Abstract
A blaKPC-9 carbapenemase variant was discovered in isolates of Klebsiella pneumoniae and Escherichia coli from a single patient. It differed from blaKPC-3 by one amino acid substitution (Val239Ala). The K. pneumoniae isolate was typed as ST258, as was the epidemic Israeli KPC-3 clone. blaKPC-9 was found on a plasmid indistinguishable from pKpQIL that carries blaKPC-3 in the epidemic clone. Compared to KPC-3, KPC-9 conferred less resistance to carbapenems and higher resistance to ceftazidime.
TEXT
Klebsiella pneumoniae carbapenemases (KPCs) are molecular class A serine β-lactamases belonging to functional group 2f. Since first reported in 2001 (18), 12 types (from KPC-2 to KPC-13) have been submitted to GenBank (http://www.lahey.org/Studies/other.asp). KPC-3-producing carbapenem-resistant K. pneumoniae is disseminating worldwide through relatively few clones. The dominant multilocus sequence type (ST) around the world is ST258, and KPC-3-producing ST258 has spread in Israel since 2005 (9, 12). This strain harbors a single blaKPC-3-positive plasmid, designated pKpQIL (10). Here we report the novel KPC-9 allele found in both K. pneumoniae and Escherichia coli isolates originating in the same patient.
(This work was presented in part at the 51st Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL, 2011.)
The strains carrying KPC-9 were isolated from a patient admitted to our hospital in November 2008 due to acute rectal bleeding. During his stay, the patient was treated with ertapenem for urinary tract infection, and a multidrug-resistant K. pneumoniae strain (designated Kp141) was isolated from a stool surveillance culture. Four weeks later, a sputum culture yielded an E. coli isolate (Ec136) which was also resistant to all the β-lactams.
Drug susceptibility assays were performed by Etest. Carbapenemase production was identified using the modified Hodge test with a 10-μg ertapenem disc (5), and carbapenem MICs were determined by a standard CLSI agar dilution method (4). All assays were repeated at least three times. Kp141 was resistant to all β-lactams but susceptible to colistin, according to EUCAST (6), and gentamicin, with MICs similar to those obtained for Kp154, the KPC-3-producing epidemic clone isolated in our hospital (16) (Table 1). Kp154 was further used as a control in all assays. As with Kp141, Ec136 also produced a carbapenemase and was susceptible to colistin, gentamicin, amikacin, co-trimoxazole, and chloramphenicol. PCR to detect blaKPC was done as previously described (2). Sequencing the blaKPC genes in both Kp141 and Ec136 revealed a novel KPC-type carbapenemase that differed from blaKPC-3 only by the transition T716C, leading to a Val239Ala change relative to KPC-3 (GenBank accession number AF395881).
Table 1.
Susceptibilities of the studied strainsa
| Strain | KPC type | MIC (μg/ml) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPM | ETP | MEM | PIP | TZP | CRO | CAZ | FEP | GEN | AMK | CIP | MXF | CST | TGC | ||
| Kp154 | KPC-3 | 128 | 64 | 64 | >256 | >256 | >32 | >256 | ≥256 | 1.5 | 32 | 24 | 12 | 1.5 | 0.5 |
| Kp141 | KPC-9 | 64 | 64 | 64 | >256 | >256 | >32 | >256 | ≥256 | 1.5 | 32 | >32 | 16 | 1.5 | 0.5 |
| Ec136 | KPC-9 | 4 | 2 | 2 | 128 | 64 | 8 | 64 | 8 | 1 | 1.5 | >32 | 12 | 1.5 | 0.125 |
| Kp154-TC | KPC-3 | 8 | 2 | 2 | 96 | 32 | 8 | 16 | 2 | 0.25 | 1.5 | 0.125 | 0.016 | 0.38 | 0.19 |
| Kp141-TC | KPC-9 | 2 | 0.5 | 1 | 96 | 32 | 12 | 64 | 4 | 0.25 | 1.5 | 0.125 | 0.016 | 0.38 | 0.19 |
| Ec136-TC | KPC-9 | 2 | 0.5 | 1 | 96 | 32 | 12 | 64 | 6 | 0.25 | 1 | 0.125 | 0.016 | 0.38 | 0.19 |
| E. coli J53 | 0.05 | 0.006 | 0.006 | 0.75 | 0.75 | 0.023 | 0.125 | 0.047 | 0.125 | 0.5 | 0.006 | 0.016 | 0.38 | 0.19 | |
TC, transconjugant; IPM, imipenem; ETP, ertapenem; MEM, meropenem; PIP, piperacillin; TZP, piperacillin-tazobactam; CRO, ceftriaxone; CAZ, ceftazidime; FEP, cefepime; GEN, gentamicin; AMK, amikacin; CIP, ciprofloxacin; MXF, moxifloxacin; CST, colistin; TGC, tigecycline.
