Abstract
Screening of a transposon insertion mutant library of Pseudomonas aeruginosa for increased susceptibility to paromomycin identified a number of genes whose disruption enhanced susceptibility of this organism to multiple aminoglycosides, including tobramycin, amikacin, and gentamicin. These included genes associated with lipid biosynthesis or metabolism (lptA, faoA), phosphate uptake (pstB), and two-component regulators (amgRS, PA2797-PA2798) and a gene of unknown function (PA0392). Deletion mutants lacking these showed enhanced panaminoglycoside susceptibility that was reversed by the cloned genes, confirming their contribution to intrinsic panaminoglycoside resistance. None of these mutants showed increased aminoglycoside permeation of the cell envelope, indicating that increased susceptibility was not related to enhanced aminoglycoside uptake owing to a reduced envelope barrier function. Several mutants (pstB, faoA, PA0392, amgR) did, however, show increased cytoplasmic membrane depolarization relative to wild type following gentamicin exposure, consistent with the membranes of these mutants being more prone to perturbation, likely by gentamicin-generated mistranslated polypeptides. Mutants lacking any two of these resistance genes in various combinations invariably showed increased aminoglycoside susceptibility relative to single-deletion mutants, confirming their independent contribution to resistance and highlighting the complexity of the intrinsic aminoglycoside resistome in P. aeruginosa. Deletion of these genes also compromised the high-level panaminoglycoside resistance of clinical isolates, emphasizing their important contribution to acquired resistance.
INTRODUCTION
Pseudomonas aeruginosa is a common nosocomial pathogen (36, 86) that causes a variety of infections, with one of the most frequent sites of P. aeruginosa infection being the lungs of cystic fibrosis (CF) patients (27). Aminoglycosides are commonly used to treat such infections, with aerosolized tobramycin and/or intravenous (i.v.) tobramycin or amikacin used to treat prechronic infections of P. aeruginosa in CF patients as well as acute exacerbations that occur following the onset of chronicity (10, 64, 82). Aminoglycoside use in treating pulmonary infections in CF patients is, however, associated with resistance development (9, 30, 58, 61), which complicates antipseudomonal chemotherapy.
Aminoglycoside antibiotics bind the 16S rRNA component of the 30S ribosomal subunit, disrupting translation and causing the accumulation of truncated/aberrant polypeptides (15). These polypeptides insert into the cytoplasmic membrane and compromise membrane integrity (16), ultimately permitting accumulation of additional drug and total inhibition of all cellular ribosomes (43) as well as activating stress responses that culminate in the production of reactive oxygen species (ROS), which appear to be responsible for the lethal effect of aminoglycosides (51). Intriguingly, initial uptake of these antimicrobials into bacterial cells is an active (i.e., energy-requiring) process, although the details are as yet unclear (81).
Bacterial resistance to aminoglycosides typically results from enzymatic modification of the drug, drug efflux, and target modification (43), with target modification involving mutation of genes for 16S rRNA and ribosomal proteins (43) or methylation of 16S rRNA by transposon-encoded methyltransferases (28). 16S rRNA methyltransferase-mediated aminoglycoside resistance is rarely seen in P. aeruginosa (19, 84), where the most common mechanism of aminoglycoside resistance involves aminoglycoside-modifying enzymes encoded by transmissible genes that are acquired by horizontal gene transfer (22, 49, 68). This is not, however, the case for CF patient isolates of P. aeruginosa, where these mechanisms are almost unknown (35, 68, 73). The predominant identified mechanism of aminoglycoside resistance in this case is efflux by the MexXY-OprM multidrug efflux system (39, 40), which is also responsible for earlier reports of so-called impermeability-type panaminoglycoside resistance that was frequently seen in CF patient P. aeruginosa isolates (68). Aminoglycoside resistance linked to lipopolysaccharide (LPS) changes in the P. aeruginosa outer membrane (OM; the site of initial binding of aminoglycosides during entry into bacterial cells [31, 46]) has also been reported (6, 25) and may, in some instances, be linked to an LPS modification locus (arn) (65) previously implicated in resistance to polycationic antimicrobials (e.g., polymyxins) (60) and regulated by the PhoPQ two-component system (TCS) (56). PhoPQ has, in fact, been implicated in aminoglycoside (including amikacin) resistance (56). Less commonly, resistance owing to ill-defined defects in aminoglycoside uptake (4, 7, 79), including defects in respiration/electron transport (5, 8, 25), and mutations in ribosomal components (25, 52) have been reported in P. aeruginosa. A recent study of P. aeruginosa transposon (Tn) insertion mutants screened for decreased susceptibility to aminoglycosides (tobramycin) identified a number of genes apparently linked to low-level aminoglycoside resistance, including a number involved in LPS biosynthesis and energy metabolism (72). An independent screen of a P. aeruginosa Tn mutant library for decreased resistance to several antimicrobial classes also identified genes linked to energy metabolism/respiration whose inactivation enhanced tobramycin susceptibility (21). Here we report a number of novel genes whose inactivation enhances susceptibility specifically to aminoglycosides. Intriguingly, all these appear to contribute independently to resistance, in some instances via an apparent impact on cytoplasmic membrane stability, highlighting the complexity of the P. aeruginosa intrinsic aminoglycoside resistome. Most importantly, these genes also contribute to the panaminoglycoside resistance of clinical (CF patient) isolates.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All bacterial strains were cultured at 37°C in Luria (L) broth or on L agar, unless otherwise indicated, with antimicrobials added as necessary. The pUT::mini-Tn5-tet plasmid was maintained in Escherichia coli with 100 μg/ml ampicillin or 10 μg/ml tetracycline. The pUC19 and pBluescript II SK (+) plasmids and their derivatives were maintained in E. coli with 100 μg/ml ampicillin. The pDSK519 plasmid and its derivatives were maintained in E. coli with 50 μg/ml kanamycin. Plasmids pEX18Tc and pRK415 and their derivatives were maintained in E. coli with 5 to 10 μg/ml tetracycline and in P. aeruginosa with 50 to 75 μg/ml tetracycline.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | ϕ80dlacZΔM15 endA1 hsdR17(rK− mK+) supE44 thi-1 recA gyrA96 relA1 F− Δ(lacZYA-argF)U169 | 1 |
| SM10(λpir) | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmr λpir | 62 |
| S17-1 | thi pro hsdR recA tra+ | 76 |
| GM2163 | F− dam-13::Tn9 (Cmr) dcm-6 hsdR2 (rk− mk+) leuB6 hisG4 thi-1 araC14 lacY1 galK2 galT22 xylA5 mtl-1 rpsL136 (Strr) fhuA31 tsx-78 glnV44 mcrA mcrB1 | NEB |
| P. aeruginosa | ||
| K767 | PAO1 wild type | 59 |
| K2153 | Panaminoglycoside-resistant clinical (CF patient) isolate | 77 |
| K2363 | K2153 amgR::mini-Tn5 tet | This study |
| K3104 | K2153 pstB::mini-Tn5 tet | This study |
| K2358 | K2153:proC::mini-Tn5 tet | This study |
| K2359 | K2153 faoA::mini-Tn5 tet | This study |
| K2365 | K2153 PA2798::mini-Tn5 tet | This study |
| K2361 | K2153 PA0392::mini-Tn5 tet | This study |
| K2413 | K767 ΔPA5471 | 63 |
| K2413-T3 | MexXY-expressing panaminoglycoside-resistant derivative of K2413 | S. Fraud, unpublished data |
| K3158 | K2413-T3 lptA::mini-Tn5 tet | This study |
| K1525 | K767 ΔmexXY | 17 |
| K3159 | K767 ΔamgR | This study |
| K3160 | K767 ΔpstB | This study |
| K3161 | K767 ΔfaoA | This study |
| K3162 | K767 ΔPA2798 | This study |
| K3163 | K767 ΔPA2797 | This study |
| K3164 | K767 ΔPA0392 | This study |
| K3165 | K767 ΔlptA | This study |
| K3166 | K767 ΔmexXY ΔamgR | This study |
| K3167 | K767 ΔmexXY ΔpstB | This study |
| K3168 | K767 ΔmexXY ΔfaoA | This study |
| K3169 | K767 ΔmexXY ΔPA2798 | This study |
| K3170 | K767 ΔmexXY ΔPA2797 | This study |
| K3171 | K767 ΔmexXY ΔPA0392 | This study |
| K3172 | K767 ΔmexXY ΔlptA | This study |
| K3173 | K767 ΔamgR ΔpstB | This study |
| K3174 | K767 ΔamgR ΔfaoA | This study |
| K3175 | K767 ΔamgR ΔPA2798 | This study |
| K3176 | K767 ΔamgR ΔPA2797 | This study |
| K3177 | K767 ΔamgR ΔPA0392 | This study |
| K3178 | K767 ΔamgR ΔlptA | This study |
| K3179 | K767 ΔpstB ΔlptA | This study |
| K3180 | K767 ΔpstB ΔfaoA | This study |
| K3181 | K767 ΔpstB ΔPA0392 | This study |
| K3182 | K767 ΔpstB ΔPA2798 | This study |
| K3183 | K767 ΔlptA ΔPA0392 | This study |
| K3184 | K767 ΔfaoA ΔPA0392 | This study |
| K3185 | K767 ΔlptA ΔfaoA | This study |
| K2160 | Panaminoglycoside-resistant clinical (CF patient) isolate | 77 |
| K3192 | K2160 ΔamgR | This study |
| K3193 | K2160 ΔpstB | This study |
| K3194 | K2160 ΔfaoA | This study |
| K3195 | K2160 ΔPA2798 | This study |
| K3196 | K2160 ΔPA0392 | This study |
| K3197 | K2160 ΔlptA | This study |
| K2162 | Panaminoglycoside-resistant clinical (CF patient) isolate | 77 |
| K3198 | K2162 ΔamgR | This study |
| K3199 | K2162 ΔpstB | This study |
| K3200 | K2162 ΔfaoA | This study |
| K3201 | K2162 ΔPA2798 | This study |
| K3202 | K2162 ΔPA0392 | This study |
| K3203 | K2162 ΔlptA | This study |
| Plasmids | Reference | |
| pUT::mini-Tn5-tet | Delivery vector for mini-Tn5 tet Apr Tcr | 18 |
| pUC19 | Cloning vector; Apr | 85 |
| pDSK519 | Broad-host-range cloning vector; Kmr | 48 |
| pBluescript II SK (+) | Cloning vector; Apr | Stratagene |
| pEX18Tc | Broad-host-range gene replacement vector; sacB Tcr | 38 |
| pRK415 | Broad-host-range cloning vector; Plac-MCS Tcr | 48 |
| pCSV05-01 | pEX18Tc::ΔmexXY | 17 |
| pCG005 | pEX18Tc::ΔamgR | This study |
| pCG006 | pEX18Tc::ΔpstB | This study |
| pCG007 | pEX18Tc::ΔfaoA | This study |
| pCG008 | pEX18Tc::ΔPA2797 | This study |
| pCG009 | pEX18Tc::ΔPA2798 | This study |
| pCG010 | pEX18Tc::ΔPA0392 | This study |
| pJDT1 | pEX18Tc::ΔlptA | This study |
| pCG017 | pEX18Tc::ΔproC | This study |
| pCG011 | pRK415::amgR | This study |
| pCG012 | pRK415::pstB | This study |
| pCG013 | pRK415::faoA | This study |
| pCG014 | pRK415::PA2797 | This study |
| pCG015 | pRK415::PA2798 | This study |
| pCG016 | pRK415::PA0392 | This study |
| pJDT3 | pRK415::lptA | This study |
Cmr, chloramphenicol resistance; Strr, streptomycin resistance; Apr, ampicillin resistance; Tcr, tetracycline resistance; Kmr, kanamycin resistance; Plac-MCS, lac promoter upstream of multicloning site.
