37) strongly decreased the propensity of the peptide to form an α-helix.
Effect of the peptides on the permeability of the bacteria.
As shown in Fig. 1, the fluorescence remained stable during the incubation of the bacteria in the presence of PI. This result confirmed the integrity of the membranes of the bacteria incubated in control conditions. When exposed to 10 μM LL-37, the cells became permeable to PI and the fluorescence increased during the first 40 min (Fig. 1). The removal of 6 amino acids from the N-terminal domain (LL7-37) decreased the activity by about 50%. Removal of the next 6 amino acids (LL13-37) abolished the activity. After the removal of 6 to 12 amino acids from the C-terminal domain (LL-31 and LL-25), the peptide still retained a significant permeabilizing activity (Fig. 2A and C). Smaller fragments (LL-19, LL-13, LL13-37, LL19-37, and LL25-37) had no activity. When truncation was performed on both sides, the fluorescence dropped by 40 to 50% for LL7-31 and LL13-31 but the effect of the fragments remained significant (Fig. 2B). These results confirmed that the smallest fragment with significant antimicrobial activity was the peptide LL13-31.
Fig 1.
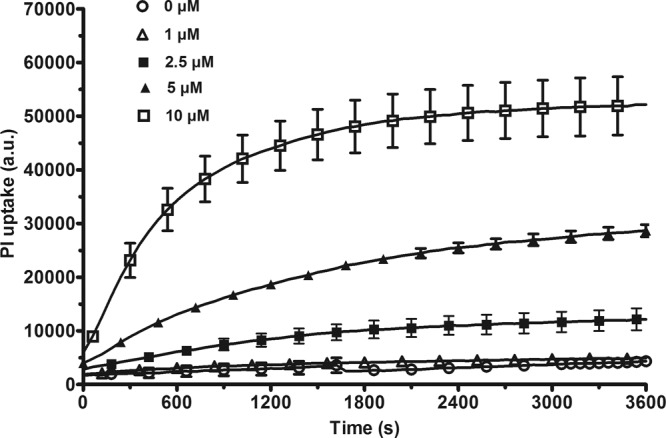
Time course of the effect of LL-37 on the permeability to PI. A P. aeruginosa PAO1 bacterial suspension was incubated in the presence of various concentrations of LL-37 for 1 h. The fluorescence was measured every min. The results are expressed as arbitrary units (a.u.) and are the means ± standard errors of the means (SEM) of three independent experiments performed in duplicate. For clarity of the graph, one in every three points has been plotted.
Fig 2.
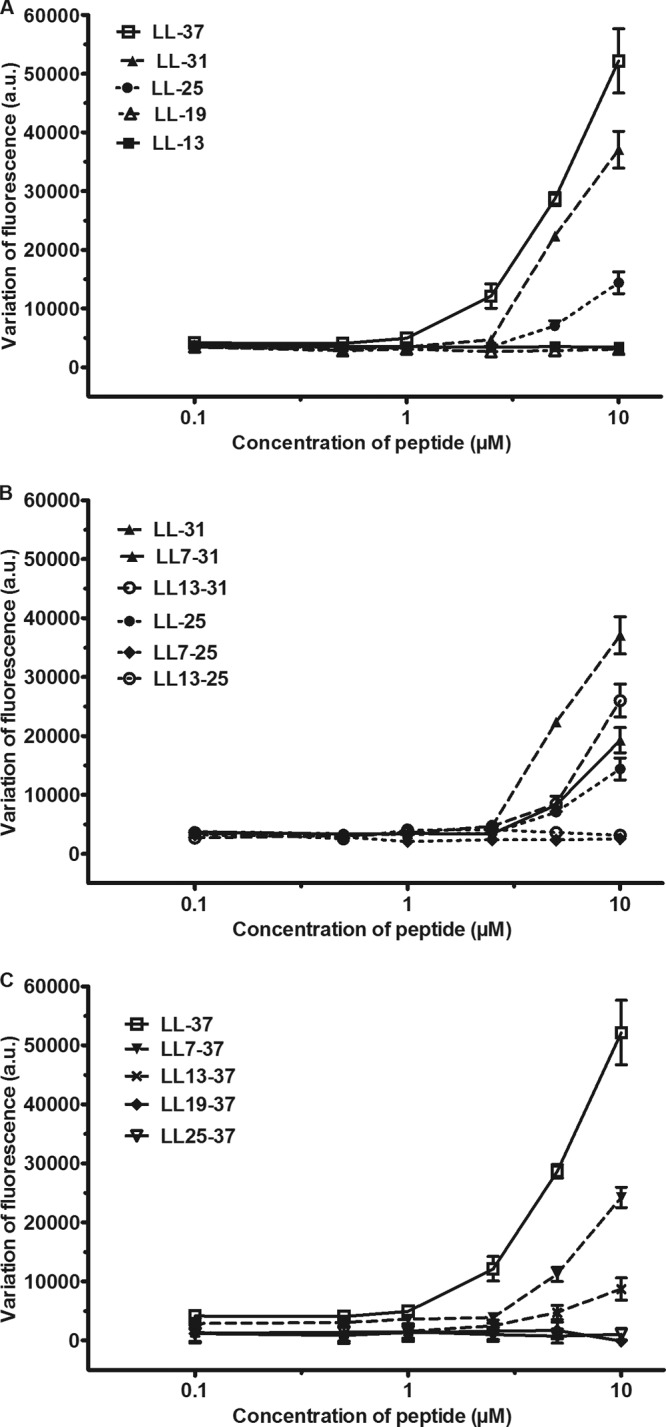
Effect of the fragments of LL-37 on the permeability to PI. A PAO1 bacterial suspension was incubated at 37°C for 1 h in the presence of 10 μM PI and various concentrations of N-terminal fragments (A), intermediate fragments (B), or C-terminal fragments (C). For comparison, LL-31 and LL-25 were also plotted in panel B. The results are the variations of the fluorescence provoked by the peptides and are expressed as arbitrary units (a.u.). They are the means ± SEM of two to four independent experiments performed in duplicate.
