Abstract
The multiprotein complexes that receive and transmit axon pathfinding cues during development are essential to circuit generation. Here, we identify and characterize the Drosophila sterile α-motif (SAM) domain-containing protein Caskin, which shares homology with vertebrate Caskin, a CASK [calcium/calmodulin-(CaM)-activated serine-threonine kinase]-interacting protein. Drosophila caskin (ckn) is necessary for embryonic motor axon pathfinding and interacts genetically and physically with the leukocyte common antigen-related (Lar) receptor protein tyrosine phosphatase. In vivo and in vitro analyses of a panel of ckn loss-of-function alleles indicate that the N-terminal SAM domain of Ckn mediates its interaction with Lar. Like Caskin, Liprin-α is a neuronal adaptor protein that interacts with Lar via a SAM domain-mediated interaction. We present evidence that Lar does not bind Caskin and Liprin-α concurrently, suggesting they may assemble functionally distinct signaling complexes on Lar. Furthermore, a vertebrate Caskin homolog interacts with LAR family members, arguing that the role of ckn in Lar signal transduction is evolutionarily conserved. Last, we characterize several ckn mutants that retain Lar binding yet display guidance defects, implying the existence of additional Ckn binding partners. Indeed, we identify the SH2/SH3 adaptor protein Dock as a second Caskin-binding protein and find that Caskin binds Lar and Dock through distinct domains. Furthermore, whereas ckn has a nonredundant function in Lar-dependent signaling during motor axon targeting, ckn and dock have overlapping roles in axon outgrowth in the CNS. Together, these studies identify caskin as a neuronal adaptor protein required for axon growth and guidance.
Introduction
The Drosophila neuromuscular system is an excellent paradigm to decipher the molecular signals orchestrating the precise matching between individual motorneurons and their muscle partners. A number of guidance cues and receptors coordinately regulate motor axon pathfinding assuring the high fidelity of this process. The axon must integrate these disparate signals as it navigates through its environment. Multidomain adaptor proteins promote such integration since they serve as platforms to facilitate communication between signal transduction cascades.
The leukocyte common antigen-related (LAR)-related subfamily of receptor protein tyrosine phosphatases (RPTPs type IIA) are conserved regulators of axon pathfinding and synaptogenesis. This subfamily includes the Drosophila receptors Lar and PTP69D, and the vertebrate receptors LAR, protein tyrosine phosphatase σ (PTPσ), and PTPδ (Johnson and Van Vactor, 2003; Ensslen-Craig and Brady-Kalnay, 2004). Family members contain a variable number of Ig and fibronectin (FN) III domains extracellularly, and two intracellular phosphatase domains. The membrane-proximal D1 phosphatase domain (D1) confers most if not all of the catalytic activity of the receptor, whereas the membrane-distal D2 domain is catalytically inactive and may contribute to LAR family function via interaction with downstream signaling components. These receptors exhibit neuronal expression patterns, and loss-of-function (LOF) mutants display defects in axon targeting and synapse formation (Krueger et al., 1996; Desai et al., 1997; Kaufmann et al., 2002). Heparan sulfate proteoglycans (HSPGs) are binding partners of LAR family members in axon pathfinding and synaptogenesis (Aricescu et al., 2002; Fox and Zinn, 2005; Johnson et al., 2006). In vertebrates, PTPσ is a neuronal receptor for chondroitin sulfate proteoglycan (CSPG) and inhibits axon regeneration after CNS injury (Shen et al., 2009; Fry et al., 2010). On the intracellular side, Lar activity in some contexts requires phosphatase activity, whereas in other contexts its function is independent of catalytic activity (Johnson et al., 2006; Hofmeyer and Treisman, 2009; Prakash et al., 2009), suggesting a diversity of downstream signaling pathways. Indeed, a number of Lar-interacting proteins have been identified (Bateman et al., 2000; Johnson and Van Vactor, 2003; Ensslen-Craig and Brady-Kalnay, 2004). Lar function in synaptic maturation requires Liprin-α, a sterile α-motif (SAM) domain-containing adaptor protein that interacts with Lar in vertebrates and invertebrates (Serra-Pagès et al., 1995, 1998; Kaufmann et al., 2002).
Drosophila Dock and vertebrate Nck are neuronal SH2/SH3-containing adaptor proteins that link guidance receptors to cytoskeletal remodeling (Li et al., 2001). Given the widespread expression of Dock in the embryonic CNS and its central position linking guidance receptors to the actin cytoskeleton, it is notable that motor axons in dock LOF mutant embryos display only subtle defects (Desai et al., 1999), raising the possibility of compensation or redundancy. Dock/Nck interact directly with a number of receptors, including Robo, DSCAM (Down syndrome cell adhesion molecule), and the insulin receptor (Schmucker et al., 2000; Fan et al., 2003; Song et al., 2003) and bind directly to cytoskeletal effectors such as p21-activated protein kinase (Pak) and WASP (Wiskott–Aldrich syndrome protein), thereby presumably linking receptor activation to cytoskeletal rearrangement (Rivero-Lezcano et al., 1995; Hing et al., 1999). Of particular relevance, vertebrate Caskin has been identified as a potential Nck interactor (Fawcett et al., 2007; Balázs et al., 2009). Caskin was first identified as a novel protein binding the CaM kinase domain of CASK (Tabuchi et al., 2002). Vertebrate Caskins are predicted scaffolding proteins with multiple ankyrin repeats, an SH3 domain, and two SAM domains, suggesting that Caskin is a component of a multiprotein complex. Here, we present genetic, cell-biological, and biochemical evidence arguing that Drosophila Caskin is a Lar-binding partner and is required for Lar signal transduction in motor axon guidance.
Materials and Methods
Ethyl methanesulfonate reversion mutagenesis.
yw; GS1205 males were mutagenized with 22 mm ethyl methanesulfonate (EMS) and crossed to homozygous yw;; elavGAL4 virgins. As ckn overexpression causes embryonic lethality, we selected viable F1 individuals, which may contain mutations in the ckn locus. The revertants were balanced over CyO and segregated from the elavGAL4 driver. The ckn genomic coding region was PCR amplified and sequenced in each mutant. In GS1205-A, C, K, and Y mutants, ckn cDNA was amplified by reverse transcription–PCR and cloned into PGEM-T Easy vector (Promega) for DNA sequencing and construct generation.
Immunohistochemistry.