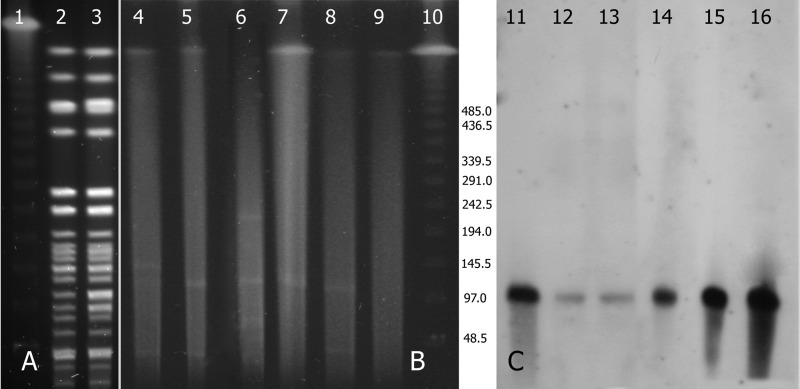
Pulsed-field gel electrophoresis (PFGE) after restriction with XbaI was performed according to a standardized protocol on Kp141 and Kp154 (http://www.cdc.gov/pulsenet/). The PFGE profiles of the two isolates were identical (Fig. 1A). Multilocus sequence typing was carried out according to the K. pneumoniae protocol described on the MLST website (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html). Kp141 belonged to ST258. The first carbapenem-resistant K. pneumoniae (CRKP) strains isolated at Hadassah in the beginning of the epidemic in 2005 were also ST258, but the local epidemic clone subsequently mutated into ST512, which then became dominant in our hospital (16). Screening a random sample of 18 isolates from the repository of the KPC-positive Enterobacteriaceae strains isolated at our hospital since 2005 for additional KPC-9 revealed one E. coli strain, isolated in September 2008, that caused intravascular catheter exit site infection in a hemodialysis patient. Thus, KPC-9 likely evolved in Kp141 from CRKP blaKPC-3 before divergence from ST258 to ST512 occurred. Analysis of a larger number of isolates is needed in order to study the prevalence of KPC-9 among other Enterobacteriaceae.
Fig 1.
(A) PFGE (XbaI restricted) of Kp154 (lane 2) and Kp141 (lane 3). Lanes 1 and 10, lambda ladder (New England BioLabs, Ipswich, MA). (B and C) PFGE of plasmids after S1 treatment and a Southern blot after blaKPC labeling. Lanes 4 and 11, Ec136; lanes 5 and 12, Ec136-TC; lanes 6 and 13, Kp141; lanes 7 and 14, Kp141-TC; lanes 8 and 15, Kp154; lanes 9 and 16, Kp154.