The uptake of PI in response to various concentrations of LL-37 was compared to the bactericidal activity of the peptide. As shown in Fig. 3, concentrations of LL-37 lower than 1 μM nonsignificantly increased the permeability to PI. At 1, 2.5, 5, and 10 μM, the peptide significantly increased the uptake of PI. In the presence of 2.5 μM LL-37, the number of viable bacteria, expressed as log CFU/ml, decreased from 7.9 to 5.4 and further dropped to 3.6 in the presence of 5 μM LL-37. The limit of detection was reached at 10 μM LL-37. These results confirmed the toxicity of LL-37 on bacteria.
Fig 3.
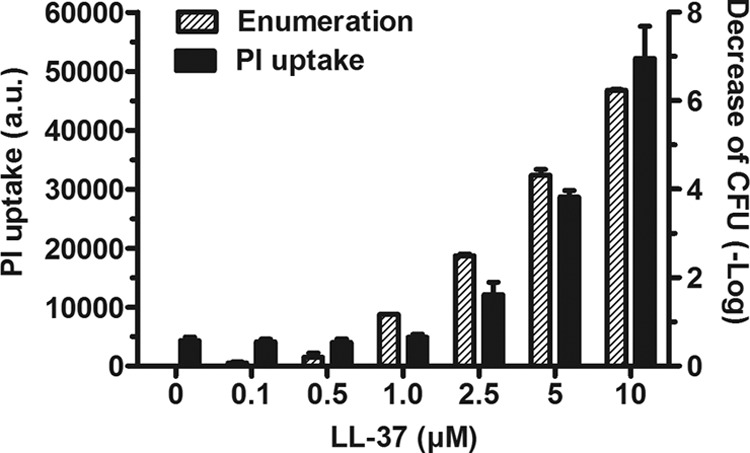
Semilogarithmic relationship between PI permeability and cell death. A PAO1 bacterial suspension was incubated at 37°C for 1 h in the presence of serial dilutions (0 to 10 μM) of LL-37. The effect of the peptide on the membranes of the bacteria and on their viability was assessed by measuring PI permeability (dark bars) and by performing an enumeration of the viable cells (hatched bars) in parallel. The results are expressed as arbitrary units (a.u.) or as logarithmic variation of the CFU and are the means ± standard deviations (SD) of two independent experiments performed in duplicate.
Study of the activity of the peptides on the formation of a biofilm.
Next, the ability of the derived fragments to prevent the formation of a biofilm after 18 h was examined (Fig. 4) based on the study by Overhage et al., who demonstrated the antibiofilm activity for LL-37 (30). As a control, the peptide LL-37 (final concentration, 10 μM) was incubated overnight in the wells of a polystyrene plate. The next day, the wells were rinsed and the development of a biofilm was compared with a control well (no preincubation with the peptide). There was no significant difference in the formation of a biofilm, confirming that the peptide did not bind to the surface of the wells.
Fig 4.
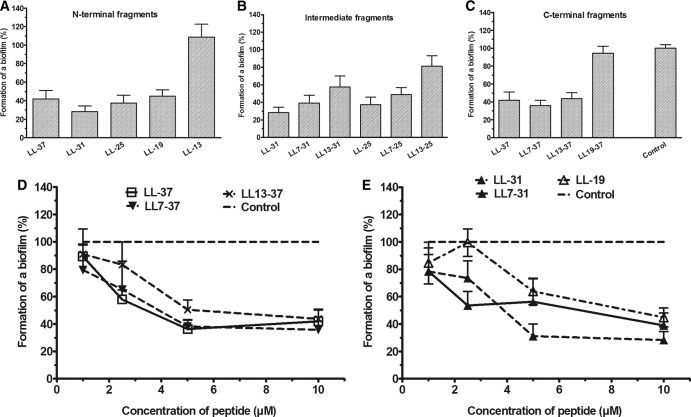
Study of the activity of the peptides on the formation of a biofilm. (A to C) The effect of the 13 peptides at 10 μM on the prevention of biofilm formation was assessed by measuring the OD590 of the wells after staining with crystal violet. (D and E) Serial dilution of the peptides LL-37, LL7-31, LL7-37, and LL13-37 (final concentrations, 1, 2.5, 5, and 10 μM) that were incubated for 18 h in the wells of a microplate containing a PAO1 bacterial suspension. The residual biofilm mass was evaluated using the crystal violet dye. The results were expressed as percentages of biofilm formation with a maximal percentage corresponding to the control well (no peptide added to bacteria). The results are the means ± SEM of three independent experiments performed in triplicate.
Eight out of 13 peptides significantly inhibited the formation of a biofilm. LL-31 and LL7-37 were the most efficient (∼70% decrease, P < 0.0001; n = 9). The activity of six peptides (LL-37, LL7-37, LL13-37, LL-31, LL7-31, and LL-19) was further investigated by measuring the percentage of biofilm formation after incubation with different concentrations of these fragments. As shown in Fig. 4, LL-37, LL7-37, and LL-31 significantly inhibited the percentage of biomass formation at 5 μM (P < 0.0001; n = 9). An enumeration of the viable cells was performed after 18 h of incubation of P. aeruginosa PAO1 with 4 peptides (10 μM). There was no significant difference in the number of viable cells for the control or for the bacteria exposed to the peptide fragments (Table 2). From these results, it could be concluded that the ability of the peptides to decrease the biomass was not related to the killing of the bacteria and a decrease of their number but probably to a decrease of the extracellular biomass.