DNA methods.
Standard protocols were used for restriction endonuclease digestions, ligations, transformations, and agarose gel electrophoresis, as previously described (71). Plasmid DNA was extracted from E. coli using a Fermentas GeneJET plasmid miniprep kit or a Qiagen plasmid midikit according to protocols provided by the manufacturers. Chromosomal DNA was extracted from P. aeruginosa using a Qiagen DNeasy blood and tissue kit according to a protocol provided by the manufacturer. PCR products and restriction endonuclease digest products requiring purification were purified using a Promega Wizard SV Gel and PCR cleanup system according to a protocol provided by the manufacturer. CaCl2-competent E. coli (71) and electrocompetent P. aeruginosa (11) cells were prepared as previously described. Oligonucleotide synthesis was performed by Integrated DNA Technologies (Coralville, IA), and nucleotide sequencing was performed by ACGT Corporation (Toronto, ON, Canada).
Transposon mutagenesis.
Aminoglycoside-resistant P. aeruginosa strain K2153 or K2413-T3 was mutagenized with mini-Tn5-tet following mobilization of mini-Tn5-tet-carrying plasmid pUT from E. coli SM10 (λpir) as described previously (78), except that K2413-derived mutants were selected on 50 μg/ml tetracycline and 5 μg/ml chloramphenicol (to counterselect donor E. coli cells) following a 6-hour incubation of the mating mixture on L-agar plates. Mini-Tn5-tet insertion mutants showing increased susceptibility to aminoglycosides were subsequently identified by first screening on plates containing paromomycin (512 μg/ml for K2153-derived mutants, 64 μg/ml for K2413-derived mutants), and the mutants that were not able to grow were subsequently examined for increased susceptibility to additional aminoglycosides (as well as paromomycin). The mini-Tn5-tet-disrupted genes from panaminoglycoside-susceptible mutants were recovered following the cloning of mini-Tn5-tet-containing PstI-derived genomic DNA fragments into pBluescript II SK (+) (selected in E. coli DH5α on 10 μg/ml tetracycline) and sequencing of the transposon-flanking chromosomal DNA as described previously (78).
Deletion strain construction.
Derivatives of P. aeruginosa strains with deletions of various genes were generated by constructing deletions in plasmid pEX18Tc and mobilizing them into these strains from E. coli S17-1 as before (63). P. aeruginosa transconjugants harboring chromosomal inserts of the deletion vectors were selected on L-agar plates containing tetracycline (50 μg/ml for all K767 derivatives except ΔlptA, where 75 μg/ml was used; 100 μg/ml for derivatives of K2160 and K2162) and chloramphenicol (5 μg/ml; to counterselect E. coli S17-1). These were subsequently streaked onto L agar containing sucrose (10% [wt/vol]), with sucrose-resistant colonies screened for the appropriate deletion using colony PCR with 2.5 U Taq polymerase in 10% (vol/vol) dimethyl sulfoxide (DMSO) (74). Colony PCR was carried out using the respective Up-F and Down-R primers for each deletion, except for ΔmexXY (mexXY-F, 5′-CTTGACCAGGGCCTCGTAG-3′; mexXY-R, 5′AAGGCCGAACTGGAGCAG-3′), with samples heated for 3 min at 95°C, followed by 34 cycles of 45 s at 95°C, 45 s at 65°C, and 3 min at 72°C, before finishing with 5 min at 75°C (except for ΔlptA, where samples were heated for 5 min at 95°C, followed by 30 cycles of 30 s at 95°C, 30 s at 65°C, and 3 min at 72°C, before finishing with 7 min at 75°C).
Gene deletions were constructed by amplifying, via PCR, 1-kb fragments upstream and downstream of the sequences being deleted and cloning these individually into plasmid pUC19 or pBluescript for sequencing (to ensure that no mutations had been introduced during PCR) and then together into pEX18Tc. PCR fragments were gel purified and digested with restriction enzymes (sites incorporated into PCR primers) prior to cloning into appropriately digested plasmids.
For ΔPA2798, the upstream and downstream fragments were amplified using primers 2798UP-F (5′-GAC TGA ATT CCC GTA CGT GAT GCT GCC GTT-3′; EcoRI site underlined) and 2798UP-R (5′-GAC TTC TAG AGA AGT TGC TGT CTT CCA AGT-3′; XbaI site underlined) and primers 2798Down-F (5′-GAC TTC TAG AGC CGG AGG CAC CCT CGA CGG-3′; XbaI site underlined) and 2798Down-R (5′-GAC TAA GCT TGG TCT CGC GCA TCT ATC GCT-3′; HindIII site underlined). The 50-μl PCR mixtures contained 1 μg of P. aeruginosa K767 chromosomal DNA as the template, 0.2 μM each primer, 0.2 mM each deoxynucleoside triphosphate (dNTP), 1× Pfu (with Mg) buffer, and 1.25 U Pfu DNA polymerase (Promega). Following an initial denaturation step at 95°C for 3 min, the mixture was subjected to 30 cycles of heating at 95°C for 45 s, 63.1°C (annealing temperature) for 45 s, and 72°C for 1 min, before finishing with a 5-min incubation at 72°C.
For ΔPA2797, the upstream and downstream fragments were amplified using primers 2797Up-F (5′-GTA CGA ATT CAC GCC GAT CAT CGT ACT TTC-3′; EcoRI site underlined) and 2797Up-R (5′-GTA CTC TAG AACTCA TGC AAG GTT CCT GCT C-3; XbaI site underlined) and primers 2797Down-F (5′-GTA CTC TAG ACATTA ACC GGT CGATCC CTT-3′; XbaI site underlined) and 2797Down-R (5′-GTA CAA GCT TCC ATG ACT TCG AAG CTG GAA C-3′; HindIII site underlined). The reaction mixture and parameters were as described above for ΔPA2798, except that 1 U Vent DNA polymerase in 1× Thermopol buffer (New England BioLabs) was used and 10% (vol/vol) DMSO was included.
For ΔpstB, the upstream and downstream fragments were amplified using primers pstBUp-F (5′-GAC TGA ATT CCA AGG TCC TGG AAG AGC AGC-3′; EcoRI site underlined) and pstBUp-R (5′-GAC TTC TAG ACA TGG GAA GCT CCC TCA ATC-3′; XbaI site underlined) and primers pstBDown-F (5′-GAC TTC TAG ATA CAT CAC CGG CCG CTA CGG-3; XbaI site underlined) and pstBDown-R (5′-GAC TAA GCT TCG TCA TCC ACC ACT AGC GCG-3′; HindIII site underlined). The reaction mixture and parameters were as described above for ΔPA2798, except that an annealing temperature of 58.6°C was used.
For ΔPA0392, the upstream and downstream fragments were amplified using primers 0392Up-F (5′-GAC TGA ATT CCG AAC TGC TGC TGG ACC TGA-3′; EcoRI site underlined) and 0392Up-R (5′-GAC TTC TAG ATC TGG AGG ATA TAG ATA GCA-3′; XbaI site underlined) and primers 0392Down-F (5′-GAC TTC TAG ACG GCA TGC CCC AGC AGT TGA-3′; XbaI site underlined) and 0392Down-R (5′-GAC TAA GCT TCG AAG TTG ACC AGG GCG TGG-3′; HindIII site underlined). The reaction mixture and parameters were as described above for ΔPA2798, except that annealing temperatures of 49°C (upstream arm) and 63.1°C (downstream arm) were used and 10% (vol/vol) DMSO was included. In the process of cloning the downstream fragment into pUC19, a Dam methylation site overlapping the XbaI site was created. In order to excise this fragment from pUC and clone it into pEX18Tc carrying the upstream fragment, it was necessary to prepare plasmid DNA from the E. coli dam mutant strain GM2163.
For ΔproC, upstream and downstream fragments were amplified using primers proCUp-F (5′-CGA TGA ATT CGA GAG GTG CAG GTC CGA AG-3′; EcoRI site underlined) and proCUp-R (5′-CG ATT CTA GAG GGT GTG CTC ATA GGG GAT T-3′; XbaI site underlined) and primers proCDown-F (5′-CGA TTC TAG ACA GCT TGG CCA ATA AGG AGT-3′; XbaL site underlined) and proCDown-R (5′-CGA TAA GCT TGT CTT TCC ATG CTC ATC CAG −3′; HindIII site underlined). The reaction mixture and parameters were as described above for ΔPA2798, except that Exact DNA polymerase (5 Prime, Inc., Gaithersburg, MD) was employed in 1× Exact buffer and 1× 5P solution, the annealing temperature was 65°C, and 40 cycles of amplification were used.
For ΔfaoA, the upstream and downstream fragments were amplified using primers faoAUp-F (5′-TGA CGA ATT CTG ACT CTA GAA GTC CAG AGC AAT GTC CTG C-3′; EcoRI site underlined) and faoAUp-R (5′-TGA CGG ATC CCA AGA GGC TTA ACC GTG ATG-3′; BamHI site underlined) and primers faoADown-F (5′-TGA CGG ATC CGT TAA TCG CGG AAC GAC TAG-3′; BamHI site underlined) and faoADown-R (5′-GAT CTC TAG ACT TGA GGT CCT TCA GCA CA-3′; XbaI site underlined). The reaction mixture was formulated as described above for ΔPA2798, except that 1 U of Phusion High-Fidelity DNA polymerase in 1× Phusion GC buffer (Fermentas) was employed, primers were present at 0.5 μM, and 5% (vol/vol) DMSO was included. Amplification of the upstream and downstream fragments was achieved using an initial denaturation step of 98°C for 30 s, followed by 30 cycles of 98°C for 30 s, 65°C for 30 s, and 72°C for 1 min, before finishing with 72°C for 5 min.