The blaKPC-carrying plasmids from Kp141, Kp154, and Ec136 were conjugated into an azide-resistant (Azr) E. coli J53 strain. Plasmids were subsequently sized by PFGE following S1 nuclease treatment, and blaKPC was identified by Southern blot analysis of donors and transconjugants as previously reported (Fig. 1B and C) (16). We showed previously that Kp154 harbored a plasmid identical to pKpQIL, the blaKPC-3-positive plasmid in the national epidemic CRKP strain. Kp141 and Ec136 carried a single blaKPC-carrying plasmid that was identical in size to pKpQIL. Additionally, BglII restriction patterns of the blaKPC-positive plasmids extracted from transconjugants of Kp141 and Ec136 were identical to that of pKpQIL. Inverse PCR followed by primer walking indicated that, as in pKpQIL, blaKPC-3 in Kp154 and blaKPC-9 in both Kp141 and Ec136 are embedded in the Tn4401a isoform, in which there is a 99-bp deletion upstream of the blaKPC gene (10). These data suggest that Kp141, derived from an ST258 strain harboring pKpQIL, and Ec136 likely acquired the plasmid by horizontal transfer.
Kp154 was 2-fold more resistant to imipenem than Kp141, whereas the MICs of ertapenem and meropenem were similar for both (Table 1). The higher resistance conferred by KPC-3 was more apparent in the transconjugants; the MICs of imipenem and ertapenem were 4-fold higher than they were for KPC-9, and the MIC of meropenem for the KPC-3 transconjugant was 2-fold higher than that for the KPC-9 transconjugant. A difference in resistance to cephalosporins was also apparent in the transconjugants. Notably, the MIC of ceftazidime was 4-fold higher for the KPC-9 transconjugant than for the KPC-3 transconjugant. There are additional antibiotic resistance genes, including blaTEM-1, in pKpQIL. To verify the different resistance phenotypes, blaKPC-3 and blaKPC-9 were cloned into the expression vector pZA24-luc (11). For cloning, their entire coding regions were amplified from strains Kp154 and Kp141 with the upstream and downstream primers KPC-F-KpnI (5′-GGGGTACCATGTCACTGTATCGCC) and KPC-R-EcoNI (5′-CCTTTATGAGGTTACTGCCCGTTGAC), respectively, which contained engineered KpnI and EcoNI sites, respectively. The PCR products were then ligated to generate pZA24KPC3 and pZA24KPC9, which were electroporated into E. coli DH5α (Gibco-BRL), and their sequences were confirmed. Under identical induction conditions (100 mM IPTG [isopropyl-β-d-thiogalactopyranoside] or 2 mg/ml d-arabinose), the MICs (Etest) of imipenem for pZA24KPC3 and pZA24KPC9 were 32 μg/ml and 12 to 16 μg/ml, respectively. The respective MICs of ertapenem were 32 μg/ml and 16 μg/ml, and the respective MICs of ceftazidime were 24 μg/ml and 128 μg/ml. To account for the slightly smaller difference in MIC for the cloned blaKPC genes alone than for the transconjugants, a chimera ofblaTEM-1 in tandem with blaKPC was cloned under a blaTEM-1 promoter. The MICs did not change. Thus, KPC-3 by itself conferred higher-order carbapenem resistance than KPC-9.
KPC-3 and KPC-9 were directly prepared from their corresponding overexpressed cloned genes as previously described (7). The extracts contained the recombinant proteins at over 90% homogeneity (Fig. 2) as determined using ImageJ 1.46r software (15). The hydrolytic activities of KPC-3 and KPC-9 were determined as previously described (1), using a Cary 300 UV-visible spectrophotometer (Agilent Technologies, Santa Clara, CA). All assays were repeated at least three times. A preparation of pZA24-luc in E. coli DH5α did not manifest catalytic activity. Overall, KPC-9 exhibited a kinetic profile similar to that of the KPC-3 enzyme for piperacillin, ceftriaxone, and ertapenem (Table 2). The catalytic efficiency of KPC-3 for imipenem was higher than that of KPC-9, which, on the other hand, hydrolyzed ceftazidime more efficiently than KPC-3; the kcat/Km ratio of KPC-9 was 6.4-fold higher than that of KPC-3. Thus, the kinetic parameters of the respective enzymes support the MIC observations: KPC-3, with its slightly higher kcat/Km ratio, provided more resistance to imipenem than KPC-9, whereas the opposite was true regarding ceftazidime.