Table 2.
Enumeration of the viable cells after exposure to the peptidesa
| Peptide | Log CFU/ml |
|---|---|
| Control | 10.1 |
| LL-37 | 9.8 |
| LL7-37 | 10.1 |
| LL13-37 | 9.7 |
| LL-31 | 9.9 |
The bacteria were grown for 18 h in the presence of 10 μM LL-37, LL7-37, LL13-37, or LL-31. An enumeration of the viable cells was then performed, and the results were expressed as the logarithm of the CFU/ml. A control was made by the incubation of the bacterial suspension without any peptides. The results are the means of two independent experiments performed in duplicate.
Study of the effect of six peptides on a pregrown biofilm using CLSM.
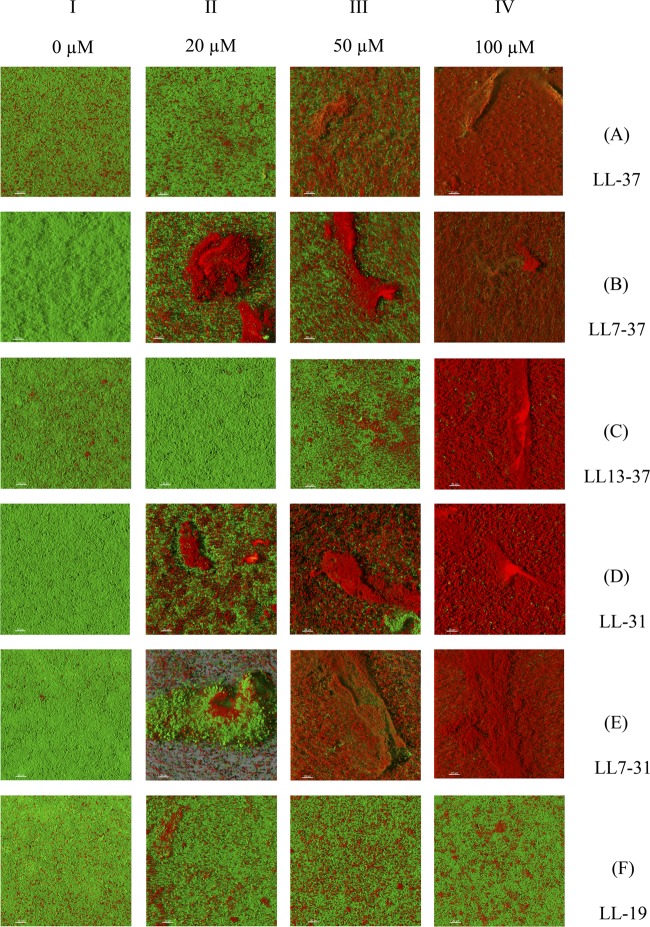
Biofilms were formed at the bottom of the wells of a microtiter plate. Bacteria expressing GFP were used for these studies. The cells were incubated for 24 h in TSB medium and then incubated for 25 min in the presence of 10 μM PI and various concentrations (from 0 to 100 μM) of the peptides. The intact cells expressing the GFP appeared green, and the dead cells, which had been permeabilized to PI, appeared red. As shown in Fig. 5A-I to F-I, the cells incubated in control conditions appeared green with very few small red spots. These results confirmed that the bacteria incubated in control conditions were not permeable to PI. Most of the bacteria exposed to 100 μM LL-37 had turned red (Fig. 5A-IV). The effect of LL-37 was dose dependent in the 20 μM to 100 μM concentration range (Fig. 5A). LL7-37 was more effective than LL-37. At 20 μM, it increased the PI uptake of a substantial fraction of the surface of the biofilm (Fig. 6B-II). The two fragments LL-31 and LL7-31 had activities similar to that of LL7-37 (Fig. 5C and D). These three fragments were the most effective in permeabilizing the bacterial membrane inside the biofilm. LL13-37 was less active than LL-37 (LL7-37 = LL-31 = LL7-31 > LL-37 > LL13-37) (Fig. 5E). The fragment LL-19 had only a marginal effect and provoked a very small increase of PI uptake at 100 μM (Fig. 5F). These results established that LL-37 and some of its derived fragments have bactericidal activity even within a biofilm.
Fig 5.
Effect of six peptides on a GFP-tagged P. aeruginosa PAO1 biofilm stained with PI. A 24-h-old GFP-tagged PAO1 biofilm, grown in the wells of a μClear microtiter plate, was exposed to increasing peptide concentrations in the presence of 10 μM PI for 25 min. After treatment, the biofilm was rinsed and observed by CLSM. Damaged cells are stained red by the entry of PI into the cell, and live cells are stained green by the expression of GFP. The micrograph images represent a tridimensional view from the top of the biofilm and are the most representative of two independent experiments. The white bars on the pictures represent a 20 μM distance.
Fig 6.

Effect of LL7-37 on a GFP-tagged P. aeruginosa PAO1 biofilm stained with PI. The peptide LL7-37 at 0 μM (A), 25 μM (B), 50 μM (C), and 100 μM (D) (0, 95, 190, and 380 mg/liter) was exposed on a 24-h-old biofilm grown in a CDC reactor for 25 min. Live cells appear green by expression of the GFP, and the damaged cells appear red by the permeability of the dead cells to PI. The confocal laser scanning micrographs show a horizontal section with two flanking images representing sections in the xz and yz planes, respectively (left column). The right column represents a tridimensional view from the top of the biofilm.
Effect of LL7-37 on bacteria within a CDC biofilm reactor.