Embryo fixation, antibody staining, and RNA in situ hybridization procedures were performed as described (Miller et al., 2008). The following primary antibodies were used: monoclonal antibody (mAb) ID4 (anti-Fasciclin II; 1:10; Developmental Studies Hybridoma Bank), mAb BP102 (1:50; Developmental Studies Hybridoma Bank), and mAb anti-β-gal (1:500; Promega). For RNA in situ hybridization, digoxigenin (DIG)-labeled RNA probe was made by Roche DIG RNA Labeling kit (SP6/T7) using full-length caskin cDNA as template. For S2R+ cell immunofluorescence, protein expression constructs were generated in Gateway-based vectors (Drosophila Gateway Collection, Carnegie Institution of Washington, Baltimore, MD) and transiently transfected into cells by FuGENE HD (Roche). After 48 h transfection, cells were fixed with 4% formaldehyde/PBS for 15 min, washed three times with PBS, and permeabilized in blocking buffer (3% BSA, 0.1% Triton X-100 in PBS) for 30 min at room temperature. Cells were incubated with primary antibodies: rat anti-hemagglutinin (HA) (3F10; Roche) at 1:500, mouse anti-Flag (Sigma-Aldrich) at 1:1000, and rabbit anti-green fluorescent protein (GFP) (Invitrogen) at 1:500 for 2 h at room temperature or 4°C overnight. Appropriate Alexa Fluor secondary antibodies (1:500) (Invitrogen) were applied for 1 h at room temperature.
Microscopy and data analysis.
Embryos were filleted in 70% glycerol using a Leica MZ9 dissecting microscope and analyzed on a Zeiss Axioplan 2 microscope with a 63× oil-immersion objective. Images were captured on an AxioCam MRc camera. Brightness and contrast were adjusted in Adobe Photoshop. Fluorescence images were obtained on a Zeiss Axio Imager.ZI confocal microscope. Statistical analyses were performed using Fisher's exact test.
Plasmids.
Full-length cDNA clones for liprin-α (LD33094), abl (GH109917), caskin (LD09801), Lar (LD45391), and ena (SD08336) were obtained from the Drosophila Genomics Resource Center. Mouse caskin-2 (clone ID 5704642), ptprD (clone ID 4223353), ptprF (clone ID 6409440), and ptprS (clone ID 6834684) expressed sequence tag clones were purchased from Open Biosystems. pDEST-GBKT7 and pDEST-GADT7 (Rossignol et al., 2007) for yeast two-hybrid assays were obtained from Arabidopsis Biological Resource Center DNA Stock Center. PRS426GPD (Alberti et al., 2007) from Addgene was used for yeast three-hybrid assay. Dock and its truncated forms fused to LexA (Fan et al., 2003) are from Greg Bashaw (University of Pennsylvania, Philadelphia, PA). To generate UAS transgenic flies, corresponding cDNA was amplified from transcription start to stop by PCR and subcloned into pUAST or pTHW vectors (HA-tagged; Carnegie Institution of Washington) by the Gateway system (Invitrogen).
For yeast two-hybrid and three-hybrid assay, GAL4BD constructs (bait) were made in PGBKT7 or in PDEST-GBKT7. Similarly, GAL4AD constructs (prey) were generated in either PACT2 or PDEST-GADT7. The bridge constructs were generated by adding nuclear localization sequence (MPKKKRK) to the N termini of Ckn or Lar and subcloned into PAG426GPD vector. For glutathione S-transferase (GST) and Ni-NTA pull-down experiments, PCR fragments of Lar-cyto (1442–2029 aa), Lar D1 (1442–1728 aa), LarD2 (1729–2029 aa), and full-length Dock were cloned into the PGEX vectors to express GST fusion protein. Full-length Caskin was cloned into pET29a vector to generate His-tagged fusion protein.
Yeast two-hybrid and three-hybrid assay.
Yeast transformation was performed by the lithium acetate method. Interactions were scored after 3 d at 30°C. In yeast two-hybrid assays, bait constructs (PGBKT7 or pDEST-GBKT7 derivatives) were transformed into strain PJ694a whereas prey constructs (PACT2 or PGADT7-DEST derivatives) were transformed into strain SL3004. Drop tests were performed by plating 5 μl of serial dilutions of mid-log-phase cultures on selective plates. For domain mapping experiments, Dock bait constructs were introduced into yeast strain PJ694a. Caskin GAL4AD and PSH18–34, a LexA-responsive LacZ reporter, were introduced into yeast strain SL3004 by cotransformation.
GST and Ni-NTA pull-down assay.
GST and His-tagged fusion proteins were expressed in Escherichia coli and purified on glutathione-Sepharose and Ni-NTA agarose beads, respectively. In the GST pull-down assay, HA-Caskin overexpression embryos were homogenized on ice in embryo lysis buffer for 15 min. The lysates were centrifuged at 15,000 × g for 20 min and the supernatant was incubated overnight at 4°C with 5 μg of GST-bound or GST-fusion protein-bound glutathione-Sepharose beads. Beads were washed three times with ice-cold embryo lysis buffer and boiled in 2× sample buffer. Protein samples were resolved by SDS-polyacrylamide gel and subjected to Western blot analysis using anti-HA antibody (12CA5; Roche). In Ni-NTA pull-down assays, 5 μg of His-tagged Caskin fusion protein immobilized on Ni-NTA agarose beads was incubated with GST-Lar in binding buffer (1% Triton X-100, 150 mm NaCl, 50 mm Tris, pH 8.0, 30 mm imidazole) at 4°C for 2 h. Beads were washed three times with cold binding buffer and subjected to Western blot analysis using anti-GST antibody (GE Healthcare).
Immunoprecipitation.
Drosophila S2 cells were transiently transfected with the indicated expression plasmids by FuGENE HD (Roche) and harvested after 48 h. Cells were lysed in immunoprecipitation (IP) buffer [50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Triton X-100, and 1× protease inhibitor (Roche)] on ice for 15 min and cleared by centrifugation. Lysates were then incubated with anti-FLAG M2 mAb (Sigma-Aldrich) prebound to protein G beads for 2 h at 4°C. Beads were washed three times with cold IP buffer. Proteins were eluted in 2× sample buffer and subjected to SDS-PAGE.
Results
Neuronal CG12424 overexpression alters CNS and motor projections
To identify genes required for neuronal development, we took advantage of the GAL4/UAS misexpression system and screened a collection of UAS-containing P-element lines for elavGAL4-dependent defects in CNS fate or axon guidance (Viquez et al., 2006; Miller et al., 2008). Axon development in these lines was characterized with 1D4 antibody, which labels a subset of ipsilateral axons within the CNS and all motor axon projections (Van Vactor et al., 1993). We identified one line, GS1205, in which 1D4-positive axons inappropriately cross the CNS midline (data not shown). Inverse PCR analysis indicated that the line contains a P-element inserted in an untranslated 5′ exon of CG12424.
