Abstract
This study aimed to determine the mechanism of action of a natural antibacterial clay mineral mixture, designated CB, by investigating the induction of DNA double-strand breaks (DSBs) in Escherichia coli. To quantify DNA damage upon exposure to soluble antimicrobial compounds, we modified a bacterial neutral comet assay, which primarily associates the general length of an electrophoresed chromosome, or comet, with the degree of DSB-associated DNA damage. To appropriately account for antimicrobial-mediated strand fragmentation, suitable control reactions consisting of exposures to water, ethanol, kanamycin, and bleomycin were developed and optimized for the assay. Bacterial exposure to the CB clay resulted in significantly longer comet lengths, compared to water and kanamycin exposures, suggesting that the induction of DNA DSBs contributes to the killing activity of this antibacterial clay mineral mixture. The comet assay protocol described herein provides a general technique for evaluating soluble antimicrobial-derived DNA damage and for comparing DNA fragmentation between experimental and control assays.
Keywords: E. coli, neutral comet assay, single cell gel electrophoresis, DNA double-strand breaks, antibacterial mineral leachate, antibiotic
1. Introduction
The threat of community-acquired methicillin-resistant Staphylococcus aureus (MRSA), emergence of additional antibiotic-resistant S. aureus strains (Chambers and Deleo, 2009), and the financial burden of antibiotic-resistant infections on the U.S. healthcare system (Roberts et al., 2009) have stressed the need for developing supplemental and alternative antimicrobial therapeutics. We have identified a natural antibacterial clay mineral mixture, designated CB, which exhibits broad-spectrum antimicrobial activity against both Gram-negative and Gram-positive bacteria, including MRSA (Cunningham et al., 2010). The soluble fraction, or leachate (CB-L), of CB maintains the antimicrobial properties of the clay, indicating a chemical, rather than physical, mechanism of induced toxicity (Cunningham et al., 2010). Because the aqueous suspensions of the CB clay minerals result in desorption of metal cations (including iron), a low pH environment, and the generation of reactive oxygen species (ROS) and hydrogen peroxide, we hypothesized that Fenton/Haber-Weiss reaction-mediated oxidative stress contributes to CB antimicrobial activity (Cunningham et al., 2010; Koehl and Haydel, unpublished results). The Fenton and Haber-Weiss reactions utilize iron and hydrogen peroxide to produce hydroxyl and superoxide radicals, which are known variations of ROS (Halliwell and Gutteridge, 1984). In oxidative stress conditions, ROS generate damage in all biological macromolecules including nucleic acids, proteins, and lipids (Imlay, 2008) with nucleic acids such as DNA being susceptible to single- and double-strand breaks (Collins et al., 1995).
The comet, or single cell gel electrophoresis, assay is a technique used to evaluate DNA strand fragmentation levels in individual cells. Following exposure to DNA-damaging agents or conditions, the assay consists of embedding cells in an agarose gel mounted on a microscope slide, lysing cells, electrophoresing the exposed cellular chromosomes, and visualizing the elongated DNA, termed “comets.” In order to differentiate single-strand breaks (SSBs) and DSBs, the relative pH of the electrophoresis buffer is altered. Alkaline comet assays (pH > 12.3) select for SSBs while lower-pH electrophoresis conditions (pH < 10), referred to as neutral comet assays, primarily detect DSBs (Fairbairn et al., 1995, Wong et al., 2005). Although the comet assay has been almost exclusively performed on eukaryotic cells (Dhawan et al., 2009), Singh et al. (1999) adapted the neutral assay for prokaryotes to assess DSB levels in Escherichia coli upon radiation treatment, using comet length to indicate the degree of DNA damage.