The biofilms formed in the wells of a microplate are rather flat. To study the effect of the peptides on thicker biofilms with a tridimensional structure and more similar to the biofilms found in vivo, an experiment with the CDC bioreactor was designed. In this experimental model, the biofilm developed under a constant flux of medium and shear forces. The biofilm formed a thick and uniform layer on the inert support. The activity of LL7-37, one of the most active fragments on biofilms, was studied. As shown in Fig. 6A, and in agreement with the previous results, PI did not stain the biofilm in control conditions. The concentrations of the peptide active on a biofilm grown in a CDC reactor were in the same range as those active on a biofilm grown in a microtiter plate. At lower concentrations, LL7-37 not only increased the permeability of some bacteria to PI but also damaged the upper layer of the biofilm (Fig. 6B and C). A high concentration (100 μM) of LL7-37 permeabilized the bacterial membrane of the entire microcolony (Fig. 6D) and disrupted the integrity of the surface of the biofilm.
Determination of the secondary structures of the peptides using ATR-FTIR.
The secondary structures of the six peptides used in the studies with CLSM were analyzed by ATR-FTIR spectroscopy (Fig. 7). The spectrum of the peptides was compared with the spectrum of myoglobin, a protein with a high content of α-helices, and with the spectrum of avidin, a peptide with β-sheets. The spectra were recorded on peptides prepared in a buffer with a low ionic strength. Out of the six peptides studied, the spectrum of five peptides (LL-37, LL7-37, LL13-37, LL-31, and LL7-31) had a narrow major peak in the 1,653- to 1,654-cm−1 region, which was also the region of the major peak of the spectrum of myoglobin (Fig. 7A) and is the region of the spectrum where most α-helical proteins have their amide I maximum (14, 27). This result suggested that these peptides formed an α-helix in aqueous buffer. This was confirmed by replacing H2O with D2O. The hydrogen bond between the nitrogen atom of an amino group involved in an amide bond and the oxygen atom of a carboxylic function in the n + 4 amide bond within an α-helix delays the exchange between this hydrogen and deuterium (Fig. 7B), whereas it has been observed that random coiled or loose secondary structures rapidly exchange their amide proton (8). The deuteration of the five peptides slightly displaced the major peak which remained in the band of the α-helix. The shortest fragment, LL-19, had different spectra. In the absence of deuterium, the major peak at 1,647 cm−1 was much wider than the peak observed with the other peptides. This result suggested that structures other than the α-helix (random coil and β-sheet) might contribute to this peak. This hypothesis was confirmed by the deuteration of the peptide. A major peak emerged at 1,616 cm−1 in the vicinity of the major peak observed with avidin, a protein used as a reference for proteins containing β-sheets. A second peak also emerged at 1,685 cm−1, which is characteristic of antiparallel β-sheet, as found in beta-amyloid oligomers and OmpF porin as opposed to parallel β-sheet (4, 5). The spectra therefore suggested that LL-19 did not form a clear α-helix in the buffer but formed another secondary structure, probably an antiparallel β-sheet aggregate.
Fig 7.
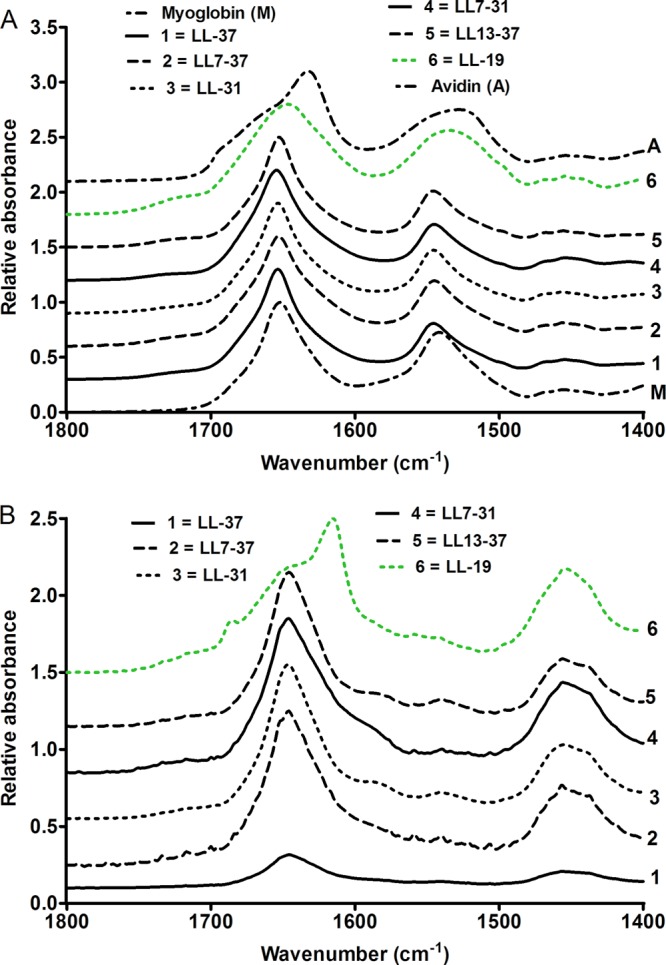
Infrared ATR spectra of six LL-37 truncated fragments. (A) Spectra of the peptides in aqueous buffer. (B) Spectra of the peptides after hydrogen/deuterium exchange.
Effect of the peptide LL-37 and some of its truncated fragments on the integrity of HUVEC.