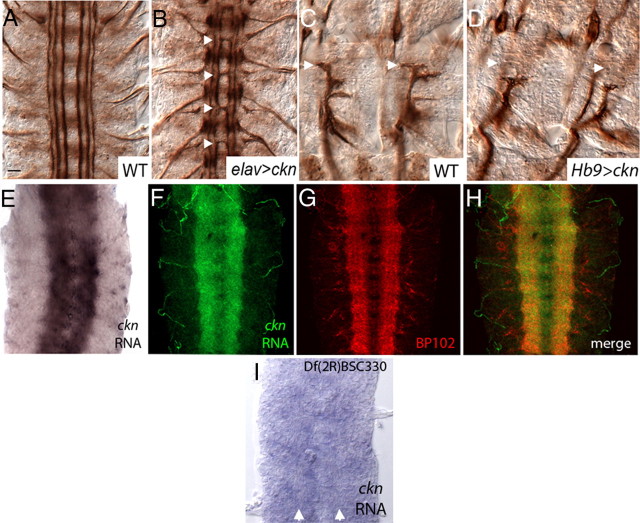
To determine whether CG12424 overexpression causes the observed midline phenotype, we generated UAS-CG12424 transgenic lines. In elavGAL4/UAS-CG12424 embryos, 1D4-positive axons frequently cross and circle the midline (Fig. 1B, arrows). These observations demonstrate that CG12424 overexpression interferes with midline repulsion. We next evaluated motor axon outgrowth in elav>CG12424 embryos. We focused this analysis on the “b” branch of the intersegmental nerve (ISN), which innervates four ventrolateral muscles in the embryo and larva. In wild type, ISNb-extending axons defasciculate from the primary ISN root at a ventral choice point called the exit junction, enter their muscle field, and grow dorsally to their stereotyped targets (Fig. 1C). In elav>CG12424 embryos, ISNb axons frequently stall in the ventrolateral muscle field and fail to innervate their targets (Table 1). Given the severity of the midline defect, however, this phenotype was difficult to interpret. To evaluate whether overexpression of CG12424 more specifically alters motor axon behavior, we characterized motor projections in Hb9GAL4/UAS-CG12424 embryos, as Hb9-GAL4 is expressed in ventrally and laterally projecting motorneurons and significant midline defects are not observed when misexpressing CG12424 with this driver (Broihier and Skeath, 2002). Hb9Gal4/UAS-CG12424 embryos also exhibit ISNb pathfinding errors. In 55% (n = 198) of hemisegments, ISNb axons stall in the ventrolateral muscle field (Fig. 1D). These experiments demonstrate that overexpression of CG12424 disrupts axon guidance in two distinct contexts, suggesting that it interacts with guidance machinery.
Figure 1.
Neuronal overexpression of CG12424 disrupts axon guidance, and CG12424 RNA localizes to the embryonic neuropil. Stage 16 wild-type embryos (A, C), stage 16 elavGAL4/UAS-CG12424 embryo (B), and Hb9Gal4/UAS-CG12424 embryo (D) labeled with mAb 1D4 recognizing all motor axons. A, B, Pan-neuronal overexpression of CG12424 causes inappropriate midline crossing by ipsilateral axons (B, arrowheads). C, D, Two hemisegments of a wild-type embryo (C) and an embryo with CG12424 overexpression in ventral and lateral motorneurons (Hb9>ckn) (D). Hb9>ckn embryos display ISNb stalls. The arrows in C and D indicate appropriate position of dorsalmost ISNb synapse. This synapse is absent in Hb9>ckn embryos. E, Stage 16 wild-type embryo hybridized with antisense CG12424 RNA probe. The embryo shown in F–H is double labeled with CG12424 antisense RNA probe and BP102 to mark the CNS axon scaffold. CG12424 RNA is apparent in CNS axons. I, Stage 16 Df(2R)BSC330 homozygous embryo hybridized with antisense CG12424 RNA probe. Although the longitudinal connectives are not labeled by CG12424 RNA, they are visible via differential interference contrast optics and are indicated by white arrows. A, B, E–I, Anterior, Up. C, D, Anterior, Left. Scale bar, 15 μm.
Table 1.
ISNb pathfinding phenotypes in caskin loss-of-function and gain-of-function mutant backgrounds
| Genotype | Total bypass [% (n)] | Partial parallel bypass (%) | Parallel bypass (%) | Fusion bypass (%) |
|---|---|---|---|---|
| Oregon R (WT) | 0 (120) | 0 | 0 | 0 |
| GS1205 parental line | 7 (180) | 5 | 2 | 0 |
| cknA/cknA | 34 (240) | 1 | 28 | 5 |
| cknA/Df(2R)BSC330 | 32 (188) | 7 | 22 | 3 |
| cknC/cknC | 27 (167) | 8 | 19 | 0 |
| cknC/Df(2R)BSC330 | 30 (199) | 9 | 16 | 5 |
| cknD/cknD | 35 (169) | 5 | 25 | 5 |
| cknD/Df(2R)BSC330 | 36 (180) | 7 | 23 | 6 |
| cknF/cknF | 39 (176) | 9 | 23 | 7 |
| cknF/Df(2R)BSC330 | 34 (155) | 6 | 20 | 8 |
| cknD/cknF | 38 (198) | 6 | 25 | 7 |
| cknK/cknK | 41 (218) | 5 | 27 | 9 |
| cknK/Df(2R)BSC330 | 38 (159) | 9 | 23 | 6 |
| cknL/cknL | 14 (230) | 5 | 6 | 3 |
| cknL/Df(2R)BSC330 | 22 (156) | 4 | 15 | 4 |
| cknY/cknY | 27 (179) | 7 | 16 | 4 |
| cknY/Df(2R)BSC330 | 27 (210) | 12 | 14 | 1 |
| Hb9>cknWTa | 5 (198) | 5 | 0 | 0 |
| elav>cknWTb | 0 (107) | 0 | 0 | 0 |
| elav>cknA | 2 (189) | 2 | 0 | 0 |
| elav>cknC | 61 (174) | 16 | 30 | 15 |
| elav>cknK | 3 (94) | 3 | 0 | 0 |
| elav>cknY | 38 (187) | 12 | 24 | 2 |
| elav>mCaskin2-SAM | 24 (179) | 14 | 10 | 0 |
All embryos were scored at late stage 16/early stage 17. Partial parallel bypass was scored if a subset of ISNb axons enter the VLM field correctly. Parallel bypass was scored if ISNb axons extend next to ISN axons in a distinct fascicle. Fusion bypass was scored if ISN and ISNb axons could not be differentiated at the light microscope level. n = number of hemisegments scored.
aHb9GAL4;UAScknWT 55% of ISNb nerves display stall phenotypes.
belavGAL4;UAScknWT 86% of ISNb nerves display stall phenotypes.