Fig 2.
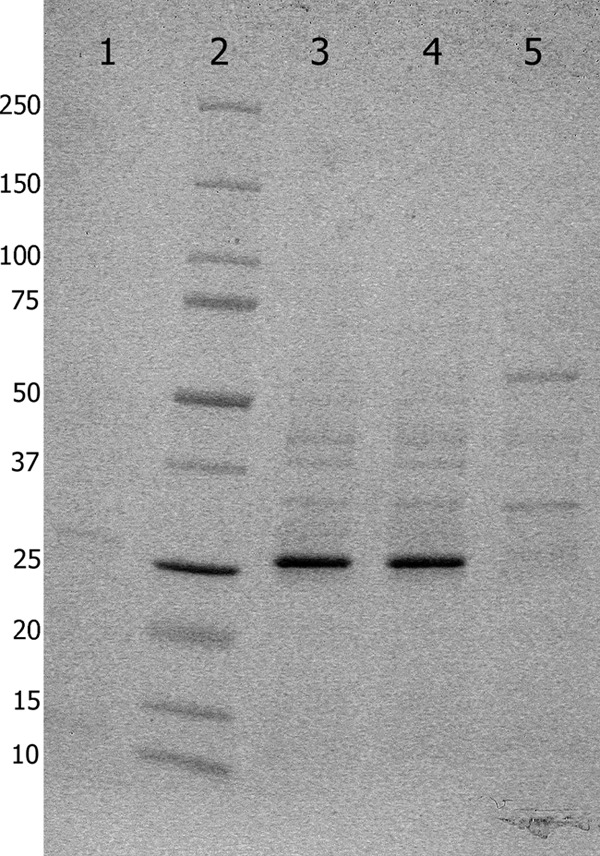
SDS-PAGE showing total proteins from freeze-thaw supernatants. Lane 1, uninduced pZA24-KPC-9 (due to its leaky expression, a faint band is seen at 28.5 kDa); lane 2, Precision Plus Protein standards (Bio-Rad, Hercules, CA); lane 3, induced pZA24-KPC-3; lane 4, induced pZA24-KPC-9; lane 5, induced pZ-Bgl (an identical plasmid expressing a higher-molecular-mass protein; no protein is seen at the size of KPC).
Table 2.
Kinetic parameters for KPC-3 and KPC-9
| Substrate | Parameters fora: |
|||||
|---|---|---|---|---|---|---|
| KPC-3 |
KPC-9 |
|||||
| Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | |
| Piperacillin | 50 ± 5 | 7.1 ± 4.3 | 0.14 ± 0.1 | 61 ± 19 | 8.5 ± 1.5 | 0.14 ± 0.07 |
| Ceftriaxone | 122 ± 19 | 29.1 ± 1 | 0.24 ± 0.05 | 104 ± 3 | 29.5 ± 1.2 | 0.28 ± 0.01 |
| Ceftazidime | 168 ± 33 | 37.9 ± 3.5 | 0.23 ± 0.03 | 32 ± 6 | 47.4 ± 5.3 | 1.47 ± 0.05 |
| Ertapenem | 177 ± 41 | 47.5 ± 10.9 | 0.27 ± 0.11 | 221 ± 56 | 52.8 ± 5.4 | 0.24 ± 0.08 |
| Imipenem | 26 ± 6 | 110.1 ± 4.2 | 4.24 ± 0.13 | 34 ± 9 | 101.5 ± 11.9 | 3.02 ± 0.7 |
Values are means and standard deviations.
Since the discoveries of the synonymous KPC-1 and KPC-2 enzymes, the conserved KPC family has grown rapidly. Except for blaKPC-12 and blaKPC-13, variations among KPCs are in four single nucleotide loci. It was previously hypothesized that blaKPC-3 evolved from blaKPC-2 (17) and that later, blaKPC-8, which evolved from blaKPC-3, was the founding sequence of blaKPC-9 (3). The isolation of blaKPC-9 strains within a dominant blaKPC-3 epidemic is, however, in agreement with an evolution directly from the latter gene variant.