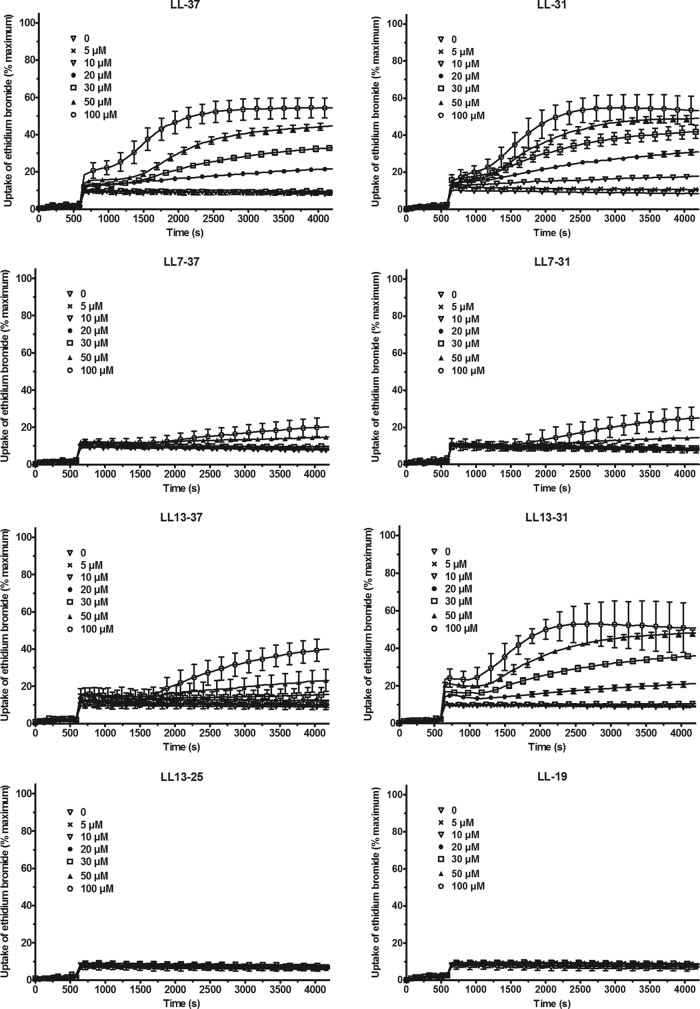
HUVEC were incubated in the presence of ethidium bromide, and the fluorescence was measured for 1 h. The cells were incubated for 10 min in the absence of the peptides (basal uptake of ethidium bromide) before being exposed to various concentrations of the various fragments. As shown in Fig. 8, 20 μM LL-37 provoked a linear increase of the fluorescence, and after 1 h incubation, the uptake of ethidium bromide increased from 7.4% ± 0.8% in the absence of the peptide to 19.8% ± 1.2% in the presence of the peptide (n = 9). The uptake of the dye sharply increased after 10 to 20 min of exposure to higher concentrations of the peptide. In the presence of 100 μM LL-37, the uptake of ethidium bromide reached 52.3% ± 2.8% (n = 9). This response was not affected by deleting 6 amino acids from the C-terminal domain (LL-31). Deleting the first 6 amino acids from the N-terminal domain of these peptides (LL7-37 and LL7-31) abolished the uptake of ethidium bromide, which was restored when the next 6 amino acids (LL13-37 and especially LL13-31) were removed. Shorter fragments (LL-19 and LL13-25) had no effect on the uptake of ethidium bromide.
Fig 8.
Effect of the peptides on the uptake of ethidium bromide by HUVEC. HUVEC were incubated at 37°C in the presence of ethidium bromide (final concentration, 25 μM) and of various concentrations of LL-37 and some of its truncated fragments. The uptake of ethidium bromide by HUVEC was followed by monitoring the fluorescence at 590 nm after excitation at 540 nm for 1 h. At the end of the experiment, 0.5% (wt/vol) Triton X-100 was added to the wells to determine the maximal uptake of ethidium bromide. The results are expressed as percentages of the maximal uptake and are the means ± SEM of three independent assays performed in triplicate.
In the next experiments, the full peptide and 7 fragments were tested on the release of the cytosolic enzyme LDH by HUVEC. Concentrations of LL-37 higher than 10 μM induced the release of LDH, which reached 50% of the total LDH content at 100 μM. Similar results were obtained with LL-31 and LL13-31. The other five tested fragments had no effect on the release of LDH (Fig. 9).
Fig 9.
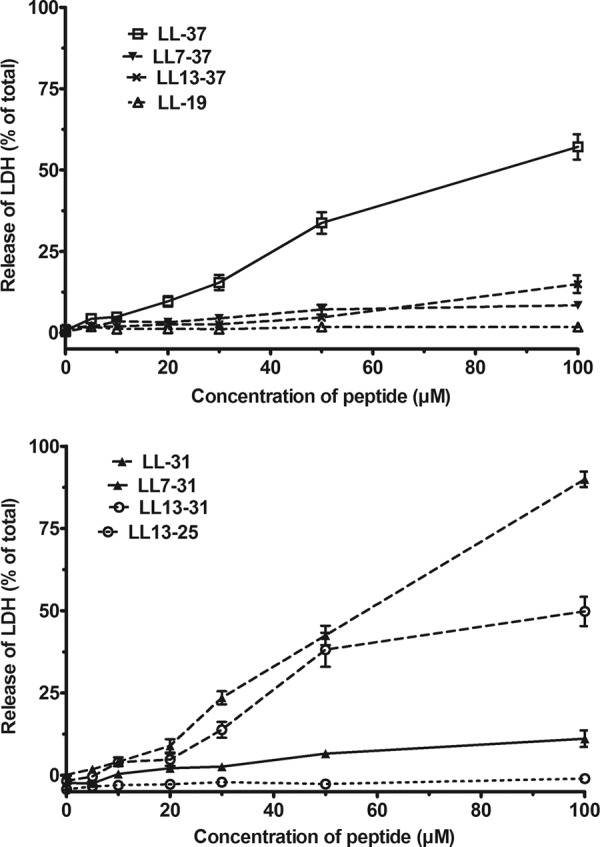
Effect of the peptides on the release of LDH by HUVEC. HUVEC were incubated at 37°C for 20 min in the presence of various concentrations of LL-37 and some of its truncated fragments. After centrifugation, the LDH present in the supernatant was assayed as described in Materials and Methods. The total activity of LDH was assayed after the lysis of HUVEC. The results are expressed as the percentages of the total LDH activity during the incubation with the peptides. They are the means ± SEM of two independent experiments performed in triplicate.