CG12424 RNA localizes to the embryonic CNS neuropil
To determine whether endogenous CG12424 might regulate axon behavior, we characterized its expression pattern. In wild type, embryonic CG12424 RNA expression appears mostly restricted to the CNS axon scaffold (Fig. 1E). We do not observe appreciable embryonic CG12424 expression outside the CNS. This expression pattern is observed with antisense CG12424 RNA probe, but not with a sense probe, suggesting the signal is specific. As an additional test of specificity, we evaluated CG12424 RNA expression in embryos homozygous for a deficiency of the CG12424 locus, Df(2R)BSC330. Neuropil expression of CG12424 RNA is not observed in these embryos (Fig. 1I), demonstrating that the signal highlights CG12424 RNA. To verify that the observed expression is axonal, embryos were colabeled with CG12424 RNA and BP102, a monoclonal antibody recognizing all CNS axons. This experiment confirms that CG12424 RNA is expressed in longitudinal and commissural axons at stage 16 (Fig. 1F–H). The presence of CG12424 RNA in neuronal processes rather than cell bodies suggests it is actively localized and is consistent with a role for CG12424 in axon guidance.
caskin gene structure and allele generation
Sequence analysis indicates that CG12424 codes for a predicted cytoplasmic adaptor protein containing two SAM domains and an EVH1-binding motif (Fig. 2A). The SAM domains in CG12424 have the highest degree of sequence homology to the SAM domains in vertebrate Caskin family members (Fig. 2A). Caskin, or CASK-interacting protein, is a neuron-specific protein identified from rat brain extract as a binding partner of the synaptic calcium/calmodulin-dependent serine kinase CASK (Tabuchi et al., 2002). CG12424 also contains a highly conserved 20 aa sequence at its C terminus present in vertebrate Caskins (Fig. 2A, CTD). In addition to two SAM domains, vertebrate Caskin homologs contain N-terminal ankyrin repeats and an SH3 domain not present in CG12424. A BLAST search of the Drosophila genome using the ankyrin repeats of vertebrate Caskin identifies CG4393 as the most closely related fly gene to the N-terminal sequences of vertebrate Caskin. To determine whether CG4393 may be functionally related to vertebrate Caskin, we assayed embryonic CG4393 expression by in situ hybridization. We do not detect expression of CG4393 in the embryonic CNS (data not shown), indicating that CG4393 is unlikely to contribute to CNS development. Based on sequence analysis and shared binding partners of vertebrate Caskins and CG12424 (see below), we have named CG12424, caskin (ckn).
Figure 2.
caskin gene structure and allele generation. A, Alignment of Drosophila Caskin and murine Caskin2. Conserved domains are labeled. Percentage amino acid similarity/identity in the two SAM domains and the CTD are indicated. B, Identity of eight caskin EMS alleles. C, cknA and cknY are missense alleles, in which conserved amino acids in the first SAM domain (cknA) or the CTD (cknY) are mutated. cknC and cknK contain small deletions. cknK returns to coding sequence in frame, whereas the cknC deletion results in a frameshift and truncated protein.
The overexpression phenotype, RNA expression profile, and sequence homology of ckn suggest that it participates in neuronal development or function. To test this hypothesis, we generated eight EMS-induced LOF alleles via reversion of lethality associated with elavGAL4-dependent ckn overexpression. All ckn alleles, including null alleles, are homozygous viable. Allele sequencing revealed mutations within coding sequence in all alleles (Fig. 2B). Notably, the two missense mutations fall in conserved domains—cknA contains an arginine-to-glutamine substitution in the first SAM domain and cknY has an alanine-to-valine substitution in the C-terminal domain (CTD) (Fig. 2C). These alleles strongly suggest these conserved sequences are required for ckn function in vivo.
caskin LOF alleles disrupt ISNb motor axon pathfinding
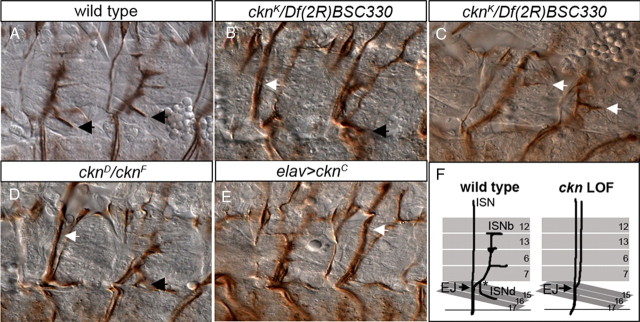
With alleles in hand, we investigated whether ckn homozygous mutant embryos exhibit defects in CNS development. After verifying that the parental GS1205 P-element line does not display appreciable CNS or motor axon guidance errors (Table 1), we asked whether the overall axonal architecture of the CNS neuropil is disrupted in ckn mutants. Overt defects in the axonal scaffold are not observed using BP102 (data not shown). In contrast, ckn mutant embryos display consistent motor axon projection defects, exhibiting classic ISNb “bypass” phenotypes, where ISNb axons fail to innervate the ventrolateral muscle (VLM) field (Fig. 3B,D,F, Table 1). ISNb-projecting axons in ckn mutants appear to defasciculate normally from the primary ISN branch at the exit junction, but fail to enter the ventral muscle field, instead bypassing their targets as they extend parallel to the primary ISN fascicle. Frequently, mistargeted ISNb axons reach back from the dorsal edge of muscle 12 to innervate the VLM field (Fig. 3C, arrows). In the majority of affected nerves, ISNb axons are visible as a distinct fascicle next to the ISN (“parallel bypass”) (Fig. 3B, arrow). Both weaker and stronger ISNb bypass phenotypes are also observed. “Partial parallel bypass” was scored when a subset of ISNb axons correctly enters the muscle field, whereas “fusion bypass” was scored if ISNb axons are tightly bundled with ISN axons and cannot be distinguished from them at the light-microscope level (Fig. 3D, arrow; Table 1). These phenotypes demonstrate that ckn participates in ISNb pathfinding, perhaps by contributing to a signaling pathway(s) mediating ISNb target recognition.
Figure 3.
ckn LOF alleles disrupt ISNb motor axon pathfinding. A–E, Two hemisegments of stage 16 embryos labeled with mAb 1D4 to mark motor axon projections. A, ISNb-projecting axons of wild-type embryos innervate four ventrolateral muscles and adopt a characteristic branched morphology. The black arrows indicate the ISNd branch (when present) in all panels. B, In cknK/Df(2R)BSC330 null embryos, ISNb axons extend in a distinct fascicle adjacent to the primary ISN branch (parallel bypass; white arrow) and do not enter their muscle field. C, cknK/Df(2R)BSC330 embryo in the plane of the VLM field. ISNb axons that bypassed the normal entry point into their muscle domain “reach back” to contact their targets (white arrows). D, In the cknD/cknK embryo, ISNb axons fail to innervate their muscle targets and extend with ISN axons in a common fascicle (fusion bypass; white arrow). E, In elavGAL4/UAS-cknC embryos, ISNb axons fail to separate from the ISN at the ventral choice point (arrow) and exhibit bypass. F, Schematic of wild-type and mutant ISNb/d nerve morphology. Exit junction (EJ) is the choice point where ISNb axons defasciculate from the ISN. The asterisk indicates the point of ISNb/d branch segregation. Anterior is to the left in all panels. Scale bar, 15 μm.