In this report, we present a modified version of the neutral comet assay with E. coli to investigate the degree of DNA DSBs generated upon CB-L exposure. Because DSBs have a higher correlation with cell death than SSBs (Olive, 1998; Singh et al., 1999), a demonstration of DSB induction by the CB clay derivative would further support the role of oxidative stress in the mechanism of antimicrobial activity. To incorporate a complete lethality control with the experimental reaction of CB-L-treated cells, we assessed comet lengths of cells exposed to 40% ethanol, which completely kills E. coli within minutes (Morton, 1977). Based on our scanning electron microscopy observations that a 40% ethanol solution does not lyse E. coli cells (Otto and Haydel, unpublished results), we expected that associated chromosomal degradation would occur following cell death. As a positive control for comet length distributions arising from DSB-mediated cell death, cells were treated with bleomycin, which forms an iron-containing complex that generates ROS, induces DNA cleavage, and inhibits DNA synthesis (Claussen and Long, 1999; Huang et al., 2012; Müller and Zahn, 1977). While bleomycin causes both double- and single-strand breaks (Huang et al., 2012; Mirabelli et al., 1985), the lower-pH electrophoresis conditions used in these neutral comet assays should primarily detect DSBs (Fairbairn et al., 1995; Wong et al., 2005). To account for strand cleavage resulting from antibiotic exposure that does not involve DNA fragmentation as a primary mechanism of action, we also analyzed comet lengths of cells exposed to kanamycin, an aminoglycoside that causes cell death by primarily inhibiting protein synthesis (Kohanski et al., 2007). Exposure of cells to water was used as a negative control.
In addition to reporting the influence of an antibacterial clay mineral mixture on DNA DSB levels in exposed E. coli and the establishment of appropriate control reactions to address this query, we demonstrated that the bacterial neutral comet assay functions as an effective, general technique to assess DNA strand fragmentation levels mediated by soluble antimicrobial compounds.
2. Materials and Methods
2.1. Strains and media
E. coli ATCC 25922, obtained from the American Type Culture Collection, was used as the model prokaryotic organism in this study. Cells were cultured in Luria-Bertani (LB) broth or agar and grown aerobically in LB at 37°C under rotary incubation to prevent sedimentation and ensure even exposure to experimental and control treatment conditions.
2.2. CB leachate preparation
CB leachates were prepared as previously described (Cunningham et al., 2010; Otto et al., 2010). Briefly, CB minerals were added to sterile, UV-irradiated, deionized H2O (1g/20mL) and stirred for 20–24 h at 300 rpm at room temperature. The mixture was then centrifuged at 30,000 × g for 3 h at 4°C. The supernatant was collected and sterilized by passage through a 0.22 μm filter.
2.3. Bacterial culture and experimental exposures
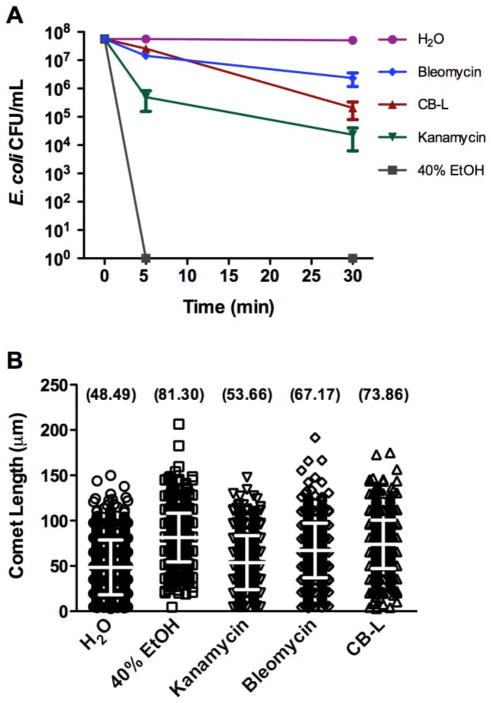
E. coli cultures were incubated with rotary agitation at 37°C until mid-logarithmic phase of growth (OD600 of ~0.6 – 0.7), and then diluted with LB broth to achieve a concentration of ~107 CFU/mL. The cells were collected by centrifugation (13,000 × g), washed with 0.1X phosphate-buffered saline (PBS), and resuspended in one of the following solutions: sterile, UV-irradiated, deionized H2O; 40% ethanol; CB-L; 3 μg/mL kanamycin (Sigma, St. Louis, MO); or 0.3 μg/mL bleomycin (Santa Cruz Biotechnology, Santa Cruz, CA). Exposed cells were then incubated for 30 min at 37°C before adding a 1 μL aliquot (~104 treated cells) to prepared slides (described below). Immediately following exposure (~5 min due to processing time) to the above conditions and following a 30-minute incubation period (described below), 100 μL aliquots from each sample were ten-fold serially diluted and plated in duplicate on LB agar. After overnight incubation at 37°C, the number of colonies was enumerated to determine viability following exposure to the experimental and control conditions (Fig. 2A).