In another experiment, the effect of the fragments on the integrity of the mitochondria of HUVEC was examined using the MTT test (Fig. 10). At all concentrations tested, the absorbance measured after incubation in the presence of the peptides was similar to the absorbance measured in their absence (Fig. 10), suggesting that the ability of the mitochondria to transfer electrons was maintained in the presence of the peptides. It could be concluded that some of the peptides permeabilized the plasma membrane of eukaryotic cells but had no deleterious effect on their mitochondria.
Fig 10.
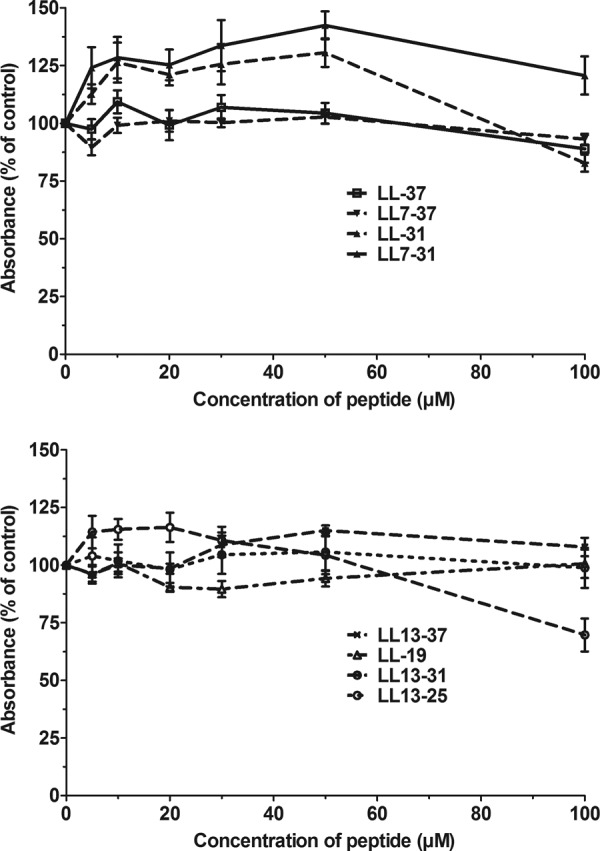
Effect of the peptides on the mitochondrial activity of HUVEC. HUVEC were incubated for 20 min in the presence of various concentrations of truncated fragments of LL-37. After removal of the medium, the mitochondrial activity was estimated with the MTT test as described in Materials and Methods. The results are expressed as the percentages of the absorbance measured in HUVEC incubated in the absence of the peptides and are the means ± SEM of two independent assays performed in triplicate.
DISCUSSION
Our results show that not only LL-37 but also some of its fragments blocked the formation of a biofilm measured with the crystal violet staining technique. The inhibition by LL-37 was previously reported by Kapoor et al. (21) and by Dean et al. (7), who showed a reduction by 43% of biofilm formation with LL-37-treated PAO1 cultures. Overhage et al. (30) demonstrated that LL-37 modified the expression of nearly 800 genes and blocked the formation of a biofilm by PAO1. Our experiments were performed in ionic conditions that inhibited the antimicrobial properties of low concentrations of the peptide (1). Hence, the decrease of the crystal violet staining was not secondary to the decrease of the cell number. Considering that the crystal violet stains not only cells but also the extracellular matrix of the biofilm, the decrease of the staining in the presence of LL-37 was probably provoked by a decrease of the extracellular matrix. We found that 7 out of the 12 truncated fragments of LL-37 also inhibited the formation of a biofilm. LL7-37 and LL-31 were the most efficient and provoked a decrease of the percentage of biomass formation at a very low concentration. These results suggest that the antibiofilm activity of LL-37 involves a rather long fragment of the peptide. Our results are at variance with the recent results of de la Fuente-Núñez et al. (9). The authors discovered that a nonapeptide derived from a fragment of LL-37 had a very strong antibiofilm activity. It should, however, have been mentioned that this active peptide (KRFRIRVRV) is not per se a fragment of LL-37, which makes the comparison between their results and ours less relevant.
LL-37 also affected PAO1 within an established biofilm. The peptide dose dependently increased the permeability of the bacteria to PI. The effect of LL-37 on existing biofilms has been demonstrated by Overage et al. (30), who reported the reduction of pregrown P. aeruginosa biofilm formation in vitro. Truncated fragments, especially LL7-37, LL-31, and LL7-31, also increased the uptake of PI by the bacteria forming the biofilm. Further truncation of LL7-37 by 6 residues (LL13-37) attenuated the response to the peptide. LL7-37 not only had an antibacterial activity but also decreased the height of the biofilm and promoted its disruption. This is the first report on the destruction of a biofilm formed by P. aeruginosa using fragments of LL-37, confirming the potentialities of these peptides for the development of new treatments of cystic fibrosis (48).
The effect of the library of LL-37 truncated fragments on a B. pseudomallei biofilm has already been investigated (20). LL-31 exhibited the strongest activity on a pregrown biofilm at a lower concentration than the concentration active on a P. aeruginosa biofilm.