We quantified the guidance errors in ckn homozygous LOF embryos and in embryos carrying each allele over Df(2R)BSC330, a small deficiency for the region (Table 1). All alleles display ISNb bypass phenotypes over the deficiency, indicating that the molecular lesions identified in the ckn locus are responsible for the observed phenotypes. Three alleles (cknK, cknD, and cknF) behave as genetic nulls as the bypass frequency does not differ between homozygotes and allele/Df (p < 0.05 for each) (Table 1). Null alleles display significant ISNb bypass phenotypes—35% of nerves exhibit bypass in cknD homozygotes, 39% in cknF, and 41% in cknK. Interestingly, cknL codes for an early nonsense mutation yet retains some activity (Table 1, Fig. 2B), suggesting there is read-through of the mutation or that an alternative downstream start methionine is used. The penetrance and nature of ISNb bypass phenotypes in null ckn alleles is similar to that of Lar mutants (Krueger et al., 1996; Desai et al., 1997), demonstrating that ckn is an important regulator of ISNb guidance and raising the possibility that ckn functions with Lar.
As the ckn alleles were generated on a chromosome with a UAS-containing GS P-element immediately upstream of the coding region, we tested whether GAL4-dependent neuronal overexpression of mutant proteins interferes with axon pathfinding. As discussed above, overexpression of wild-type Ckn leads to ISNb stall in the ventrolateral muscle field but does not give bypass phenotypes (Table 1). We analyzed motor axon behavior in embryos with neuronal overexpression of four missense mutants: Ckn-A, Ckn-C, Ckn-K, and Ckn-Y. Interestingly, overexpression of these mutants has quite different effects on motor axon guidance. Neuronal overexpression of either Ckn-C or Ckn-Y drives ISNb bypass (Table 1). Ckn-C is particularly potent, as overexpression gives 61% ISNb bypass (Fig. 3E), suggesting that Ckn-C blocks the function of protein(s) essential for ISNb pathfinding. In contrast, neither of the N-terminal SAM domain mutants, Ckn-A or Ckn-K, interferes with ISNb guidance when overexpressed, suggesting that an intact SAM domain is required for the Ckn overexpression ISNb stall phenotypes. Furthermore, this analysis demonstrates that Ckn-K, the LOF allele giving the highest frequency of ISNb bypass, does not have dominant-negative-like activity and is a functional null.
caskin and Lar interact genetically
Lar LOF mutants display ISNb bypass phenotypes similar to those displayed by ckn LOF alleles (Table 2, Fig. 4B,C) (Krueger et al., 1996; Desai et al., 1997). We tested for dose-dependent genetic interactions between Lar and ckn as genes acting in common molecular pathways often interact genetically (Wills et al., 1999; Fan et al., 2003). For this analysis, the Lar5.5 and Lar13.2 alleles were used, both of which contain nonsense mutations in the extracellular domain and are presumed nulls (Fig. 4C) (Krueger et al., 1996). Neither ckn nor Lar heterozygous embryos have ISNb guidance defects, whereas Lar5.5 cknK double heterozygotes display bypass phenotypes with 39% penetrance (Fig. 4D, Table 2), which is similar to the severity observed when either gene alone is homozygous mutant (Table 2, Fig. 4B,C). We next asked whether embryos homozygous mutant for both cknK and Lar5.5 have an increased frequency of pathfinding errors. Previous genetic analysis indicates that maternal Lar product contributes to ISNb pathfinding (Fox and Zinn, 2005). Thus, if ckn and Lar function in a common pathway, loss of ckn might compromise residual Lar activity present in Lar zygotic mutants. Consistently, we observe increased expressivity and penetrance of ISNb bypass when we remove ckn function in Lar zygotic LOF embryos. Whereas Lar5.5 zygotic mutants have an ISNb bypass frequency of 31%, in Lar5.5 cknK embryos the frequency of ISNb bypass rises to 49% (Table 2, Fig. 4E). This interaction is not allele specific as it is also observed in Lar13.2 cknA double homozygotes (Table 2). The enhanced bypass phenotypes observed in Lar ckn double-homozygous embryos are consistent with genetic dependence of maternal Lar product on ckn function. However, ckn may also participate in a parallel genetic pathway(s) that contributes to the increased bypass frequency observed in Lar ckn embryos. Together, the phenotypic similarity between ckn and Lar LOF alleles, as well as dominant genetic interactions, suggest ckn and Lar may operate in a common signaling pathway.
Table 2.
Genetic interactions between caskin, Lar, and PTP69D loss-of-function mutants
| Genotype | Total bypass [% (n)] | Partial parallel bypass (%) | Parallel bypass (%) | Fusion bypass (%) |
|---|---|---|---|---|
| Lar5.5/+ | 5 (90) | 5 | 0 | 0 |
| cknK/+ | 4 (137) | 4 | 0 | 0 |
| Lar5.5/Lar5.5 | 31 (209) | 7 | 19 | 5 |
| ptp69D1/ptp69D7 | 16 (185) | 12 | 4 | 0 |
| cknK/cknK | 37 (202) | 5 | 25 | 7 |
| Lar5.5 cknK/+ + | 39 (165) | 30 | 7 | 2 |
| Lar5.5 cknK/Lar5.5 cknK | 49 (233) | 12 | 17 | 20 |
| Lar13.2 cknA/Lar13.2 cknA | 46 (181) | 21 | 20 | 5 |
| cknk/+; ptp69D1/+ | 5 (58) | 5 | 0 | 0 |
| cknk/+; ptp69D7/+ | 3 (110) | 3 | 0 | 0 |
| cknk/cknk; ptp69D1/ptp69D1 | 35 (151) | 10 | 19 | 6 |
All embryos were scored at late stage 16/early 17. Bypass phenotypes were scored as detailed in the Table 1 legend. n = total hemisegments scored.
Figure 4.
ckn and Lar interact genetically. A–E, Two hemisegments of stage 16 embryos of the indicated genotypes labeled with mAb 1D4 to mark motor axons. A, ISNb nerve of a wild-type embryo. B, cknK mutant embryo displaying an intermediate level of ISNb bypass (arrow). C, Lar5.5 mutant embryo with parallel bypass phenotype in one hemisegment (arrow). D, Lar5.5 cknK/+ + embryo with moderate frequency of ISNb bypass (arrow). E, Lar5.5 cknK double-homozygous embryo exhibiting increased penetrance of ISNb bypass (arrows). Anterior is to the left in all panels.