For ΔlptA, the upstream and downstream fragments were amplified using primers lptAUp-F (5′-GAC TGA ATT CCT GTA GAA GTC CTG GCG GT-3′; EcoRI site underlined) and lptAUp-R (5′-GAC TTC TAG ATG GCC TGC ACT GTC GAC AT 3′; BamHI site underlined) and primers lptADown-F (5′-GAC TTC TAG AGC CGT CGG TGG TCT CGT GA-3′; BamHI site underlined) and lptADown-R (GAC TCT GCA GCG GCG CTG GAG AAA CTG GT; PstI site underlined). The reaction mixture and parameters were as described above for ΔfaoA, except that primers were used at 0.6 μM and the 30 cycles of amplification involved 10 s at 98°C, 30 s at 65°C, and 25 s at 72°C, before finishing with 7 min at 72°C.
For ΔamgR, the upstream and downstream fragments were amplified using primers amgRUp-F (5′-GAC TGA ATT CCT GTA GAA GTC CTG GCG GT-3′; EcoRI site underlined) and amgRUp-R (5′-GAC TTC TAG ATG GCC TGC ACT GTC GAC AT 3′; XbaI site underlined) and primers amgRDown-F (5′-GAC TTC TAG AGC CGT CGG TGG TCT CGT GA-3′; XbaI site underlined) and AmgRDown-R (5′-GAC TCT GCA GCG GCG CTG GAG AAA CTG GT-3′; PstI site underlined). The reaction mixture and parameters were as described above for amplification of the ΔPA2798 deletion fragments.
For ΔPA0006, the upstream and downstream fragments were amplified using primers 0006UP-F (5′-GAC TGA ATT CCC TTG ATG TTG TCG GG-3′; EcoRI site underlined) and 0006UP-R (GAC TTC TAG AAT CGT CGG AGT CGA GG-3′; XbaI site underlined) and primers 0006Down-F (5′-GAC TTC TAG AAG TCG CCA GCG CAT TA-3′; XbaI site underlined) and 0006Down-R (5′-GAC TCT GCA GTC GGT TCG GTC GAT CG-3′; PstI site underlined). The reaction mixture and amplification parameters were as described above for ΔfaoA, except that the extension time at 72°C was 30 s.
For construction of ΔmexXY mutants of P. aeruginosa, plasmid pCSV05-01 (pEX18Tc:: ΔmexXY) was employed as described previously (77).
Gene cloning.
Various genes were amplified from P. aeruginosa K767 chromosomal DNA using primers tagged with restriction sites to facilitate their cloning into plasmid pRK415, which was selected in CaCl2-competent E. coli DH5α. Following sequencing of the PCR-amplified and cloned genes to ensure that no errors had been introduced during PCR, plasmids were electroporated into P. aeruginosa. The PA2798 gene was amplified using primers 2798-F (5′-GCA TAA GCT TTA GGA CCC ACT GTT TCC GG-3′; HindIII site underlined) and 2798-R (5′-GAT CGA ATT CGA CCT CAC CGA CGA ATT TCA-3′; EcoRI site underlined) in a reaction mixture formulated as described above for amplification of the PA2798 deletion fragments, with the exception that 10% (vol/vol) DMSO was included. PCR parameters were also as described above for the PA2798 deletion fragments, except that an annealing temperature of 60°C was used and extension at 72°C was for 2 min 45 s rather than 1 min. The PA2797 gene was amplified using primers 2797-F (5′-GAT CAA GCT TCG GAG ATG CCG GAT GAT AT-3′; HindIII site underlined) and 2797-R (5′-GAT CGG ATC CCA GCG GAG GAT TCA GCA G-3′; BamHI site underlined) using Exact DNA polymerase (2.5 U) in a reaction mixture containing 1 μg of P. aeruginosa K767 chromosomal DNA as the template, 1 μM each primer, and 0.3 mM each dNTP in 1× Exact buffer. Following an initial denaturation step at 95°C for 5 min, the mixture was subjected to 30 cycles of heating at 94°C for 45 s, 45.5°C for 45 s, and 72°C for 1 min, before finishing with a 5-min incubation at 72°C. The pstB and amgR genes were amplified using primers pstB-F (5′-CTA GAA GCT TCT GCG CGA GAA GTA CAA GGC-3′; HindIII site underlined) and pstB-R (5′-CTA GGA ATT CAA CGA GCT ACG GAG AGC GC-3′; EcoRI site underlined) and primers amgR-F (5′-CTA GAA GCT TGC CGC TAC CTG GGC GAT AA-3′; HindIII site underlined) and amgR-R (5′-CTA GGA ATT CCC TGT TGC GGG TAA GAC GAC-3′; EcoRI site underlined), respectively, in reaction mixtures formulated as described above for amplification of the PA2797 deletion fragments using the parameters described for amplification of the PA2798 deletion fragments. The PA0392 gene was amplified using primers 0392-F (5′-CTA GAA GCT TGT GGA GCA GGC CCT GAA C-3′; HindIII site underlined) and 0392-R (5′-CTA GGA ATT CGT GGC AAT CGA GGA GCA G-3′; EcoRI site underlined) in a reaction mixture formulated as described above for amplification of the faoA deletion fragments. PCR parameters were as described above for the ΔfaoA fragments, except that the extension time at 72°C was 30 s as opposed to 1 min. The faoA gene was amplified using primers faoA-F (5′-GAC TGG ATC CGG CGT CCG ATG TGT AAG TTC-3′; BamHI site underlined) and faoA-R (5′-GAT CGA ATT CGT CTC TCG GAT TCA GGC TC-3′; EcoRI site underlined) in a reaction mixture formulated as described above for amplification of the faoA deletion fragments. PCR parameters were also as described above for the ΔfaoA fragments, except that an annealing temperature of 63.5°C was used and annealing was carried out for 30 s. The lptA gene was amplified using primers lptA-F (5′-GACTAAGCTTTCGACGATCTGGCGGCAGT-3′; HindIII site underlined) and lptA-R (5′-GACTGAATTCCGAATCGGCGCACAGTACG-3′; EcoRI site underlined) in a reaction mixture formulated as described above for amplification of the lptA deletion fragments. PCR parameters were also as described above for amplification of the lptA deletion fragments.
Antimicrobial susceptibility testing.
The susceptibility of bacterial strains to various antimicrobials was assessed using the 2-fold serial dilution technique in 96-well microtiter plates as previously described (44).
NPN assay.
Aminoglycoside (gentamicin) promotion of 1-N-phenylnaphthylamine (NPN) uptake into P. aeruginosa, as a measure of aminoglycoside interaction with and permeation of membranes, was assessed as described previously (55), except that cells were treated with carbonyl cyanide m-chlorophenylhydrazone (CCCP; 5 μM) rather than KCN.
Membrane depolarization assay.
The assay for measuring cytoplasmic membrane (CM) depolarization promoted by gentamicin treatment of P. aeruginosa was loosely based on a previously described assay (51). Overnight L-broth cultures of P. aeruginosa were diluted 1:99 in L broth (100 ml) and grown until early logarithmic phase (optical density at 600 nm [OD600] = 0.3 to 0.5), at which time the culture was divided in two and gentamicin (final concentration, 5 μg/ml) was added to one of the cultures. Samples (5 ml) of the gentamicin-treated and untreated control cultures were taken immediately and then hourly over 3 h and exposed to the membrane potential-sensitive dye bis-(1,3-dibutylbarbituric acid) trimethine oxonol [DIBAC4(3); Invitrogen] at 37°C for 5 min in the dark at a final concentration of 10 μg/ml. Bacteria were then pelleted by centrifugation (10 min at 3,000 rpm in a microcentrifuge) and resuspended in phosphate-buffered saline to a final OD600 of 0.1. Membrane depolarization-dependent fluorescence emitted by cells was then measured using a Varian Cary Eclipse fluorescent spectrophotometer with excitation and emission wavelengths of 490 and 518, respectively.
Gentamicin killing assay.
P. aeruginosa cultures were grown as described above for the membrane depolarization assay, samples were taken from the gentamicin-containing cultures at the indicated time points, and dilutions were plated on L agar and incubated overnight at 37°C in order to determine viable cell numbers over time following gentamicin exposure.
RESULTS
Identification of genes contributing to intrinsic aminoglycoside resistance in P. aeruginosa.
Random transposon mutagenesis of panaminoglycoside-resistant P. aeruginosa strains K2153 and K2413-T3 was undertaken to identify a gene(s) contributing to aminoglycoside resistance. Plate screening for mutants showing increased susceptibility first to paromomycin and then to other aminoglycosides identified several showing a panaminoglycoside susceptibility phenotype (data not shown). Cloning and sequencing of the transposon-flanking chromosomal DNA identified several genes whose disruption correlated with increased aminoglycoside susceptibility, including faoA, proC, pstB, amgR, lptA, PA0392, and PA2798. proC occurs upstream of PA0392 in a probable 3-gene operon, while PA2798 occurs upstream of PA2797 in a probable 2-gene operon, such that Tn insertions in these genes might impact aminoglycoside susceptibility as a result of polar effects on the downstream genes and not as a result of the gene disruptions themselves. Individual in-frame deletions of each these genes were engineered into the chromosome of the P. aeruginosa prototroph strain K767 to assess their link to panaminoglycoside susceptibility. With the exception of the mutant with the proC deletion (data not shown), all of the resultant mutant strains were panaminoglycoside susceptible, and this was reversed by cloned wild-type copies of the corresponding genes (Table 2). Of note, none of the deletion mutants showed any change in susceptibility to nonaminoglycosides, including carbenicillin, norfloxacin, nalidixic acid, chloramphenicol, and tetracycline (data not shown), while spectinomycin MICs were reduced ≤2-fold. These genes thus contribute to intrinsic aminoglycoside resistance in P. aeruginosa.
Table 2.