Fig. 2.
Viability (A) and comet lengths (B) of E. coli subjected to experimental and control exposures. A) Viability of logarithmic-phase E. coli was assessed following exposure to the indicated conditions. Error bars represent the standard error of the mean from three independent experiments. B) E. coli comet length distributions resulting after 30-minute experimental and control exposures. Data collected represent comet length distributions from three independent replicates with a total of ~750 comets scored per experimental sample. Error bars represent the mean and standard deviation. The mean comet lengths for each sample distribution are indicated in parentheses above each scatter plot. Length measurements were subject to errors of ±5 μm. Statistical analysis of the negative control (H2O) values compared to the values of 40% ethanol-, kanamycin-, bleomycin-, and CB-L-exposed samples yielded p values of <0.0001, 0.0008, <0.0001, and <0.0001, respectively. The p value of the CB-L-exposed sample compared to the kanamycin-exposed sample was <0.0001.
2.4. Neutral comet assay
The neutral comet assay was adapted from Singh et al. (Singh et al., 1999) and modified as described below.
2.4.1. Slide preparation
To facilitate adhesion of the stratified agarose microgel (described below) to the slide, frosted microscope glass slides with a clear window (Thermo Scientific, Portsmouth, NH) were pre-coated immediately prior to use by dipping briefly in a 1% agarose solution (prepared with sterile H2O). Pre-coated slides were dried in an incubator at 60–70°C for approximately 15 min to completely evaporate the agarose, thus producing a thin coating to facilitate adhesion.
2.4.2. Microgel formation and processing
To form a stratified microgel in which to embed cells, agarose suspensions (described below) were pipetted onto the clear window region of the slide surface before the addition of a 22 mm × 50 mm coverglass. After incubating the slide at 4°C for 10 min to allow the initial layer of agarose to cool, the coverslip was removed and a subsequent agarose layer was added in the same manner. The first layer consisted of 200 μL of 0.5% agarose (SR 3:1 blend, Apex BioResearch Products, Research Triangle Park, NC) prepared in 0.1X PBS or 0.85% NaCl and maintained at 55–60°C. For the second layer, 2 μL of exposed cells were mixed thoroughly with 200 μL of the same 0.5% agarose solution and 100 μL of this mixture was transferred to the slide. A third layer was comprised of the 0.5% agarose solution containing 5 μg/ml RNase A (5 Prime, Gaithersburg, MD), 1 mg/mL lysozyme (Sigma, St. Louis, MO), and 0.25% sodium N-lauroyl sarcosine. Slides were refrigerated for 10 min at 4°C and incubated for 30 min at 37°C. Embedded cells were then lysed by immersing slides in a solution containing 2.5 M NaCl, 100 mM EDTA, 10 mM Tris pH 10, 1% sodium lauroyl sarcosine, and 1% Triton X-100 for 1 h at room temperature. Triton X-100 was added fresh to the lysis solution by stirring for at least 10 min at room temperature. Following lysis, slides were immersed in an enzyme digestion solution prepared with 2.5 M NaCl, 10 mM EDTA, 10 mM Tris pH 7.4, and 0.5–0.6 mg/mL of proteinase K (Ambion, Austin, TX) for 2 h at 37°C. Lysis and enzyme digestion steps were carried out in opaque microscope slide transporters to prevent additional strand fragmentation due to light exposure.
2.4.3. Electrophoresis and slide processing
Following enzyme digestion, slides were transferred to an opaque electrophoresis tank (Cleaver Scientific, Rugby, UK) and equilibrated for 20 min in buffer containing 300 mM sodium acetate and 100 mM Tris, pH 9. Slides were electrophoresed in this buffer for 50 min at 12 V. Following electrophoresis, slides were sequentially immersed in 1 M ammonium acetate prepared in ethanol for 20 min, absolute ethanol for 30 min, and 70% ethanol for 10 min. Slides were then allowed to dry in ambient air for complete evaporation of the microgel.