The N-terminal SAM domain of Caskin binds Lar
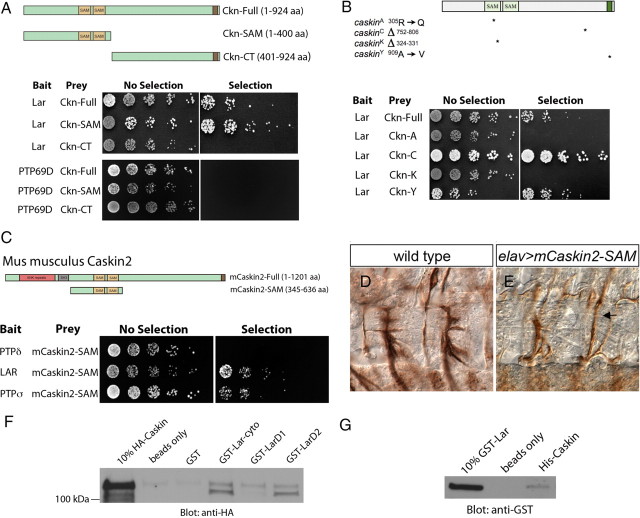
To test for a physical interaction between Ckn and Lar, we assayed whether they interact in a yeast two-hybrid assay. Both full-length Ckn (Ckn-full) and an N-terminal fragment including both SAM domains (Ckn-SAM) interact with the Lar cytoplasmic domain (Lar-cyto) containing both D1 and D2 phosphatase domains (Fig. 5A).
Figure 5.
Ckn and Lar interact physically. A, Yeast two-hybrid interactions indicate a direct physical interaction between Ckn and Lar, but not Ckn and PTP69D. The cytoplasmic domain of Lar was fused to the Gal4-DNA binding domain (bait). Schematics of the Ckn constructs that were fused to the Gal4-activation domain (prey) are shown. Both full-length Ckn and Ckn-SAM interact with Lar-cyto. In contrast, none of the Ckn constructs interacts with PTP69D. B, Schematics of four caskin mutant variants are shown. These mutant cDNAs were fused to the Gal4-activation domain (prey) and tested for binding with the Lar-cyto domain fused to the Gal4-DNA binding domain (bait). Both of the SAM domain mutations, Ckn-A and Ckn-K, abrogate Lar binding, whereas both Ckn-C and Ckn-Y maintain Lar interactions. C, The cytoplasmic domains of the three vertebrate type IIA RPTPs were fused to the Gal4-DNA binding domain (bait) and tested against full-length mCaskin2 and a fragment of mCaskin2 containing both SAM domains fused to the Gal4-activiation domain (prey). Lar and PTPσ interact with mCaskin-SAM. D, E, Two hemisegments of stage 16 wild-type (D) and elavGAL4/UAS-mCaskin2-SAM (E) embryos labeled with mAb 1D4. Neuronal overexpression of mCaskin2-SAM yields ISNb bypass (arrow) suggesting that it interferes with Lar signal transduction in a dominant-negative fashion. F, A GST-Lar-cyto fusion protein precipitates HA-Caskin from elavGAL4/UAS-HA-Caskin embryonic lysates, whereas GST alone does not. GST-Lar D2 pulls HA-Caskin out of embryonic lysates arguing that Ckn binds the D2 domain. G, A bacterially expressed and purified HA-Caskin fusion protein precipitates purified GST-Lar, whereas Ni-NTA beads alone cannot.
The Drosophila genome contains two type IIA RPTPs—Lar and PTP69D (Johnson and Van Vactor, 2003). Whereas PTP69D single mutant embryos have subtle ISNb phenotypes, the role of PTP69D in ISNb guidance is uncovered unambiguously in Lar; PTP69D double-mutant embryos, which display levels of ISNb bypass significantly elevated over that seen in Lar single mutants (Desai et al., 1996, 1997). The intracellular domains of Lar and PTP69D share extensive sequence similarities, raising the possibility that Ckn binds PTP69D. However, ckn and PTP69D do not interact in the yeast two-hybrid assay (Fig. 5A), and we do not observe dominant genetic interactions between ckn and PTP69D alleles (Table 2). Interestingly, the penetrance of ISNb bypass in ckn; PTP69D double homozygotes is not elevated over that observed in ckn homozygotes (Table 2). These data raise the possibility that ckn plays a specific role in the Lar signaling pathway.
The physical interaction between Ckn and Lar suggests that the pathfinding phenotype in ckn LOF mutant embryos results from defective Lar signal transduction. Thus, we asked whether Ckn mutant proteins corresponding to four alleles: Ckn-A, Ckn-C, Ckn-K, and Ckn-Y, disrupt the Ckn/Lar association. Ckn-A and Ckn-K, the two variants that alter the N-terminal SAM domain, abrogate binding to Lar, whereas Ckn-C and Ckn-Y, which have intact SAM domains, retain Lar association (Fig. 5B). These results indicate that the N-terminal SAM domain of Ckn is necessary for Lar interaction and are in agreement with our in vivo data as cknA and cknK are the two alleles that do not promote ISNb bypass when overexpressed (Table 1), suggesting that the Ckn-A and Ckn-K mutant proteins are innocuous when overexpressed in vivo as they lack Lar-binding activity and cannot compete with wild-type Ckn for Lar binding. Together, these studies implicate deficits in Lar signal transduction in ckn axon pathfinding defects.
We next asked whether the molecular relationship between Lar and Ckn is conserved. In support of this hypothesis, both full-length mouse Caskin2 and a Caskin2 fragment containing both SAM domains (mCaskin2-SAM) interact with two of three murine LAR receptor family members, LAR and PTPσ, in a yeast interaction assay (Fig. 5C). These data suggest that Caskins contribute to Lar signaling in vertebrate systems and raise the possibility that murine Caskins interact preferentially with distinct RPTP type IIA family members—similar to the preference of Drosophila Ckn for Lar over RPTP69D. Mouse Caskin2 also interacts with Drosophila Lar in yeast two-hybrid experiments (data not shown), supporting the model that the pathway is well conserved. The in vitro interaction led us to ask whether the proteins interact in vivo. To this end, we tested whether vertebrate Caskin alters motor axon pathfinding when overexpressed in Drosophila neurons. We generated a UAS line to drive overexpression of the Lar-binding region of mouse Caskin2. elavGAL4/UAS-mCaskin2-SAM embryos display ISNb bypass phenotypes with a frequency of 24% (Table 1, Fig. 5D,E). Hence, mCaskin2 overexpression impairs Drosophila motor axon pathfinding, presumably by binding to the Lar receptor and blocking downstream signal transduction.
To advance the hypothesis that Ckn and Lar associate biochemically, we tested whether GST-Lar fusion proteins precipitate HA-Ckn from embryonic extracts. GST fusions to the full-length Lar cytoplasmic domain or to the membrane-distal inactive D2 phosphatase domain pull down HA-Ckn from embryo extracts (Fig. 5F), indicating that Lar and Ckn can form a complex. If Ckn and Lar interact directly, the two purified proteins would be expected to maintain the ability to form a complex. Consistent with a direct physical interaction, bacterially expressed and purified Lar and Ckn associate with each other (Fig. 5G). In sum, these data demonstrate that Ckn binds the Lar RPTP D2 phosphatase domain via its N-terminal SAM domain.