Panaminoglycoside susceptibilitya of P. aeruginosa mutants
| Strain | Plasmid | Relevant property | MIC (μg/ml)b |
||||||
|---|---|---|---|---|---|---|---|---|---|
| GEN | PAR | SPC | KAN | TOB | AMI | STR | |||
| K767 | Wild type | 2 | 256 | 512 | 64 | 1 | 2 | 32 | |
| K3163 | PA2797− | 0.25 (8) | 16 (16) | 128 (4) | 16 (4) | 0.125 (8) | 0.25 (8) | 8 (4) | |
| K3162 | PA2798− | 0.5 (4) | 32 (8) | 256 (2) | 8 (8) | 0.25 (4) | 0.5 (4) | 8 (4) | |
| K3162 | pRK415 | PA2798− | 0.25 | 16 | 128 | 16 | ND | ND | ND |
| K3162 | pCG014 | PA2798+ | 2 | 128 | 512 | 64 | ND | ND | ND |
| K3160 | PstB− | 0.5 (4) | 32 8) | 256 (2) | 8 (8) | 0.5 (2) | 0.5 (4) | 8 (4) | |
| K3160 | pRK415 | PstB− | 0.5 | 32 | 128 | 16 | ND | ND | ND |
| K3160 | pCG012 | PstB+ | 1 | 64 | 512 | 32 | ND | ND | ND |
| K3164 | PA0392− | 0.5 (4) | 32 (8) | 256 (2) | 16 (4) | 0.5 (2) | 1 (2) | 8 (4) | |
| K3164 | pRK415 | PA0392− | 0.5 | 32 | 256 | 16 | ND | ND | ND |
| K3164 | pCG015 | PA0392+ | 2 | 128 | 512 | 64 | ND | ND | ND |
| K3159 | AmgR− | 1 (2) | 64 (4) | 512 (1) | 16 (4) | 0.5 (2) | 0.5 (4) | 8 (4) | |
| K3159 | pRK415 | AmgR− | 1 | 64 | 512 | 16 | ND | ND | ND |
| K3159 | pCG011 | AmgR+ | 4 | 512 | 1024 | 128 | ND | ND | ND |
| K3161 | FaoA− | 0.5 (4) | 16 (16) | 512 (1) | 16 (4) | 0.5 (2) | 1 (2) | 8 (4) | |
| K3161 | pRK415 | FaoA− | 0.5 | 16 | 512 | 16 | ND | ND | ND |
| K3161 | pCG013 | FaoA+ | 1 | 128 | 512 | 32 | ND | ND | ND |
| K3165 | LptA− | 0.5 (4) | 64 (4) | 512 (1) | 16 (4) | 0.5 (2) | 0.5 (4) | 8 (4) | |
| K3165 | pRK415 | LptA− | 0.5 | 64 | 512 | 16 | 0.5 | 0.5 | 8 |
| K3165 | pJDT03 | LptA+ | 1 | 256 | 512 | 64 | ND | ND | ND |
| K1525 | MexXY− | 0.5 (4) | 16 (16) | 64 (8) | 32 (2) | 0.5 (2) | 0.5 (4) | 2 (16) | |
Aminoglycoside susceptibility of P. aeruginosa mutants with deletions of genes linked to intrinsic aminoglycoside resistance in a transposon mutagenesis screen. The impact of the corresponding cloned genes and vector controls on susceptibility was also assessed.
Fold changes in MIC for the mutants relative to that for wild type are indicated in parentheses. GEN, gentamicin; PAR, paromomycin; SPC, spectinomycin; KAN, kanamycin; TOB, tobramycin; AMI, amikacin; STR, streptomycin.
Transposon insertion mutants showing a panaminoglycoside-susceptible phenotype and disrupted in pstB (encoding a component of a high-affinity phosphate transport system [66]), amgR (encoding the response regulator component of the amgRS TCS homologue of the Escherichia coli OmpR-EnvZ envelope stress-response regulators [54]), faoAB (encoding a multienzyme complex involved in degradative fatty acid [FA] β-oxidation [47]), and PA0392 (encoding a conserved hypothetical product of unknown function [29, 54]) have been reported previously. Of the novel intrinsic aminoglycoside resistance genes, lptA encodes a homologue of the E. coli PlsC lysophosphatidic acid acyltransferase (LPA), responsible for adding the second FA to glycerol-3-phosphate in the synthesis of phospholipids (PLs) (13), while PA2798 is part of a 2-gene operon that includes PA2797, where PA2797 is annotated as an anti-anti-sigma factor whose activity is dictated by its phosphorylation state (inactive upon phosphorylation), and PA2798 encodes a probable sensor phosphatase that acts on the PA2797 gene product, thus activating it.
Intrinsic aminoglycoside resistance genes contribute to aminoglycoside resistance independently of AmgR(S) and MexXY.
A previous study revealed that mexXY is not regulated by the AmgRS two-component putative stress-response regulatory protein pair and that these genes likely contribute independently to intrinsic aminoglycoside resistance (54). Results presented here confirm the additive effects of eliminating amgR and mexXY. A mutant lacking both, K3166, was more susceptible to aminoglycosides than either single-knockout strain (Table 3). Given the previously shown link of AmgRS to membrane stress, the possibility that AmgRS contributed to aminoglycoside resistance as a result of protection against aberrant polypeptide accumulation in aminoglycoside-treated cells and their disruption of membranes (54), and the identification of a number of lipid/membrane-related genes as determinants of aminoglycoside resistance (i.e., lptA, faoAB), it was reasoned that AmgRS and MexXY might represent two distinct mechanisms of aminoglycoside resistance, with one (involving AmgRS and lipid/membrane-related genes) based on limiting aberrant polypeptide-mediated membrane damage. To assess this, deletions in each of pstB, faoA, lptA, PA0392, and PA2798 were coupled with deletions in mexXY and amgR. Interestingly, all double knockouts showed increased panaminoglycoside susceptibility relative to single mexXY and amgR knockouts (Table 3), indicating that all worked additively or synergistically and, apparently, independently in contributing to intrinsic aminoglycoside resistance. Still, amgR double knockouts were generally more aminoglycoside susceptible than the mexXY double knockouts; i.e., loss of amgR(S) rendered cells more sensitive to aminoglycosides upon loss of a second intrinsic aminoglycoside resistance determinant. Of note, while most double knockouts were markedly more susceptible to aminoglycosides than the ΔamgR single-knockout strain (at least 4-fold), the impact of lptA loss in this background was minimal (≤2-fold), suggesting that LptA might be in some way linked to the AmgRS-regulated stress-response pathway.
Table 3.
Impact on aminoglycoside susceptibility of intrinsic aminoglycoside resistance gene knockouts in combination with ΔmexXY and ΔarmR
| Strain | Relevant property | MIC (μg/ml)a |
||||||
|---|---|---|---|---|---|---|---|---|
| GEN | PAR | SPC | KAN | TOB | AMI | STR | ||
| K767 | Wild type | 2 | 256 | 512 | 64 | 1 | 2 | 32 |
| K1525 | MexXY− | 0.5 (4) | 16 (16) | 64 (8) | 32 (2) | 0.5 (2) | 0.5 (4) | 2 (16) |
| K3166 | MexXY−, AmgR− | 0.125 (16, 4) | 4 (64, 4) | 32 (16, 2) | 8 (8, 4) | 0.125 (8, 4) | 0.125 (16, 4) | 0.5 (64, 4) |
| K3169 | MexXY−, PA2798− | 0.0625 (32, 8) | 4 (64, 4) | 32 (16, 2) | 8 (8, 4) | 0.125 (8, 4) | 0.125 (16, 4) | 1 (32, 2) |
| K3167 | MexXY−, PstB− | 0.25 (8, 2) | 4 (64, 4) | 64 (8, 1) | 8 (8, 4) | 0.25 (4, 2) | 0.25 (8, 2) | 1 (32, 2) |
| K3171 | MexXY−, PA0392− | 0.25 (8, 2) | 4 (64, 4) | 32 (16, 2) | 8 (8, 2) | 0.25 (4, 2) | 0.25 (8, 2) | 1 (32, 2) |
| K3168 | MexXY−, FaoA− | 0.25 (8, 2) | 4 (64, 4) | 64 (8, 1) | 8 (8, 4) | 0.25 (4, 2) | 0.25 (8, 2) | 1 (32, 2) |
| K3172 | MexXY−, LptA− | 0.125 (16, 4) | 16 (16, 1) | 64 (8, 1) | 16 (4, 2) | 0.125 (8, 4) | 0.125 (16, 4) | 1 (32, 2) |
| K3159 | AmgR− | 0.5 (4) | 32 (8) | 256 (2) | 16 (4) | 0.25 (4) | 0.25 (8) | 8 (4) |
| K3175 | AmgR−, PA2798− | 0.03125 (64, 16) | 2 (128, 16) | 32 (16, 8) | 2 (32, 8) | 0.03125 (32, 8) | 0.03125 (64, 8) | 0.5 (64, 16) |
| K3173 | AmgR−, PstB− | 0.03125 (64, 16) | 4 (64, 8) | 64 (8, 4) | 4 (16, 4) | 0.0625 (16, 4) | 0.0625 (32, 4) | 0.5 (64, 16) |
| K3177 | AmgR−, PA0392− | 0.0625 (32, 8) | 4 (64, 8) | 64 (8, 4) | 4 (16, 4) | 0.0625 (16, 4) | 0.0625 (32, 4) | 0.5 (64, 16) |
| K3174 | AmgR−, FaoA− | 0.125 (16, 4) | 8 (32, 4) | 128 (4, 2) | 8 (8, 2) | 0.0625 (16, 4) | 0.0625 (32, 4) | 1 (32, 8) |
| K3178 | AmgR−, LptA− | 0.25 (8, 2) | 16 (16, 2) | 256 (2, 1) | 16 (4, 1) | 0.25 (4, 1) | 0.125 (16, 2) | 2 (16, 4) |
Fold changes in MIC for the mutants relative to that for wild type are indicated in parentheses. Where two numbers are indicated, the first is the fold change in MIC for the indicated mutant relative to that for wild type, and the second is the fold change in MIC for the indicated double mutant relative to that for its parent MexXY− or AmgR− single mutant. GEN, gentamicin; PAR, paromomycin; SPC, spectinomycin; KAN, kanamycin; TOB, tobramycin; AMI, amikacin; STR, streptomycin.
Cumulative effects of intrinsic aminoglycoside resistance genes.
Since the various resistance genes operated nominally independently of amgRS and mexXY, it was of interest to assess whether they also operate independently of one another. To assess this, various combinations of knockouts were engineered in individual strains, and the impact on resistance was determined. Again, all double knockouts examined showed enhanced aminoglycoside susceptibility compared with their counterpart single knockouts (Table 4), indicating that they have independent and cumulative effects on resistance.
Table 4.
Impact of loss of multiple intrinsic aminoglycoside resistance genes on aminoglycoside susceptibility in P. aeruginosa
| Strain | Relevant property | MIC (μg/ml)a |
||||||
|---|---|---|---|---|---|---|---|---|
| GEN | PAR | SPC | KAN | TOB | AMI | STR | ||
| K767 | Wild type | 2 | 256 | 512 | 64 | 1 | 2 | 32 |
| K3160 | PstB− | 0.5 (4) | 32 (8) | 256 (2) | 16 (4) | 0.5 (2) | 0.5 (4) | 8 (4) |
| K3179 | PstB−, LptA− | 0.25 (8) | 16 (16) | 256 (2) | 16 (4) | 0.25 (4) | 0.25 (8) | 2 (16) |
| K3180 | PstB−, FaoA− | 0.25 (8) | 16 (16) | 256 (2) | 8 (8) | 0.125 (8) | 0.25 (8) | 2 (16) |
| K3181 | PstB−, PA0392− | 0.25 (8) | 8 (32) | 128 (4) | 8 (8) | 0.125 (8) | 0.25 (8) | 2 (16) |
| K3182 | PstB−, PA2798− | 0.125 (16) | 4 (64) | 64 (8) | 4 (16) | 0.0625 (16) | 0.125 (16) | 2 (16) |
| K3161 | FaoA− | 0.5 (4) | 16 (16) | 512 (1) | 16 (4) | 0.5 (2) | 1 (2) | 8 (4) |
| K3165 | LptA− | 0.5 (4) | 64 (4) | 512 (1) | 16 (4) | 0.5 (2) | 0.5 (4) | 8 (4) |
| K3164 | PA0392− | 0.5 (4) | 32 (8) | 256 (2) | 16 (4) | 0.5 (2) | 1 (2) | 8 (4) |
| K3185 | FaoA−, LptA− | 0.125 (16) | 8 (32) | 256 (2) | 8 (8) | 0.0625 (16) | 0.25 (8) | 2 (16) |
| K3184 | FaoA−, PA0392− | 0.25 (8) | 16 (16) | 128 (4) | 8 (8) | 0.125 (8) | 0.25 (8) | 4 (8) |
| K3183 | LptA−, PA0392− | 0.25 (8) | 16 (16) | 256 (2) | 16 (4) | 0.125 (8) | 0.25 (8) | 4 (8) |
Fold changes in MIC for the mutants relative to that for wild type are indicated in parentheses. GEN, gentamicin; PAR, paromomycin; SPC, spectinomycin; KAN, kanamycin; TOB, tobramycin; AMI, amikacin; STR, streptomycin.