LL-37, LL7-37, LL13-37, LL-31, and LL7-31, the 5 peptides with antibacterial activity on sessile bacteria, formed an α-helix in an aqueous solution. This secondary structure was confirmed by deuteration. LL-19, the N-terminal half of the peptide, was unique among the 6 tested peptides: it was not toxic against bacteria within the biofilm and formed secondary structures distinct from an α-helix, probably a β-sheet. The Agadir score (Table 1) differentiated LL-19 from the other 5 peptides (from less than 1 for LL-19 to 4 to 5 for the other peptides). This is in agreement with the results of Sigurdardottir et al. (37), who also noted the important difference of the Agadir score between the N-terminal fragment and the original peptide. These authors also reported that the score for LL17-37, the C-terminal fragment of LL-37, was close to 3. In line with this result, the C-terminal fragment of LL-37 formed an α-helix (36).
Cathelicidin-derived peptides disrupt not only bacterial membranes but also the plasma membrane of eukaryotic cells. This side effect has been a major drawback in the development of these peptides as anti-infective drugs. Our results confirm that LL-37 increases the permeability of the membrane of HUVEC to ethidium bromide and to LDH. LL-37 is also a cell-penetrating peptide which has been used to transfer DNA to the nucleus (32) and might thus have some intracellular toxicity. The activity of the mitochondrial dehydrogenases measured with the MTT test was not affected by the AMP. This result was unexpected, considering that LL-37 has been previously reported to affect the MTT test in keratinocytes (3). This toxicity might differ among various cells: Wong et al. (46) recently reported that LL-37 had no toxicity on the mitochondria of peripheral blood monocytes. Removal of the first 6 residues at the N-terminal side of the molecule greatly attenuated the toxicity of the peptide: LL7-37 and LL7-31 were much less toxic than LL-37 and LL-31. LL7-37 and LL7-31 also showed the most important increase of the uptake of PI by the bacteria within an established biofilm, confirming that these two fragments were the most promising peptides described in this study.
ACKNOWLEDGMENTS
We thank M. Franklin (Center for Biofilm Engineering, University of Montana) for giving us the GFP-tagged P. aeruginosa PAO1 strain and S. Pochet (Faculty of Pharmacy, Université libre de Bruxelles) for providing HUVEC.
C.N. is a research fellow of the Fonds National de la Recherche Scientifique (FNRS) in Belgium and was awarded a travel grant by the French Community Wallonie-Bruxelles of Belgium. This work was supported in part by the FNRS (grant 3457710 to J.-P.D. and M.V.) and by a grant from the University of Amsterdam for research into the focal point Oral Infections and Inflammation to J.G.B. and K.N.
Confocal microscopy was performed in the Imaging Shared Resource of the Center for Biofilm Engineering in Bozeman, MT. C.N., J.-P.D., M.V., and P.S.S. designed the experiments which were conducted by C.N. (all experiments), M.V. (IR spectroscopy), P.S.S., and B.P. (CLSM). J.G.B. and K.N. synthesized all the peptides. C.N. and J.-P.D. wrote the initial draft of the manuscript which was then edited by all the authors.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 20 August 2012
REFERENCES
- 1. Bals R, Wang X, Zasloff M, Wilson JM. 1998. The peptide antibiotic LL-37/hCAP-18 is expressed in epithelia of the human lung where it has broad antimicrobial activity at the airway surface. Proc. Natl. Acad. Sci. U. S. A. 95:9541–9546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bolscher JG, et al. 2011. Sortase A as a tool for high-yield histatin cyclization. FASEB J. 25:2650–2658 [DOI] [PubMed] [Google Scholar]
- 3. Braff MH, et al. 2005. Structure-function relationships among human cathelicidin peptides: dissociation of antimicrobial properties from host immunostimulatory activities. J. Immunol. 174:4271–4278 [DOI] [PubMed] [Google Scholar]
- 4. Cerf E, et al. 2009. Antiparallel β-sheet: a signature structure of the oligomeric amyloid β-peptide. Biochem. J. 421:415–423 [DOI] [PubMed] [Google Scholar]
- 5. Chirgadze YN, Nevskaya NA. 1976. Infrared spectra and resonance interaction of amide-I vibration of the antiparallel-chain pleated sheet. Biopolymers 15:607–625 [DOI] [PubMed] [Google Scholar]
- 6. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 7. Dean SN, Bishop BM, van Hoek ML. 2011. Susceptibility of Pseudomonas aeruginosa biofilm to alpha-helical peptides: D-enantiomer of LL-37. Front. Microbiol. 2:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Jongh HHJ, Goormaghtigh E, Ruysschaert JM. 1997. Amide-proton exchange of water-soluble proteins of different structural classes studied at the submolecular level by infrared spectroscopy. Biochemistry 36:13603–13610 [DOI] [PubMed] [Google Scholar]
- 9. de la Fuente-Núñez C, et al. 2012. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 56:2696–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. den Hertog AL, et al. 2006. The human cathelicidin peptide LL-37 and truncated variants induce segregation of lipids and proteins in the plasma membrane of Candida albicans. Biol. Chem. 387:1495–1502 [DOI] [PubMed] [Google Scholar]
- 11. Drenkard E, Ausubel FM. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740–743 [DOI] [PubMed] [Google Scholar]
- 12. Emerson J, Rosenfield M, McNamara S, Ramsey B, Gibson RL. 2002. Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr. Pulmonol. 34:91–100 [DOI] [PubMed] [Google Scholar]
- 13. Goeres DM, et al. 2005. Statistical assessment of a laboratory method for growing biofilms. Microbiology 151:757–762 [DOI] [PubMed] [Google Scholar]