Ckn is sufficient to redistribute the cytoplasmic domain of Lar in S2R+ cells
We turned to adherent S2R+ cells to visualize the intracellular distribution of epitope-tagged Ckn protein. We first compared the subcellular localization of Ckn and Liprin-α. Like Ckn, Liprin-α is a SAM-domain protein that binds the D2 domain of the cytoplasmic tail of Lar via a SAM domain interaction (Serra-Pagès et al., 1995, 1998). Liprin-α and Lar have been shown to colocalize with focal adhesion markers in Drosophila and vertebrate cell lines (Serra-Pagès et al., 1995; Hofmeyer et al., 2006). In agreement with these studies, we find that Flag-Liprin-α exhibits punctate localization (Fig. 6A). Notably, HA-Ckn displays a similar pattern of subcellular localization (Fig. 6B), and Ckn and Liprin-α colocalize when cotransfected (Fig. 6D–F)—consistent with the presence of Ckn, Liprin-α, and Lar in focal adhesions. Whereas full-length Lar colocalizes with Liprin-α in S2R+ cells, we found that a tagged Lar cytoplasmic domain construct (GFP-Larcyto) exhibits a diffuse cytosolic distribution (Fig. 6C). The cytoplasmic localization of GFP-Larcyto presented us with an opportunity to test whether Ckn can recruit Larcyto to Ckn-positive punctae. In fact, when HA-Ckn is cotransfected with GFP-Larcyto, its distribution shifts to Ckn-positive punctae (Fig. 6G–I), consistent with a Ckn/Lar complex. We next asked whether the ability of Ckn to relocalize Lar in cotransfection experiments relies on its N-terminal SAM domain. In support of this hypothesis, Ckn-C shifts the distribution of Larcyto to Ckn-positive punctae, whereas Ckn-A and Ckn-K do not (Fig. 6J–O) (data not shown). These localization studies argue that Ckn and Lar interact via the N-terminal SAM domain of Ckn and raise the question of the physical relationship between Lar, Ckn, and Liprin-α.
Figure 6.

Ckn redistributes Lar-cyto in S2R+ cells, and Ckn and Liprin-α may not simultaneously bind Lar. A–C, S2R+ cells transfected singly with Flag-Liprin-α (A), HA-Ckn (B), or GFP-Lar-cyto (C). Flag-Liprin-α and HA-Ckn have punctate localization pattern, whereas GFP-Lar-cyto has an even cytosolic distribution. D–F, S2R+ cell cotransfected with HA-Ckn and Flag-Liprinα. The proteins colocalize. G–I, S2R+ cell cotransfected with HA-Ckn and GFP-Lar-cyto. HA-Ckn recruits GFP-Lar-cyto to Ckn-positive punctae. J–L, S2R+ cell cotransfected with HA-Ckn-K and GFP-Lar-cyto. HA-Ckn-K does not relocalize Lar-cyto. M–O, S2R+ cell cotransfected with Ckn-C and GFP-Lar-cyto. HA-Ckn-C relocalizes Lar-cyto to Ckn-positive punctae. P, Cotransformation of yeast with a Lar-cyto/Gal4-DNA binding domain fusion (bait) and either Ckn-SAM or Liprin-α Gal4-DNA activation domain fusions (prey) yield yeast two-hybrid interactions. Full-length Ckn could not be used in this assay as it activates transcription in the absence of the Gal4-activation domain. Ckn-SAM/Gal4-DNA binding domain fusion (bait) and Liprin-α/Gal4-DNA activation domain fusions do not interact. If Ckn and Liprin-α bind Lar simultaneously, Lar is predicted to serve as a bridge in a yeast three-hybrid assay. A nuclear localization signal (NLS) was added to the N terminus of Lar to promote nuclear localization. No interaction between Ckn and Liprin-α is detected in the presence of Lar, arguing that Ckn and Liprin-α cannot bind Lar simultaneously.
Ckn and Liprin-α may not simultaneously bind Lar
The molecular similarities between Ckn and Liprin-α and the evidence that they function at distinct developmental stages (Ckn in guidance and Liprin-α in synaptogenesis) raise the possibility that Liprin-α and Ckn occupy a shared/overlapping binding site(s) on the Lar receptor. Hence, we assayed for concurrent binding of Ckn and Liprin-α to Lar in a yeast three-hybrid assay (Fig. 6P). Whereas Lar and Liprin-α interact in a yeast two-hybrid test, Ckn-SAM and Liprin-α do not. We first asked whether a Lar/Gal4 DNA-binding domain fusion still interacts with a Ckn/Gal4 DNA-activation domain fusion in the presence of Liprin-α on a bridge vector. Liprin-α blocks the Ckn–Lar interaction (data not shown), consistent with a shared/overlapping binding site(s) for Ckn and Liprin-α. As a more direct test, if Ckn and Liprin-α can co-occupy the Lar receptor, Lar is expected to serve as a bridge between Ckn and Liprin-α. The three proteins are not detected in a complex (Fig. 6P), suggesting that Lar associates either with Ckn or with Liprin-α, but not both. These data are consistent with a common Ckn/Liprin-α binding site on Lar and argue that the two proteins may compete for Lar binding. Together, Ckn and Liprin-α are well situated to differentially regulate the interaction of Lar with downstream proteins that mediate the roles of Lar in axon outgrowth and synaptic development, respectively.
Caskin and Dock interact physically
Recently, vertebrate Caskin proteins have been identified in screens for Nck-binding proteins (Fawcett et al., 2007; Balázs et al., 2009). Nck/Dock proteins are SH2/SH3-containing scaffolding proteins that participate in the signaling cascades of multiple guidance receptors (Garrity et al., 1996; Schmucker et al., 2000; Li et al., 2001; Fan et al., 2003; Song et al., 2003). Thus, we investigated the possibility of Dock/Ckn interactions. Full-length Ckn and Dock interact in yeast (Fig. 7A). Neither Ckn-SAM nor Ckn-C-terminal (Ckn-CT) constructs interact with Dock, implicating both N- and C-terminal Ckn sequences in the interaction. We further mapped the interacting domain(s) in Ckn using four mutant variants. Whereas Ckn-A and Ckn-Y retain Dock binding, neither Ckn-C nor Ckn-K interacts with Dock (Fig. 7B). Mapping the Ckn interaction domain within Dock using a series of deletion constructs (Fan et al., 2003) reveals that the second SH3 (SH3–2) domain of Dock is sufficient to bind Ckn (Fig. 7C).
Figure 7.