Contribution of intrinsic aminoglycoside resistance genes to the panaminoglycoside resistance of clinical isolates.
To determine whether genes linked to intrinsic aminoglycoside resistance in a prototroph strain were significant contributors to the panaminoglycoside resistance of clinical (i.e., CF patient) isolates, deletions of these genes were individually engineered into two representative panaminoglycoside-resistant CF patient isolates, K2160 and K2162 (Table 5). The mechanism(s) of aminoglycoside resistance in these isolates is not known, although a contribution of MexXY-OprM has been reported (77). Elimination of any of these genes increased susceptibility to multiple aminoglycosides, particularly to those most commonly used to treat P. aeruginosa infections (e.g., gentamicin, up to 8- to 32-fold; amikacin, up to 8- to 16-fold; tobramycin, up to 8-fold). Moreover, the impact of these gene deletions on the susceptibility of the clinical isolates to amikacin, gentamicin, and tobramycin was generally greater than that of the same deletions in wild-type P. aeruginosa strain K767 (compare Tables 5 and 3), arguing that they not only contribute to intrinsic resistance but also are important for the acquired aminoglycoside resistance of these isolates. In many cases, the effect of the gene deletions on the susceptibility of the clinical isolates to these aminoglycosides was equal to or greater than that seen when the mexXY efflux genes were deleted, indicating that amgR, pstB, PA2798, faoA, lptA, and PA0392 are as important as this well-known efflux contributor to aminoglycoside resistance in clinical strains (68). The impact of these deletions was less noteworthy for paromomycin and, intriguingly, spectinomycin in the clinical strains, where only the mexXY knockout had any appreciable impact on resistance (Table 5).
Table 5.
Impact of loss of intrinsic aminoglycoside resistance genes on aminoglycoside susceptibility of clinical P. aeruginosa isolates
| Strain | Relevant property | MIC (μg/ml)a |
|||||
|---|---|---|---|---|---|---|---|
| GEN | PAR | SPC | KAN | TOB | AMI | ||
| K2160 | Panaminoglycoside resistant | 32 | 1,024 | 512 | 256 | 8 | 32 |
| K3192 | AmgR− | 8 (4) | 512 (2) | 256 (2) | 64 (4) | 1 (8) | 4 (8) |
| K3193 | PstB− | 8 (4) | 512 (2) | 256 (2) | 64 (4) | 2 (4) | 16 (2) |
| K3194 | FaoA− | 4 (8) | 256 (4) | 256 (2) | 64 (4) | 2 (4) | 8 (4) |
| K3195 | PA2798− | 4 (8) | 256 (4) | 256 (2) | 64 (4) | 1 (8) | 4 (8) |
| K3196 | PA0392− | 4 (8) | 256 (4) | 256 (2) | 64 (4) | 2 (4) | 8 (8) |
| K3197 | LptA− | 8 (4) | 512 (2) | 512 (1) | 128 (2) | 2 (4) | 8 (8) |
| K2168 | MexXY− | 8 (4) | 128 (8) | 64 (8) | 128 (2) | 4 (2) | 8 (8) |
| K2162 | Panaminoglycoside resistant | 256 | >4,096 | 2,048 | 1,024 | 32 | 128 |
| K3198 | AmgR− | 64 (4) | 4,096 (2)b | 1,024 (2) | 512 (2) | 8 (4) | 32 (4) |
| K3199 | PstB− | 16 (16) | 2,048 (4)b | 1,024 (2) | 256 (4) | 16 (2) | 16 (8) |
| K3200 | FaoA− | 64 (4) | 1,024 (8)b | 2,048 (1) | 512 (2) | 16 (2) | 32 (4) |
| K3201 | PA2798− | 8 (32) | 1,024 (8)b | 1,024 (2) | 512 (2) | 4 (8) | 8 (16) |
| K3202 | PA0392− | 16 (16) | 1,024 (8)b | 1,024 (2) | 256 (4) | 8 (4) | 16 (8) |
| K3203 | LptA− | 64 (4) | 4,096 (2)b | 2,048 (1) | 512 (2) | 8 (4) | 32 (4) |
| K2170 | MexXY− | 16 (16) | 256 (32)b | 128 (16) | 256 (4) | 8 (4) | 16 (8) |
Fold changes in MIC for the mutants relative to that for the corresponding wild-type clinical parent strain are indicated in parentheses. GEN, gentamicin; PAR, paromomycin; SPC, spectinomycin; KAN, kanamycin; TOB, tobramycin; AMI, amikacin.
Because a precise paromomycin MIC was not determined for the K2162 parent of these mutants, the fold change reported in parentheses is a minimum.
Impact of intrinsic aminoglycoside resistance gene deletions on aminoglycoside permeation of membranes.
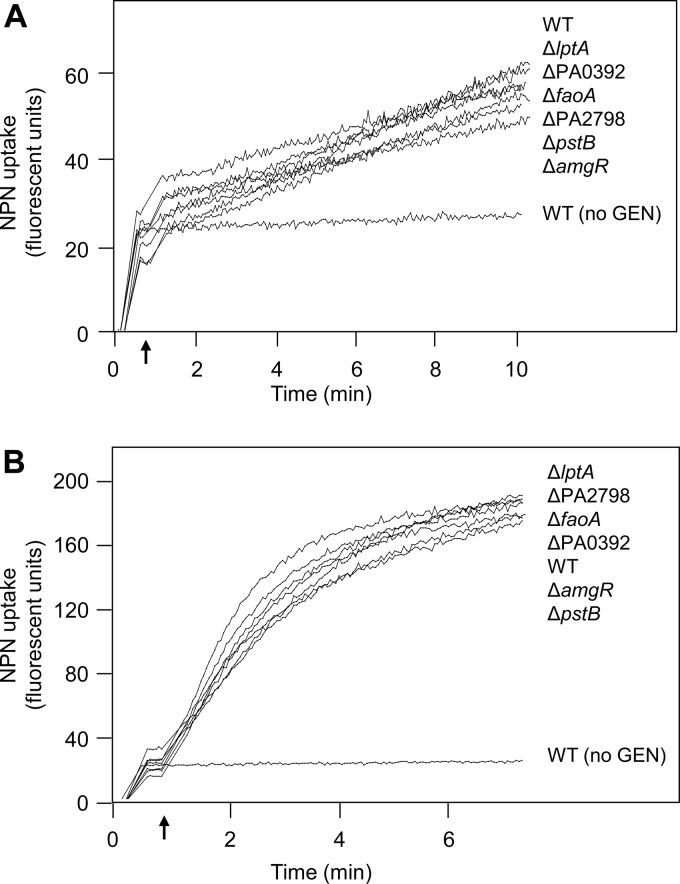
Aminoglycosides enter bacterial cells, including those of P. aeruginosa, via a so-called self-promoted uptake route (33) that involves their permeation of the OM (55). One possibility for the enhanced panaminoglycoside susceptibility of the aforementioned mutants is increased aminoglycoside interaction with/permeation of membranes and subsequent uptake into P. aeruginosa. Aminoglycoside interaction with and promotion of permeation of membranes were assessed in wild-type (strain K767) and mutant cells using enhanced NPN fluorescence following exposure to gentamicin. In the absence of gentamicin, no increase in NPN fluorescence was observed in K767, while at low (5 μg/ml) and high (20 μg/ml) concentrations of this aminoglycoside, NPN fluorescence increased over time (Fig. 1), indicative of gentamicin association with membranes and promotion of membrane permeation. Gentamicin-promoted membrane permeation was also observed for the mutant strains at both gentamicin concentrations, and this was indistinguishable from that seen for K767 (Fig. 1). Thus, the enhanced aminoglycoside susceptibility of these mutants is not explained by their increased membrane association/permeation and, so, uptake into the cell.
Fig 1.
Impact of intrinsic aminoglycoside resistance gene knockouts on gentamicin-promoted outer membrane permeability. Membrane permeability, as assessed by NPN fluorescence, was measured over time following exposure of wild-type (WT; strain K767) and mutant P. aeruginosa strains to 5 μg/ml (A) and 20 μg/ml (B) of gentamicin (added at the time indicated by the arrow). The strains listed at the right (top to bottom) correspond to the traces at the left (top to bottom, respectively) at the final time points. Data for wild-type strain K767 not treated with gentamicin (no GEN) are also shown. The data are representative of at least 2 independent experiments for each strain.
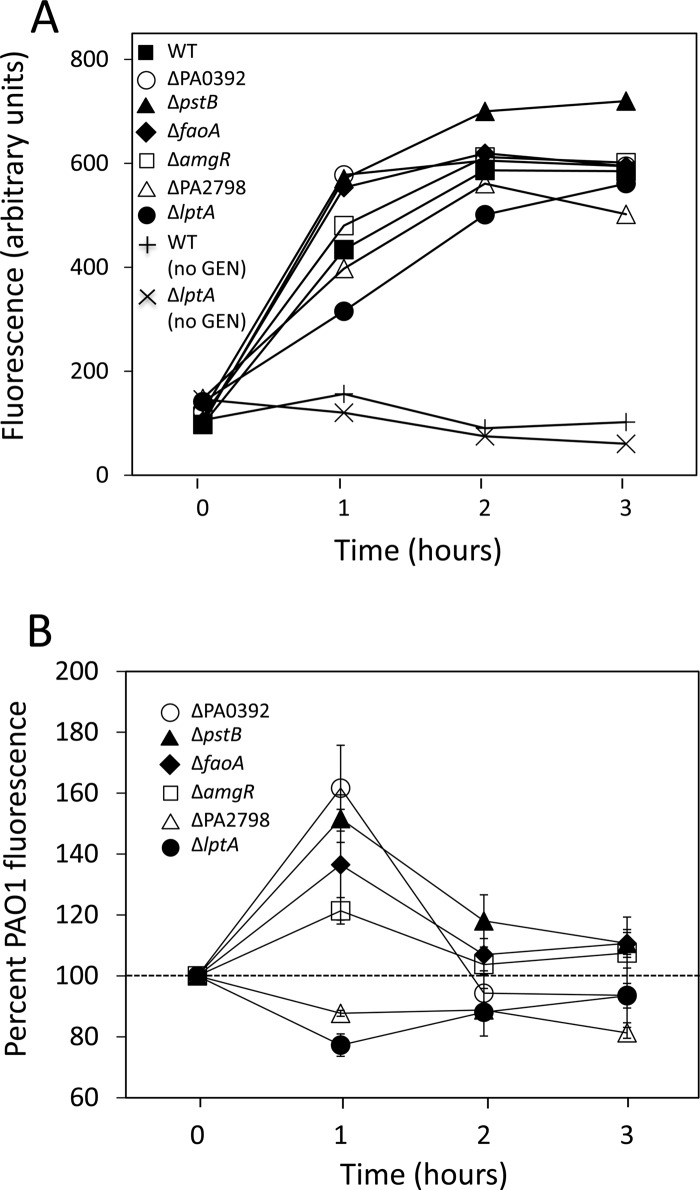
Impact of intrinsic aminoglycoside resistance gene deletions on aminoglycoside-promoted cytoplasmic membrane depolarization.