2.4.4. Staining and visualization
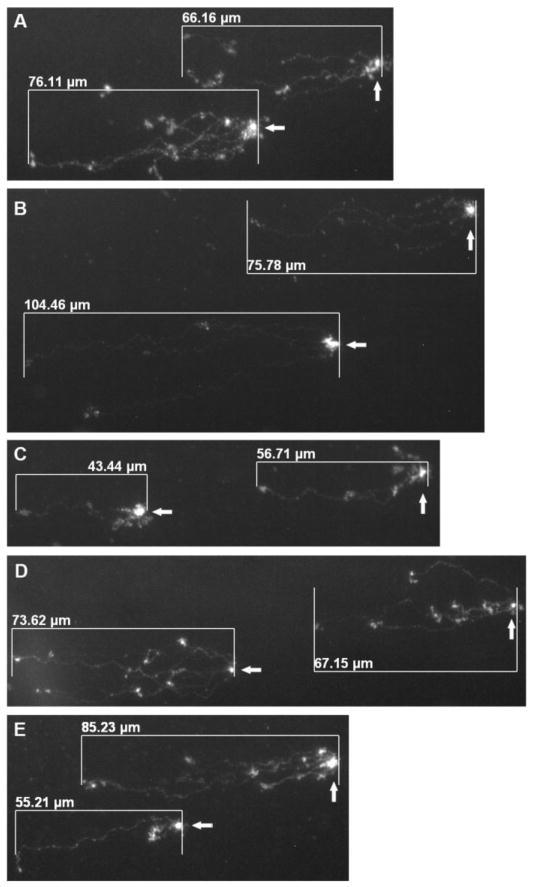
Prior to staining, slides were pretreated with a freshly prepared solution of 5% DMSO and 10 mM NaH2PO4. Pre-treatment comprised of pipetting 50 μL of the solution in a dropwise fashion over the clear window. While the slides were still wet, DNA was stained with 50 μL of 1 μM YOYO-1 (Molecular Probes, Eugene, OR) in 5% DMSO and visualized under an Axioskop 40 fluorescent microscope (Zeiss, Göttingen, Germany) at 40× magnification (Achroplan and EC Plan-Neofluar objectives) with the appropriate filter set for YOYO-1 (excitation 491 nm and emission 509 nm). Comets were imaged and comet lengths were measured (Fig. 1) using AxioVision software version 4.7.2 (Zeiss).
Fig. 1.
Representative images acquired from neutral bacterial comet assays following E. coli exposure to CB leachate (A), bleomycin (B), kanamycin (C), ethanol (D), and water (E). The head (arrows) of each comet represents the chromosomal mass of a single cell or a small cluster of overlapping cells, and the tail signifies the migrated DNA due to strand fragmentation. Comet lengths were measured as the horizontal distance from the outer edge of the head (non-migrated portion of the chromosome) to the visible end of the tail (migrated region) using scaling software (described in Materials and Methods).
2.5. Statistical analyses
DSB levels in the experimental sample, relative to the controls, were evaluated by comparing average comet lengths across samples and using two-tailed, unpaired Student’s t tests on the length distributions to confirm statistical significance. A p value of <0.05 was considered statistically significant.
3. Results and discussion
3.1. Assessment of appropriate controls
In this study, we performed the neutral comet assay to determine if exposure to the antibacterial clay mineral mixture CB is linked to higher DSB levels in E. coli cells. To prepare for the assay, we first assessed positive and antibacterial control reactions at varying concentrations to determine the effectiveness of each control and the optimal antibiotic concentrations to utilize. For this study, optimal antibiotic concentrations were those that consistently exhibited killing rates similar to, but not exceeding, that of CB, so as to exclude any additional strand fragmentation in the control samples that may arise following cell death. It should be noted that the optimal concentrations for these antibiotic control reactions may vary depending on the killing activity of the particular antimicrobial compound being investigated. For all screening assays, exposure to water was used as a negative control.
The antibiotic, bleomycin, was tested as a positive control for generating DNA strand breaks. E. coli cells were treated for 20 min with concentrations ranging from 0.1–0.3 μg/mL. Exposure of E. coli to increasing concentrations of bleomycin resulted in consistent, statistically significant increases in neutral comet assay-detected DSB levels relative to the negative control (data not shown), thus verifying the suitability of using bleomycin for strand fragmentation positive control reactions.