Residue 237 is necessary to maintain carbapenemase activity in KPC (14). Based on the solved structure of KPC-2 (8), residue 239, which is one of the four hot spots of KPC variations, is in proximity to the active site and probably affects enzymatic activity. Amino acid changes between KPCs also appear to influence the kinetic properties of the enzymes. It has previously been shown that the MICs of carbapenems for the KPC-4 (Arg103, Gly239) transformant are lower than those for the KPC-2 (Pro103, Val239) transformant, whereas the MIC of ceftazidime was higher for the KPC-4 transformant; the MICs for the KPC-5 (Arg103, Val239) transformant gave intermediate susceptibility (17). Our finding of higher-order ceftazidime resistance for the KPC-9 (Ala239) transformant than for the KPC-3 (Val239) transformant and a reverse effect for imipenem are in agreement with these findings. Further structure work is required to examine the importance of residue changes in KPC enzymes on resistance.
Nucleotide sequence accession number.
The GenBank accession number for blaKPC-9 is FJ624872.
ACKNOWLEDGMENT
This work was supported in part by grant Morasha 1833/07 from the Israel Science Foundation to J.S.
Footnotes
Published ahead of print 10 September 2012
REFERENCES
- 1. Alba J, Ishii Y, Thomson K, Moland ES, Yamaguchi K. 2005. Kinetics study of KPC-3, a plasmid-encoded class A carbapenem-hydrolyzing β-lactamase. Antimicrob. Agents Chemother. 49:4760–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bradford PA, et al. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 beta-lactamases in New York City. Clin. Infect. Dis. 39:55–60 [DOI] [PubMed] [Google Scholar]
- 3. Chen L, et al. 2011. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (blaKPC) variants. J. Clin. Microbiol. 49:579–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. CLSI 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. CLSI 2012. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement. CLSI M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. EUCAST 2012. Breakpoint tables for interpretation of MICs and zone diameters, version 2.0. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_2.0_120221.pdf
- 7. Johnson BH, Hecht MH. 1994. Recombinant proteins can be isolated from E. coli cells by repeated cycles of freezing and thawing. Biotechnology 12:1357–1360 [DOI] [PubMed] [Google Scholar]
- 8. Ke W, Bethel CR, Thomson JM, Bonomo RA, van den Akker F. 2007. Crystal structure of KPC-2: insights into carbapenemase activity in class A beta-lactamases. Biochemistry 46:5732–5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kitchel B, et al. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53:3365–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leavitt A, Chmelnitsky I, Ofek I, Carmeli Y, Navon-Venezia S. 2010. Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug-resistant epidemic Klebsiella pneumoniae strain. J. Antimicrob. Chemother. 65:243–248 [DOI] [PubMed] [Google Scholar]
- 11. Lutz R, Bujard H. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Navon-Venezia S, et al. 2009. First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob. Agents Chemother. 53:818–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reference deleted.
- 14. Papp-Wallace KM, et al. 2010. Substrate selectivity and a novel role in inhibitor discrimination by residue 237 in the KPC-2 β-lactamase. Antimicrob. Agents Chemother. 54:2867–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9:671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Warburg G, et al. 2012. A carbapenem-resistant Klebsiella pneumoniae epidemic clone in Jerusalem: sequence type 512 carrying a plasmid encoding aac(6′)-Ib. J. Antimicrob. Chemother. 67:898–901 [DOI] [PubMed] [Google Scholar]
- 17. Wolter DJ, et al. 2009. Phenotypic and enzymatic comparative analysis of the novel KPC variant KPC-5 and its evolutionary variants, KPC-2 and KPC-4. Antimicrob. Agents Chemother. 53:557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yigit H, et al. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]