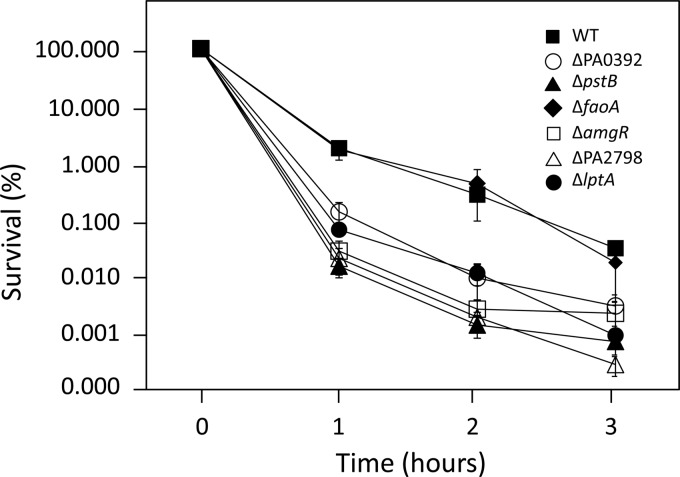
Aminoglycosides are known to promote mistranslation, with mistranslated polypeptides inserting into and damaging cytoplasmic membranes (16). In E. coli, this aminoglycoside-dependent membrane perturbation initiates a cascade of events, including activation of the CpxRA envelope stress-response pathway, which leads to the production of ROS and, ultimately, cell death (51). Disruption of the intrinsic aminoglycoside resistance genes in P. aeruginosa might, therefore, increase susceptibility to aminoglycosides by increasing the susceptibility of membranes to perturbation by aminoglycoside-generated mistranslated polypeptides. One measure of cytoplasmic membrane perturbation is membrane depolarization, and, indeed, aminoglycoside-promoted membrane depolarization has been demonstrated in E. coli and correlates well with aminoglycoside-promoted cell killing (51). Moreover, E. coli mutants showing enhanced aminoglycoside susceptibility showed higher levels of gentamicin-promoted membrane depolarization than wild type (51), consistent with the membranes of these mutants being more susceptible to perturbation by aminoglycoside-generated mistranslated polypeptides. As seen in Fig. 2, exposure of wild-type P. aeruginosa strain K767 to a supra-MIC of gentamicin (5 μg/ml) promoted membrane depolarization, which increased over the 3-hour exposure period, in contrast to unexposed K767, where no increase was seen. Gentamicin-promoted membrane depolarization that increased over time was also seen for the mutant strains, with the ΔpstB, ΔfaoA, and ΔPA0392 mutants and, to a lesser extent, the ΔamgR mutant showing an increase in gentamicin-promoted membrane depolarization relative to strain K767 after 1 h of exposure to the drug (Fig. 2). This correlated with the period of maximal killing of the cells by gentamicin (≥99%; Fig. 3), suggesting that the enhanced susceptibility of these mutants to aminoglycosides results from enhanced aminoglycoside-promoted membrane perturbation. The ΔPA2798 (at all time points) and ΔlptA (at the 1-hour time point) mutants, in contrast, appeared to be less susceptible to gentamicin-promoted membrane depolarization than the wild type (Fig. 2). As such, the enhanced aminoglycoside susceptibility of these mutants did not result from enhanced susceptibility of their membranes to aminoglycoside-promoted perturbation.
Fig 2.
Impact of intrinsic aminoglycoside resistance gene knockouts on gentamicin-promoted cytoplasmic membrane depolarization. Cytoplasmic membrane depolarization, as assessed by DIBAC4(3) fluorescence, was measured over time following exposure of wild-type (WT; strain K767) and mutant P. aeruginosa strains to 5 μg/ml of gentamicin (added at time zero). (A) Results of a representative experiment showing increased gentamicin-promoted fluorescence over time. Background fluorescence in the absence of gentamicin is shown for wild-type strain K767 but is representative of all strains, with the exception of the indicated ΔlptA strain. (B) Gentamicin-promoted fluorescence over time for the indicated mutants as a percentage of gentamicin-promoted fluorescence for wild-type strain K767. The data are the mean ± standard deviation of 3 to 4 independent experiments where the fluorescence data were adjusted for the background fluorescence of gentamicin-unexposed cells prior to calculating percentages.
Fig 3.
Gentamicin-promoted killing of P. aeruginosa. The impact of gentamicin (5 μg/ml) on the viability of wild-type (WT; strain K767) and mutant P. aeruginosa was assessed over time by determining viable cell counts at the indicated time points. Results are presented as percent survival and are the mean ± standard deviation of 3 independent experiments.
DISCUSSION
Aminoglycoside uptake into and action on bacterial cells are complex processes that involve LPS binding/OM permeation (31, 32, 46), CM traversal using the membrane potential (8, 32), and ribosome disruption, leading to production of membrane-damaging mistranslated polypeptides (16). Polypeptide disruption of the CM triggers a cell envelope stress response (51) that ultimately leads to the generation of ROS (i.e., hydroxyl radicals), whose oxidation of key macromolecules appears to be responsible for the lethal effects of these agents (24, 51). As such, mutations leading to alterations in LPS binding/OM permeation (6), CM potential (67), envelope stress responses (51, 54), and ROS production (50) can influence aminoglycoside susceptibility. The current study identifies several apparently unrelated genes whose disruption enhances aminoglycoside susceptibility in P. aeruginosa, confirming them to be determinants of intrinsic aminoglycoside resistance in this organism. The amgRS genes, encoding a TCS that apparently regulates an envelope stress response, have previously been identified to be determinants of intrinsic aminoglycoside resistance (54), as have the faoA, pstB, and PA0392 genes (29, 54). While the amgRS link to resistance is explained, at least in part, by their control of protease genes whose products may well function to turn over aminoglycoside-generated mistranslated polypeptides, thereby ameliorating their perturbation of the CM (37), the nature of the link of these other genes was undefined. This work highlights the probable contribution of several of these genes to maintenance of CM integrity and/or stability, with their mutational loss compromising this and so rendering the CM more susceptible to aminoglycoside-promoted perturbation, likely by aminoglycoside-generated mistranslated polypeptides. In agreement with this, susceptibility to spectinomycin, a ribosome-targeting agent related to the aminoglycosides but known not to generate mistranslated protein products (14), was only minimally enhanced (≤2-fold increase) in the amgR, faoA, pstB, and PA0392 deletion strains. Indeed, only the mexXY deletion substantially increased susceptibility to spectinomycin, suggesting that the increased aminoglycoside susceptibility of a mexXY knockout is not explained by an increased sensitivity to CM perturbation by mistranslated polypeptides and, thus, that MexXY-OprM contributes to aminoglycoside resistance via some mechanism other than alleviation of mistranslated polypeptide-mediated CM stress. While antimicrobial export is the simplest explanation for its contribution to resistance, a mexZ mutant expressing MexXY and showing elevated aminoglycoside MICs did not exhibit any change in aminoglycoside (tobramycin) accumulation relative to its MexZ+ parent strain in a recent study (57).
Intriguingly, the enhanced susceptibility to aminoglycoside-promoted CM perturbation in the amgR, faoA, pstB, and PA0392 deletion strains typically occurs 1 hour after initial exposure to an aminoglycoside only, during the period of maximal killing (≥2-log-unit decline in viable cell numbers), suggesting that the two are linked. In E. coli there is also a spike of membrane depolarization (and hydroxyl radical formation) 1 h after initial aminoglycoside exposure, and this, too, parallels the period of maximal cell killing (51). Moreover, aminoglycoside-sensitive E. coli hflKC mutants show enhanced gentamicin-promoted membrane depolarization at 1 h postexposure (51), reminiscent of what we saw here with the pstB, faoA, and PA0392 mutants. HflKC regulates a protease, FtsH, which is responsible, in part, for turning over aberrant/misfolded membrane-associated polypeptides (41), suggesting that the increased susceptibility and membrane depolarization of the mutants result from increases in the levels of aminoglycoside-generated mistranslated polypeptides and, so, membrane perturbation. Consistent with this and with enhanced aminoglycoside-promoted membrane depolarization in P. aeruginosa being an indicator of enhanced CM perturbation by aminoglycoside-generated mistranslated polypeptides, an ftsH mutant of P. aeruginosa also exhibits enhanced susceptibility to aminoglycosides (37).
FaoAB/FadBA-mediated FA oxidation is typically associated with growth on/metabolism of exogenously provided FAs (3), although the observation that its mutational loss provides for enhanced aminoglycoside-promoted membrane perturbation and aminoglycoside susceptibility suggests that it may also play a role in membrane FA turnover in response to envelope stress. In the face of membrane perturbation by aminoglycoside-generated mistranslated polypeptides, for example, P. aeruginosa might normally replace shorter and/or unsaturated FAs with longer and/or saturated FAs so as to enhance membrane stability and, possibly, limit CM perturbation by mistranslated polypeptides. Certainly, there are many examples of bacteria modulating the FA composition of their membranes in response to membrane perturbants (e.g., organic solvents) to stabilize these structures and, so, limit membrane perturbation (23, 69, 83).