- 14. Goormaghtigh E, Ruysschaert JM, Raussens V. 2006. Evaluation of the information content in infrared spectra for protein secondary structure determination. Biophys. J. 90:2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gordon YJ. 2005. Human cathelicidin (LL-37), a multifunctional peptide, is expressed by ocular surface epithelia and has potent antibacterial and antiviral activity. Curr. Eye Res. 30:385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hancock RE, Sahl HG. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24:1551–1557 [DOI] [PubMed] [Google Scholar]
- 17. Høiby N, Koch C. 1990. Pseudomonas aeruginosa infection in cystic fibrosis and its management. Thorax 45:881–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johansson J, Gudmundsson GH, Rottenberg ME, Berndt KD, Agerberth B. 1998. Conformation-dependent antibacterial activity of the naturally occurring human peptide LL-37. J. Biol. Chem. 273:3718–3724 [DOI] [PubMed] [Google Scholar]
- 19. Kanthawong S, et al. 2010. Antimicrobial activities of LL-37 and its truncated variants against Burkholderia thailandensis. Int. J. Animicrob. Agents 36:447–452 [DOI] [PubMed] [Google Scholar]
- 20. Kanthawong S, et al. 2012. Antimicrobial and antibiofilm activity of LL-37 and its truncated variants against Burkholderia pseudomallei. J. Antimicrob. Agents 39:39–44 [DOI] [PubMed] [Google Scholar]
- 21. Kapoor R, et al. 2011. Antimicrobial peptoids are effective against Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 55:3054–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lacroix E, Viguera AR, Serrano L. 1998. Elucidating the folding problem of α-helices: local motifs, long-range electrostatics, ionic-strength dependence and prediction of NMR parameters. J. Mol. Biol. 284:173–191 [DOI] [PubMed] [Google Scholar]
- 23. Moncla BJ, Pryke K, Rohan LC, Graebing PW. 2011. Degradation of naturally occurring and engineered antimicrobial peptides by proteases. Adv. Biosci. Biotechnol. 2:404–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nagant C, et al. 2010. Interaction between tobramycin and CSA-13 on clinical isolates of Pseudomonas aeruginosa in a model of young and mature biofilms. Appl. Microbiol. Biotechnol. 88:251–263 [DOI] [PubMed] [Google Scholar]
- 25. National Committee for Clinical Laboratory Standards 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed Approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, PA [Google Scholar]
- 26. Nguyen LT, Haney EF, Vogel HJ. 2011. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 29:464–472 [DOI] [PubMed] [Google Scholar]
- 27. Oberg KA, Ruysschaert JM, Goormaghtigh E. 2003. Rationally selected basis proteins: a new approach to selecting proteins for spectroscopic secondary structure analysis. Protein Sci. 12:2015–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reference deleted.
- 29. O'Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49–79 [DOI] [PubMed] [Google Scholar]
- 30. Overhage J, et al. 2008. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 76:4176–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Porcelli F, et al. 2008. NMR structure of the cathelicidin-derived human antimicrobial peptide LL-37 in dodecylphosphocholine micelles. Biochemistry 47:5565–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sandgren S, et al. 2004. The human antimicrobial peptide LL-37 transfers extracellular DNA plasmid to the nuclear compartment of mammalian cells via lipid rafts and proteoglycan-dependent endocytosis. J. Biol. Chem. 279:17951–17956 [DOI] [PubMed] [Google Scholar]
- 33. Reference deleted.
- 34. Reference deleted.
- 35. Seil M, El Ouaaliti M, Dehaye JP. 2012. Secretion of IL-1β triggered by dynasore in murine peritoneal macrophages. Innate Immun. 18:241–249 [DOI] [PubMed] [Google Scholar]
- 36. Seil M, et al. 2010. Regulation by CRAMP of the responses of murine peritoneal macrophages to extracellular ATP. Biochim. Biophys. Acta 1798:569–578 [DOI] [PubMed] [Google Scholar]
- 37. Sigurdardottir T, et al. 2006. In silico identification and biological evaluation of antimicrobial peptides based on human cathelicidin LL-37. Antimicrob. Agents Chemother. 50:2983–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Singh PK, et al. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762–764 [DOI] [PubMed] [Google Scholar]
- 39. Sørensen OE, et al. 2001. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood 97:3951–3959 [DOI] [PubMed] [Google Scholar]
- 40. Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. 2000. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 40:175–179 [DOI] [PubMed] [Google Scholar]
- 41. Takahashi D, Shukla SK, Prakash O, Zhang G. 2010. Structural determinants of host defense peptides for antimicrobial activity and target cell selectivity. Biochimie 92:1236–1241 [DOI] [PubMed] [Google Scholar]
- 42. Tomasinsig L, et al. 2009. Structure dependence of biological activities for primate cathelicidins. J. Pept. Sci. 15:576–582 [DOI] [PubMed] [Google Scholar]
- 43. Turner J, Cho Y, Dinh NN, Waring AJ, Lehrer RI. 1998. Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob. Agents Chemother. 42:2206–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vigano C, Manciu L, Buyse F, Goormaghtigh E, Ruysschaert JM. 2000. Attenuated total reflection IR spectroscopy as a tool to investigate the structure, orientation and tertiary structure changes in peptides and membrane proteins. Biopolymers 55:373–380 [DOI] [PubMed] [Google Scholar]
- 45. Wang G. 2008. Structures of human host defense cathelicidin LL-37 and its smallest antimicrobial peptide KR-12 in lipid micelles. J. Biol. Chem. 283:32637–32643 [DOI] [PubMed] [Google Scholar]
- 46. Wong JH, et al. 2011. Antifungal action of human cathelicin fragment (LL13-37) on Candida albicans. Peptides 32:1996–2002 [DOI] [PubMed] [Google Scholar]
- 47. Zanetti M, Gennaro R, Skerlavaj B, Tomasinsig L, Circo R. 2002. Cathelicidin peptides as candidates for a novel class of antimicrobials. Curr. Pharm. 8:779–793 [DOI] [PubMed] [Google Scholar]
- 48. Zhang L, et al. 2005. Antimicrobial peptide therapeutics for cystic fibrosis. Antimicrob. Agents Chemother. 49:2921–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]