Kanamycin, a protein synthesis inhibitor, was evaluated as an antibacterial control to account for DSB levels corresponding to antibiotic-associated cell death. Cells were treated for 25 min with kanamycin concentrations ranging from 2–4 μg/mL. Corresponding DNA DSB levels were not consistently higher than that of the negative control, nor did they rise with increasing antibiotic concentrations (data not shown). However, the killing rate was enhanced with increasing concentration, confirming that kanamycin does not generate DNA DSBs as its primary mechanism of antimicrobial activity, and thereby allowing kanamycin to serve as an appropriate antibacterial control.
3.2. CB clay-derived leachate generates DSBs in E. coli
Following optimization of the positive and antibacterial control reactions, bleomycin and kanamycin were incorporated into the experimental design to evaluate DSB induction by the CB clay-derived leachate. Treatment with the bleomycin positive control produced a statistically significant increase in comet length relative to the negative control of cells exposed to sterilized water (Fig. 2B). Comparing the antibacterial and negative control reactions, comet length values, though closer in scale, were also significantly different (Fig. 2B). Exposure of E. coli to CB-L for 30 min significantly increased the average comet length of the sample compared to a similar incubation in sterilized water (Fig. 2B), indicating that the clay-derived leachate generates DNA DSBs in bacterial cells. CB-L treatment also yielded a comet length value significantly higher than the kanamycin antibacterial control, demonstrating higher DSBs levels upon CB-L exposure compared to incubation with an aminoglycoside (Fig. 2B). CB-L treatment exerted a 10-fold stronger killing effect than bleomycin (Fig. 2A), yet the average comet length values of 73.86 μm vs. 67.17 μm, respectively (Fig. 2B) were comparable, suggesting that additional mechanisms are likely contributing to CB-L bactericidal activity.
3.3. Bacterial comet assays require the use of dimeric cyanine DNA stains
Our study has further confirmed the necessity of dimeric cyanine dyes, such as YOYO-1, for bacterial comet assay DNA staining. Although widely used nucleic acid stains, such as ethidium bromide and propidium iodide, have been used extensively to stain comets of eukaryotic cells (Fairbairn et al., 1995), these dyes lack the sensitivity to detect electrophoresed DNA from prokaryotes. Throughout the course of these experiments, we tested ethidium bromide, propidium iodide, and SYTO 9 nucleic acid stains for their ability to detect and distinguish individual untreated and electrophoresed chromosomes from lysed E. coli cells, but were unable to visualize bacterial chromosomes using these stains. YOYO-1, and structurally related cyanine dyes (e.g. TOTO-1), derive their improved sensitivities from their dimeric nature, which allows for enhanced binding affinity and the ability to detect single DNA molecules (Armitage, 2005).
3.4. Modifications to the bacterial comet assay
The modified protocol for comet analysis described in this report confers specific benefits over conventional techniques. After performing the assay, various parameters can be used to assess DNA damage, including comet length, visual appearance, or a combination of length and fluorescence intensity in the comet tail, which often requires the use of specific imaging software in order to quantify intensity (Fairbairn et al., 1995). When conducting the neutral assay on irradiated E. coli cells, Singh et al. (Singh et al., 1999) determined comet length as a sensitive indicator of DNA damage and measured the length using a lens piece micrometer. Alternatively, in this study, we photographed comet images from various regions along the slide window and measured comet lengths using the imaging software associated with the fluorescent microscope. This approach would aid laboratories without access to eyepiece micrometers, preserves the data in the form of saved digital images, and allows for the comparison of comet appearance across multiple samples (Fig. 1).
3.5. Limitations to the bacterial comet assay
One analytical limitation to the bacterial comet assay is the variability associated with using comet length as an indicator of DSB levels. Because the structure of bacterial chromosomes in replicating cells is such that the length of migration will vary depending on the location of the break along the DNA strand, two chromosomes that possess the same number of DSBs may exhibit different lengths (Singh et al., 1999). Since we experienced difficulty in determining whether an observed comet represents single or multiple bacterial chromosomes due to frequent overlap in a given focal plane during visualization (Fig. 1), counting the number of DSBs per cell was not considered for this study. We instead used comet length as an indicator of strand fragmentation levels. Although we acknowledge that length does not proportionally correlate to the number of DSBs in a chromosome, it is an effective relative indicator of strand fragmentation as supported by the significantly higher average comet length in the sample treated with bleomycin, whose mechanism of action by DNA cleavage is extensively supported by scientific literature, compared to the negative water control (Fig. 2B). In addition, the large sample size measured in multiple replicates further validates the reliability of this observed trend (Fig. 2B).