A link between pstB function and the CM is obvious, given the importance of phosphate for synthesis of a key CM component, PLs. Loss of the Pst inorganic phosphate transporter effectively renders cells phosphate limited, even in rich media, and it is well-known that phosphate limitation has a negative impact on PL incorporation into bacterial membranes. In Pseudomonas fluorescens, for example, increased incorporation of the non-PL ornithine amide lipid occurs in membranes of cells grown under phosphate-limiting conditions, at the expense of PLs (20), although it is not clear whether this would impact membrane stability. pst mutations have also been reported in E. coli, where they are linked to membrane FA alterations, with a tendency toward an increase in unsaturated FAs (53). This would tend to make the CM more fluid and, possibly, more susceptible to perturbation by mistranslated polypeptides. In light of the information presented above, it is likely that a pstB mutation impacts the lipid composition of P. aeruginosa so as to enhance CM perturbation by aminoglycoside-generated mistranslated polypeptides. PA0392 has no known function at present, though its homologue in E. coli, YggT (36.7% amino acid identity), appears to play a role in osmotic stress tolerance (42). Given the impact of a PA0392 deletion on aminoglycoside-dependent CM perturbation and reports that membrane PL and FA changes occur in response to osmotic stress in several bacteria and may contribute to osmotic stress tolerance (70), PA0392 may well influence the CM lipid composition in ways that stabilize this structure, contributing to aminoglycoside resistance and, perhaps, tolerance to osmotic stress. Of particular note is the greater increase in aminoglycoside susceptibility seen upon loss of faoA, pstB, or PA0392 in a mutant already lacking amgR relative to that seen upon loss of these genes in an otherwise wild-type strain. This presumably reflects the heightened impact of the absence of an envelope stress response in mutants with compromised and, so, more stress-sensitive CMs.
The link between lptA and the CM is readily apparent, yet mutation of lptA does not provide for any obvious increase in aminoglycoside-promoted CM perturbation. A homologue of E. coli plsC, lptA is capable of complementing a plsC(Ts) mutant (2), consistent with LptA functioning as an LPA. Its closest homologue, however, is Neisseria meningitidis NlaB (36% identity), a second LPA in this organism (the first is encoded by nlaA [80]). Like N. meningitidis (and unlike E. coli), P. aeruginosa apparently expresses two LPAs (PA4351 is a close homologue of NlaA; 54% similarity and 33% identity), which likely explains the ability to derive an lptA knockout in this organism when knockouts of E. coli plsC are lethal (12). A previously reported lptA mutant of P. aeruginosa PAO1 showed alterations in membrane FAs (decrease in C16:0, increase in C18:0, and loss of C17:0 cyclo) and evidence for reduced membrane fluidity (2). This is consistent with the reduced aminoglycoside-promoted membrane perturbation seen in this mutant and, as such, is unlikely to explain its enhanced aminoglycoside susceptibility. Possibly, given that elimination of lptA in a strain already lacking amgR has a minimal additional impact on aminoglycoside susceptibility, loss of lptA somehow compromises activation of the AmgRS TCS by aminoglycoside-generated mistranslated polypeptides. In light of the reduced membrane fluidity and reduced aminoglycoside-promoted membrane perturbation in the lptA mutant, one possibility is that this results from a reduced mistranslated polypeptide perturbation of the lptA mutant membranes (i.e., reduced membrane stress). If true, however, there would then be less need for the AmgRS-regulated envelope stress response, a major component of which are proteases that likely turn over aberrant polypeptides. If these aberrant polypeptides are less able to perturb the membranes of an lptA mutant, there would be less need to turn them over. Still, it may be that aminoglycoside-generated mistranslated polypeptides have additional deleterious effects on the cell, independent of CM perturbation, such that their turnover by AmgRS-controlled proteases would still be beneficial and their loss (in the absence of AmgRS activation) would compromise aminoglycoside resistance.
Interestingly, N. meningitidis NlaB is encoded as part of an operon with gmhX, whose product is required for incorporation of l-glycero-d-manno-heptose into lipooligosaccharide (75), and intriguingly, the closest homologue to gmhX in P. aeruginosa is the PA0006 gene (51% identical, 67% overall similarity), which occurs as part of a probable 2-gene operon with lptA. A homologue of gmhX and PA0006 in E. coli, YaeD (also known as GmhB), is also linked to heptose synthesis, though as part of LPS biosynthesis. Given this possible link to LPS biosynthesis, the operon organization of PA0006 and lptA, and the known connection between LPS and aminoglycoside uptake and resistance, it was of interest to assess the impact of a PA0006 knockout on aminoglycoside susceptibly. Unfortunately, and despite repeated attempts, a deletion mutant lacking PA0006 could not be recovered, suggesting that this gene may well be essential. While the link to a probable LPS biosynthetic gene suggests that lptA may well play a role (nonessential) in the synthesis of this macromolecule and that its loss produces aminoglycoside resistance as a result of some defect in LPS, deletion of lptA did not adversely impact LPS binding/permeation of the OM. Thus, it may also operate independently of PA0006, at least as regards its contribution to aminoglycoside resistance. Despite the probably operonic link between proC and PA0392, for example, only a ΔproC mutant displays proline auxotrophy (data not shown), while only the PA0392 deletion impacts aminoglycoside resistance.
PA2797 and PA2798 are the respective homologues of the Bacillus SpoIIAA and SpoIIE proteins that regulate the SpoIIAB anti-sigma factor of the sporulation sigma factor σF (26) and the Bacillus RbsV and RbsU proteins that regulate the RbsW anti-sigma factor of the stress-response sigma factor σB (34). One possibility, then, is that these proteins ultimately regulate a, possibly, stress-response sigma factor in P. aeruginosa whose lack (in the PA27987 and PA2798 mutants, which would be defective in acting on/sequestering their presumed anti-sigma factor target to promote sigma factor activation) enhances susceptibility to aminoglycoside-promoted stress. This would appear to be independent of aminoglycoside-promoted membrane perturbation, which is not enhanced and is, in fact, reduced in the PA2798 mutant. Strikingly, loss of PA2798 in an amgR background has a marked and greater impact on aminoglycoside susceptibility than loss of this gene in an otherwise wild-type background. As such, these two systems may regulate overlapping stress responses such that the full impact of a stress-response defect vis-à-vis aminoglycoside susceptibility is not felt until both are absent. While the probable sigma factor controlled by PA2797-PA2798 is unknown, disruption of this locus has recently been linked to increased colistin tolerance (45), although a mechanism was not elucidated.
Significantly, the intrinsic aminoglycoside resistance genes identified here were also important contributors to the panaminoglycoside resistance of clinical strains, including strains where MexXY-OprM has been implicated in resistance (77). Given their apparent link to membrane stability and the not unexpected role of membrane perturbability in aminoglycoside susceptibility, this is not surprising. These results do suggest, however, that undermining CM integrity and/or compromising adaptive responses to CM perturbation can enhance the susceptibility of P. aeruginosa to aminoglycosides, and thus, targeting components of CM biosynthesis or these adaptive responses might be useful in promoting aminoglycoside susceptibility.
ACKNOWLEDGMENT
This work was supported by an operating grant from Cystic Fibrosis Canada.
Footnotes
Published ahead of print 20 August 2012
REFERENCES
- 1. Ausubel FM, et al. 1992. Short protocols in molecular biology, 2nd ed John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 2. Baysse C, et al. 2005. Modulation of quorum sensing in Pseudomonas aeruginosa through alteration of membrane properties. Microbiology 151:2529–2542 [DOI] [PubMed] [Google Scholar]
- 3. Black PN, DiRusso CC. 1994. Molecular and biochemical analyses of fatty acid transport, metabolism, and gene regulation in Escherichia coli. Biochim. Biophys. Acta 1210:123–145 [DOI] [PubMed] [Google Scholar]
- 4. Bryan LE, Haraphongse R, Van den Elzen HM. 1976. Gentamicin resistance in clinical-isolates of Pseudomonas aeruginosa associated with diminished gentamicin accumulation and no detectable enzymatic modification. J. Antibiot. (Tokyo) 29:743–753 [DOI] [PubMed] [Google Scholar]
- 5. Bryan LE, Nicas T, Holloway BW, Crowther C. 1980. Aminoglycoside-resistant mutation of Pseudomonas aeruginosa defective in cytochrome c552 and nitrate reductase. Antimicrob. Agents Chemother. 17:71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bryan LE, O'Hara K, Wong S. 1984. Lipopolysaccharide changes in impermeability-type aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 26:250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bryan LE, Van den Elzen HM. 1976. Streptomycin accumulation in susceptible and resistant strains of Escherichia coli and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 9:928–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bryan LE, Van den Elzen HM. 1977. Effects of membrane-energy mutations and cations on streptomycin and gentamicin accumulation by bacteria: a model for entry of streptomycin and gentamicin in susceptible and resistant bacteria. Antimicrob. Agents Chemother. 12:163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burkhardt O, et al. 2006. Once-daily tobramycin in cystic fibrosis: better for clinical outcome than thrice-daily tobramycin but more resistance development? J. Antimicrob. Chemother. 58:822–829 [DOI] [PubMed] [Google Scholar]
- 10. Canton R, et al. 2005. Antimicrobial therapy for pulmonary pathogenic colonisation and infection by Pseudomonas aeruginosa in cystic fibrosis patients. Clin. Microbiol. Infect. 11:690–703 [DOI] [PubMed] [Google Scholar]
- 11. Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391–397 [DOI] [PubMed] [Google Scholar]
- 12. Coleman J. 1990. Characterization of Escherichia coli cells deficient in 1-acyl-sn-glycerol-3-phosphate acyltransferase activity. J. Biol. Chem. 265:17215–17221 [PubMed] [Google Scholar]
- 13. Coleman J. 1992. Characterization of the Escherichia coli gene for 1-acyl-sn-glycerol-3-phosphate acyltransferase (plsC). Mol. Gen. Genet. 232:295–303 [DOI] [PubMed] [Google Scholar]
- 14. Davies J, Anderson P, Davis BD. 1965. Inhibition of protein synthesis by spectinomycin. Science 149:1096–1098 [DOI] [PubMed] [Google Scholar]
- 15. Davis BD. 1987. Mechanism of bactericidal action of aminoglycosides. Microbiol. Rev. 51:341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davis BD, Chen LL, Tai PC. 1986. Misread protein creates membrane channels: an essential step in the bactericidal action of aminoglycosides. Proc. Natl. Acad. Sci. U. S. A. 83:6164–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Kievit TR, et al. 2001. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 45:1761–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Lorenzo V, Herrero M, Jakubzik U, Timmis KN. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in Gram-negative eubacteria. J. Bacteriol. 172:6567–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Doi Y, Arakawa Y. 2007. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 45:88–94 [DOI] [PubMed] [Google Scholar]
- 20. Dorrer E, Teuber M. 1977. Induction of polymyxin resistance in Pseudomonas fluorescens by phosphate limitation. Arch. Microbiol. 114:87–89 [DOI] [PubMed] [Google Scholar]
- 21. Dötsch A, et al. 2009. Genomewide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:2522–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dubois V, et al. 2008. β-Lactam and aminoglycoside resistance rates and mechanisms among Pseudomonas aeruginosa in French general practice (community and private healthcare centres). J. Antimicrob. Chemother. 62:316–323 [DOI] [PubMed] [Google Scholar]
- 23. Dubois-Brissonnet F, Naitali M, Mafu AA, Briandet R. 2011. Induction of fatty acid composition modifications and tolerance to biocides in Salmonella enterica serovar Typhimurium by plant-derived terpenes. Appl. Environ. Microbiol. 77:906–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dwyer DJ, Kohanski MA, Collins JJ. 2009. Role of reactive oxygen species in antibiotic action and resistance. Curr. Opin. Microbiol. 12:482–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. El'Garch F, Jeannot K, Hocquet D, Llanes-Barakat C, Plesiat P. 2007. Cumulative effects of several nonenzymatic mechanisms on the resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 51:1016–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Errington J, et al. 1996. Control of the cell-specificity of sigma F activity in Bacillus subtilis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 351:537–542 [DOI] [PubMed] [Google Scholar]
- 27. Foweraker J. 2009. Recent advances in the microbiology of respiratory tract infection in cystic fibrosis. Br. Med. Bull. 89:93–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Galimand M, Lambert T, Courvalin P. 2005. Emergence and dissemination of a new mechanism of resistance to aminoglycosides in Gram-negative bacteria: 16S rRNA methylation. Euro Surveill. 10(1):E050127.2. [DOI] [PubMed] [Google Scholar]
- 29. Gallagher LA, Shendure J, Manoil C. 2011. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2(1):e00315–10 doi:10.1128/mBio.00315-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Geller DE. 2009. Aerosol antibiotics in cystic fibrosis. Respir. Care 54:658–670 [DOI] [PubMed] [Google Scholar]
- 31. Hancock RE, Farmer SW, Li ZS, Poole K. 1991. Interaction of aminoglycosides with the outer membranes and purified lipopolysaccharide and OmpF porin of Escherichia coli. Antimicrob. Agents Chemother. 35:1309–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hancock REW. 1981. Aminoglycoside uptake and mode of action with specific reference to streptomycin and gentamicin. II. Effects of aminoglycosides on cells. J. Antimicrob. Chemother. 8:429–445 [DOI] [PubMed] [Google Scholar]
- 33. Hancock REW, Bell A. 1988. Antibiotic uptake into Gram-negative bacteria. Eur. J. Clin. Microbiol. Infect. Dis. 7:713–720 [DOI] [PubMed] [Google Scholar]
- 34. Hecker M, Pane-Farre J, Volker U. 2007. SigB-dependent general stress response in Bacillus subtilis and related Gram-positive bacteria. Annu. Rev. Microbiol. 61:215–236 [DOI] [PubMed] [Google Scholar]
- 35. Henrichfreise B, Wiegand I, Pfister W, Wiedemann B. 2007. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob. Agents Chemother. 51:4062–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hidron AI, et al. 2008. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect. Control Hosp. Epidemiol. 29:996–1011 [DOI] [PubMed] [Google Scholar]
- 37. Hinz A, Lee S, Jacoby K, Manoil C. 2011. Membrane proteases and aminoglycoside antibiotic resistance. J. Bacteriol. 193:4790–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 39. Hocquet D, et al. 2008. Relationship between antibiotic use and incidence of MexXY-OprM overproducers among clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52:1173–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Islam S, et al. 2009. Chromosomal mechanisms of aminoglycoside resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Clin. Microbiol. Infect. 15:60–66 [DOI] [PubMed] [Google Scholar]
- 41. Ito K, Akiyama Y. 2005. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59:211–231 [DOI] [PubMed] [Google Scholar]
- 42. Ito T, et al. 2009. The implication of YggT of Escherichia coli in osmotic regulation. Biosci. Biotechnol. Biochem. 73:2698–2704 [DOI] [PubMed] [Google Scholar]
- 43. Jana S, Deb JK. 2006. Molecular understanding of aminoglycoside action and resistance. Appl. Microbiol. Biotechnol. 70:140–150 [DOI] [PubMed] [Google Scholar]
- 44. Jo JT, Brinkman FS, Hancock RE. 2003. Aminoglycoside efflux in Pseudomonas aeruginosa: involvement of novel outer membrane proteins. Antimicrob. Agents Chemother. 47:1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jochumsen N, Liu Y, Molin S, Folkesson A. 2011. A Mig-14-like protein (PA5003) affects antimicrobial peptide recognition in Pseudomonas aeruginosa. Microbiology 157:2647–2657 [DOI] [PubMed] [Google Scholar]
- 46. Kadurugamuwa JL, Lam JS, Beveridge TJ. 1993. Interaction of gentamicin with the A band and B band lipopolysaccharides of Pseudomonas aeruginosa and its possible lethal effect. Antimicrob. Agents Chemother. 37:715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kang Y, Nguyen DT, Son MS, Hoang TT. 2008. The Pseudomonas aeruginosa PsrA responds to long-chain fatty acid signals to regulate the fadBA5 β-oxidation operon. Microbiology 154:1584–1598 [DOI] [PubMed] [Google Scholar]
- 48. Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191–197 [DOI] [PubMed] [Google Scholar]
- 49. Kim JY, et al. 2008. Occurrence and mechanisms of amikacin resistance and its association with β-lactamases in Pseudomonas aeruginosa: a Korean nationwide study. J. Antimicrob. Chemother. 62:479–483 [DOI] [PubMed] [Google Scholar]
- 50. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810 [DOI] [PubMed] [Google Scholar]
- 51. Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. 2008. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135:679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kono M, O'Hara K. 1976. Mechanisms of streptomycin (SM)-resistance of highly SM-resistant Pseudomonas aeruginosa strains. J. Antibiot. (Tokyo) 29:169–175 [DOI] [PubMed] [Google Scholar]
- 53. Lamarche MG, Harel J. 2010. Membrane homeostasis requires intact pst in extraintestinal pathogenic Escherichia coli. Curr. Microbiol. 60:356–359 [DOI] [PubMed] [Google Scholar]
- 54. Lee S, et al. 2009. Targeting a bacterial stress response to enhance antibiotic action. Proc. Natl. Acad. Sci. U. S. A. 106:14570–14575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Loh B, Grant C, Hancock RE. 1984. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 26:546–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Macfarlane EL, Kwasnicka A, Hancock RE. 2000. Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146:2543–2554 [DOI] [PubMed] [Google Scholar]
- 57. MacLeod DL, et al. 2012. Fosfomycin enhances the active transport of tobramycin in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 56:1529–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Manno G, et al. 2005. Antimicrobial use and Pseudomonas aeruginosa susceptibility profile in a cystic fibrosis centre. Int. J. Antimicrob. Agents 25:193–197 [DOI] [PubMed] [Google Scholar]
- 59. Masuda N, Ohya S. 1992. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:1847–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McPhee JB, Lewenza S, Hancock RE. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50:205–217 [DOI] [PubMed] [Google Scholar]
- 61. Merlo CA, et al. 2007. Incidence and risk factors for multiple antibiotic-resistant Pseudomonas aeruginosa in cystic fibrosis. Chest 132:562–568 [DOI] [PubMed] [Google Scholar]
- 62. Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Morita Y, Sobel ML, Poole K. 2006. Antibiotic inducibility of the MexXY multidrug efflux system of Pseudomonas aeruginosa: involvement of the antibiotic-inducible PA5471 gene product. J. Bacteriol. 188:1847–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Moskowitz SM, et al. 2008. Shifting patterns of inhaled antibiotic use in cystic fibrosis. Pediatr. Pulmonol. 43:874–881 [DOI] [PubMed] [Google Scholar]
- 65. Mulcahy H, Charron-Mazenod L, Lewenza S. 2008. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 4:e1000213 doi:10.1371/journal.ppat.1000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nikata T, et al. 1996. Molecular analysis of the phosphate-specific transport (pst) operon of Pseudomonas aeruginosa. Mol. Gen. Genet. 250:692–698 [DOI] [PubMed] [Google Scholar]
- 67. Parr TR, Jr, Bayer AS. 1988. Mechanisms of aminoglycoside resistance in variants of Pseudomonas aeruginosa isolated during treatment of experimental endocarditis in rabbits. J. Infect. Dis. 158:1003–1010 [DOI] [PubMed] [Google Scholar]
- 68. Poole K. 2005. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ramos JL, et al. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56:743–768 [DOI] [PubMed] [Google Scholar]
- 70. Romantsov T, Guan Z, Wood JM. 2009. Cardiolipin and the osmotic stress responses of bacteria. Biochim. Biophys. Acta 1788:2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 72. Schurek KN, et al. 2008. Novel genetic determinants of low-level aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52:4213–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shawar RM, et al. 1999. Activities of tobramycin and six other antibiotics against Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob. Agents Chemother. 43:2877–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sheu DS, Wang YT, Lee CY. 2000. Rapid detection of polyhydroxyalkanoate-accumulating bacteria isolated from the environment by colony PCR. Microbiology 146:2019–2025 [DOI] [PubMed] [Google Scholar]
- 75. Shih GC, Kahler CM, Carlson RW, Rahman MM, Stephens DS. 2001. gmhX, a novel gene required for the incorporation of l-glycero-d-manno-heptose into lipooligosaccharide in Neisseria meningitidis. Microbiology 147:2367–2377 [DOI] [PubMed] [Google Scholar]
- 76. Simon R, Priefer U, Puehler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784–791 [Google Scholar]
- 77. Sobel ML, McKay GA, Poole K. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 47:3202–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sobel ML, Poole K, Neshat S. 2005. Mutations in PA2491 (mexS) promote MexT-dependent mexEF-oprN expression and multidrug resistance in a clinical strain of Pseudomonas aeruginosa. J. Bacteriol. 187:1246–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Stefani S, Russo G. 1989. Aminoglycoside resistance due to alterations of transport mechanisms. J. Chemother. 1:369–370 [PubMed] [Google Scholar]
- 80. Swartley JS, Balthazar JT, Coleman J, Shafer WM, Stephens DS. 1995. Membrane glycerophospholipid biosynthesis in Neisseria meningitidis and Neisseria gonorrhoeae: identification, characterization, and mutagenesis of a lysophosphatidic acid acyltransferase. Mol. Microbiol. 18:401–412 [DOI] [PubMed] [Google Scholar]
- 81. Taber HW, Mueller JP, Miller PF, Arrow AS. 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 51:439–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Taccetti G, Campana S, Neri AS, Boni V, Festini F. 2008. Antibiotic therapy against Pseudomonas aeruginosa in cystic fibrosis. J. Chemother. 20:166–169 [DOI] [PubMed] [Google Scholar]
- 83. Torres S, Pandey A, Castro GR. 2011. Organic solvent adaptation of Gram positive bacteria: applications and biotechnological potentials. Biotechnol. Adv. 29:442–452 [DOI] [PubMed] [Google Scholar]
- 84. Yamane K, et al. 2007. 16S rRNA methylase-producing, Gram-negative pathogens, Japan. Emerg. Infect. Dis. 13:642–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yanisch-Perron C, Vieira J, Messing J. 1985. M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]
- 86. Zhanel GG, et al. 2010. Prevalence of antimicrobial-resistant pathogens in Canadian hospitals: results of the Canadian Ward Surveillance Study (CANWARD 2008). Antimicrob. Agents Chemother. 54:4684–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]