Another potential consideration is whether exposure to 40% ethanol functions as an appropriate complete lethality control to exclusively account for post-mortem chromosomal degradation when assessing antimicrobial-mediated DNA strand fragmentation. The average comet length detected in ethanol-exposed cells (~80 μm) was consistently higher than other samples (Fig. 2B). Although ethanol and kanamycin both exhibit bactericidal activity, but neither induces strand fragmentation as their primary mechanism of action, there is a substantial discrepancy in comet lengths associated with the ethanol- and kanamycin-exposed samples (~81 μm vs. ~54 μm, respectively, Fig. 2B). These data suggest that additional mechanisms, such as the induction of bacterial programmed cell death involving DNA fragmentation (Aertsen and Michiels, 2004; Engelberg-Kulka et al., 2006; Yarmolinsky, 1995), may be responsible for the sizable comet length increase in cells exposed to ethanol. Additionally, the ability of ethanol to permeabilize the cell membrane by disrupting hydrophobic interactions allows for leakage of small molecules into and out of cells (Ingram, 1990). As a result, the increased exposure of cellular material to the extracellular environment could attribute to enhanced DNA degradation, indicated by longer comet lengths. As an alternative, because of its 99% percent lethality upon initial exposure, the kanamycin-treated sample in this assay could potentially function as a control to account for both antimicrobial-mediated cell death and post-mortem DNA degradation (Fig. 2).
4. Conclusions
A modified version of the bacterial neutral comet assay was developed and performed to assess DNA DSBs in E. coli upon exposure to an aqueous derivative of the CB antibacterial clay mineral mixture. Incubation of cells with the derived leachate of the clay in three independent replicates corresponded to significantly higher DSB levels than the negative (H2O) and antibacterial (kanamycin) controls. The increased killing strength of CB-L relative to the bleomycin positive control (Fig. 2A), despite similarities in their average comet lengths (Fig. 2B), suggests that additional cytotoxic mechanisms associated with CB-L exposure account for the enhanced bactericidal activity. Although further research is required to fully characterize the mechanism of action of the CB clay, data presented herein support the role of DNA damage as a contributor to its bactericidal activity. The incorporation of additional in situ studies, such as diffusion-based investigations of DNA fragmentation (Fernandez et al., 2008), with E. coli and other bacteria would be valuable in further implicating DNA damage as an antibacterial mechanism. We also attempted an alkaline version of the assay to detect DNA SSBs, but the resulting comets exhibited low fluorescence, had unclear boundaries, or were undetectable. This alkaline comet assay, developed to evaluate CB-mediated DNA damage, can be easily implemented to primarily assess double-strand fragmentation in bacterial cells exposed to soluble antimicrobial compounds.
Highlights.
We adapted the bacterial neutral comet assay for use with soluble antimicrobials.
The assay was employed to investigate the mechanism of action of an antibacterial clay mineral mixture.
Longer comets and thus, higher DNA double-strand break levels, were detected in antibacterial clay leachate-exposed E. coli.
Appropriate control reactions and nucleic acid stains for the assay were established.
Acknowledgments
We would like to thank Caitlin Otto and Andrea Loes for their assistance in the experimental design process and for providing helpful discussions and critical evaluation of the manuscript. This research was supported by Public Health Service grant AT004690 (awarded to S.E.H.) from the NIH National Center for Complementary and Alternative Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aertsen A, Michiels CW. Stress and how bacteria cope with death and survival. Crit Rev Microbiol. 2004;30:263–273. doi: 10.1080/10408410490884757. [DOI] [PubMed] [Google Scholar]
- Armitage BA. Cyanine dye–DNA interactions: intercalation, groove binding, and aggregation. In: Waring MJ, Chaires JB, editors. DNA binders and related subjects. Vol. 253. Springer; Berlin/Heidelberg: 2005. pp. 55–76. [Google Scholar]
- Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussen CA, Long EC. Nucleic acid recognition by metal complexes of bleomycin. Chem Rev. 1999;99:2797–2816. doi: 10.1021/cr980449z. [DOI] [PubMed] [Google Scholar]
- Collins AR, Ma AG, Duthie SJ. The kinetics of repair of oxidative DNA damage (strand breaks and oxidised pyrimidines) in human cells. Mutat Res. 1995;336:69–77. doi: 10.1016/0921-8777(94)00043-6. [DOI] [PubMed] [Google Scholar]
- Cunningham TM, Koehl JL, Summers JS, Haydel SE. pH-Dependent metal ion toxicity influences the antibacterial activity of two natural mineral mixtures. PLoS One. 2010;5:e9456. doi: 10.1371/journal.pone.0009456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan A, Bajpayee M, Parmar D. Comet assay: a reliable tool for the assessment of DNA damage in different models. Cell Biol Toxicol. 2009;25:5–32. doi: 10.1007/s10565-008-9072-z. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2:e135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbairn DW, Olive PL, O’Neill KL. The comet assay: a comprehensive review. Mutat Res. 1995;339:37–59. doi: 10.1016/0165-1110(94)00013-3. [DOI] [PubMed] [Google Scholar]
- Fernandez JL, Cartelle M, Muriel L, Santiso R, Tamayo M, Goyanes V, Gosalvez J, Bou G. DNA fragmentation in microorganisms assessed in situ. Appl Environ Microbiol. 2008;74:5925–5933. doi: 10.1128/AEM.00318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JM. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SX, Feng Z, Wang L, Galm U, Wendt-Pienkowski E, Yang D, Tao M, Coughlin JM, Duan Y, Shen B. A designer bleomycin with significantly improved DNA cleavage activity. J Am Chem Soc. 2012;134:13501–13509. doi: 10.1021/ja3056535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram LO. Ethanol tolerance in bacteria. Crit Rev Biotechnol. 1990;9:305–319. doi: 10.3109/07388558909036741. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- Mirabelli CK, Huang CH, Fenwick RG, Crooke ST. Quantitative measurement of single- and double-strand breakage of DNA in Escherichia coli by the antitumor antibiotics bleomycin and talisomycin. Antimicrob Agents Chemother. 1985;27:460–467. doi: 10.1128/aac.27.4.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton HE. Alcohols. In: Block SS, editor. Disinfection, sterilization, and preservation. Lea & Febiger; Philadelphia: 1977. pp. 301–308. [Google Scholar]
- Müller WE, Zahn RK. Bleomycin, an antibiotic that removes thymine from double-stranded DNA. Prog Nucleic Acid Res Mol Biol. 1977;20:21–57. doi: 10.1016/s0079-6603(08)60469-9. [DOI] [PubMed] [Google Scholar]
- Olive PL. The role of DNA single- and double-strand breaks in cell killing by ionizing radiation. Radiat Res. 1998;150:S42–51. [PubMed] [Google Scholar]
- Otto CC, Cunningham TM, Hansen MR, Haydel SE. Effects of antibacterial mineral leachates on the cellular ultrastructure, morphology, and membrane integrity of Escherichia coli and methicillin-resistant Staphylococcus aureus. Ann Clin Microbiol Antimicrob. 2010;9:26. doi: 10.1186/1476-0711-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RR, Hota B, Ahmad I, Scott RD, 2nd, Foster SD, Abbasi F, Schabowski S, Kampe LM, Ciavarella GG, Supino M, Naples J, Cordell R, Levy SB, Weinstein RA. Hospital and societal costs of antimicrobial-resistant infections in a Chicago teaching hospital: implications for antibiotic stewardship. Clin Infect Dis. 2009;49:1175–1184. doi: 10.1086/605630. [DOI] [PubMed] [Google Scholar]
- Singh NP, Stephens RE, Singh H, Lai H. Visual quantification of DNA double-strand breaks in bacteria. Mutat Res. 1999;429:159–168. doi: 10.1016/s0027-5107(99)00124-4. [DOI] [PubMed] [Google Scholar]
- Wong VWC, Szeto YT, Collins AR, Benzie I. The comet assay: a biomonitoring tool for nutraceutical research. Curr Top Nutraceutical Res. 2005;3:1–14. [Google Scholar]
- Yarmolinsky MB. Programmed cell death in bacterial populations. Science. 1995;267:836–837. doi: 10.1126/science.7846528. [DOI] [PubMed] [Google Scholar]