Ckn exhibits physical and genetic interactions with Dock. A, Full-length Ckn interacts with Dock in yeast two-hybrid. Ckn-Full, Ckn-SAM, and Ckn-CT constructs were fused to Gal4-DNA activation domain (prey). Full-length Dock was fused to the Gal4-DNA binding domain (bait). Only full-length Ckn interacts with Dock. B, Mutant cDNAs for four ckn alleles were fused to the GAL4-DNA activation domain and tested against a Dock-Gal4-DNA binding domain bait plasmid. Neither Ckn-C nor Ckn-K bind Dock, whereas both Ckn-A and Ckn-Y maintain the interaction. C, A series of LexA-Dock fusions were tested for interaction with full-length Ckn-activation domain fusions. + +, Medium blue; −, white (no interaction) after 1 h. D, Schematic outlining the yeast three-hybrid interactions between Caskin, Lar, and Dock. Yeast were cotransformed with a Lar-cyto/Gal4-DNA binding domain fusion (bait) and a full-length Dock/Gal4-activation domain fusion (prey). Dock and Lar do not interact. After 2 d, the yeast were mated to yeast transformed with Ckn-WT, Ckn-A, Ckn-C, and Ckn-K. A nuclear localization signal (NLS) was added to the N terminus of Ckn. Only wild-type Ckn couples Lar and Dock. Ckn-Y could not be tested in this assay as it activates transcription in the absence of the Gal4-activation domain. E, Flag-tagged Dock coimmunoprecipitates Caskin-HA from S2 cells. F, A Dock-GST fusion protein precipitates Ckn-HA from elavGal4/UAS-Caskin-HA embryos, whereas GST alone does not. G–K, Two hemisegments of stage 16 embryos labeled with mAb 1D4 to mark motor axon projections. Anterior is to the left. Scale bar, 15 μm. G, ISNb nerve exhibits characteristic branched morphology in wild type. The black arrow indicates ISNd. H, Axons in dock3 cknK/dock3 + embryos are still en route to their targets at late stage 16. I, ISNb axons in dock3 cknK double homozygotes are loosely fasciculated and extend multiple projections. J–L, ISNb/d axons do not separate in ckn dock double mutants. Whereas in wild type, ISNb and ISNd axons are tightly bundled and cleanly separated (J), ISNb/d axons remain tangled in the double mutant (K). Schematic of the ISNb/d branch segregation defect is shown (L). M, N, The CNS midline of wild-type (M) and dock ckn (N) embryos labeled with ID4. Axons in the lateral two connectives are poorly fasciculated and stall in the double mutant.
Together, these mapping data argue that Ckn interacts with Lar and Dock via separable domains, as Ckn-A binds Dock but not Lar, and Ckn-C binds Lar but not Dock. To obtain additional support for this hypothesis, we asked whether Ckn simultaneously interacts with Dock and Lar in a yeast three-hybrid assay. Dock does not interact with the Lar cytoplasmic domain in a yeast two-hybrid assay (Fig. 7D). However, with the addition of full-length Ckn as a bridging protein in a yeast three-hybrid assay, Dock and Lar are found in a complex (Fig. 7D). The Ckn mutant variants that do not bind either Lar or Dock individually cannot couple Dock to Lar demonstrating the specificity of the three-hybrid interaction. These results are consistent with our mapping data and indicate that Ckn interacts with Lar and Dock via distinct domains. We further investigated the relationship between Ckn and Dock in S2 cells. In support of a physical interaction between Ckn and Dock, Dock can coimmunoprecipitate Ckn from S2 cells (Fig. 7E). Finally, we tested whether this interaction holds in embryos and find that GST-Dock pulls down Ckn-HA from embryonic lysates (Fig. 7F). These results argue that Ckn directly binds Dock and raise the possibility that the two proteins interact in axon development.
caskin and dock have redundant functions in axon outgrowth
To investigate whether these biochemical interactions have in vivo relevance, we asked whether ckn and dock interact genetically. In agreement with published work (Desai et al., 1999), we find that dock mutants display a delay in neuromuscular junction (NMJ) formation by the RP3 motorneuron on the muscle 6/7 pair, and no defects are associated with loss of one copy of dock. The absence of pathfinding defects in zygotic dock LOF mutants is not the result of maternally derived Dock as motor axon pathfinding is also normal in embryos lacking both maternal and zygotic Dock (Desai et al., 1999). Given that Dock is strongly expressed in embryonic axons (Desai et al., 1999), normal motor axon pathfinding observed in dock LOF raises the possibility of redundancy. Therefore, we tested whether dock mutants are sensitive to ckn dosage. In support of the model that ckn and dock have shared functions in axon outgrowth, dose-sensitive interactions are observed between alleles of the two genes. In wild-type embryos, ISNb pathfinding is complete by late stage 16, whereas in 23% of dock3 cknk double heterozygotes, motor axons are still en route to their targets at this stage. This dominant interaction suggests that dock and ckn contribute jointly to the efficacy of motor axon outgrowth. To expand this analysis, we investigated the effect of halving ckn dosage in a dock homozygous mutant background. Loss of one copy of ckn has no effect on wild-type embryos but has a striking effect on ISNb outgrowth in dock homozygotes. In dock3 cknK/dock3 + embryos, ISNb outgrowth is incomplete in 43% (n = 187) of hemisegments, compared with 6% of dock3 homozygotes (Fig. 7H) (n = 110). In dock3 cknK double homozygotes, the frequency of the “immature ISNb” phenotype rises to 65% (Fig. 7I) (n = 212). This interaction is not allele specific as 59% of dockP cknC hemisegments exhibit delayed outgrowth (n = 178). Motor axons in the affected nerves are loosely organized with multiple projections and resemble wild-type axons at earlier stages. Furthermore, the ISNd branch is frequently absent or reduced in size. Examination of the ISNb/d choice point revealed defects in ISNb/d branch segregation (Fig. 7J–L). Whereas in wild-type stage 16 embryos, ISNb and ISNd branches are tightly bundled and cleanly separated—a subset of ISNb/d axons are entangled and remain clustered near the choice point in dock ckn double mutants. Finally, we characterized the development of FasII-positive longitudinal connectives. In late-stage 16 wild-type embryos, the connectives are tightly fasciculated and continuous (Fig. 7M). In contrast, the lateral two longitudinals in dock ckn double mutants are poorly fasciculated and discontinuous (Fig. 7N), demonstrating the activities of ckn and dock in axon outgrowth are not limited to the periphery.
The pervasive defects in axonogenesis found in dock ckn mutants argue that ckn activity is not limited to the Lar pathway, but rather that ckn likely participates in signaling downstream of multiple receptors. Notably, we do not detect appreciable ISNb bypass in dock ckn double homozygotes. However, the absence of penetrant bypass phenotypes in dock ckn embryos may be attributable to early pleiotropic defects in fasciculation and outgrowth and does not exclude a role for dock in Lar-dependent signal transduction. Together, the data presented here identify caskin as a conserved adaptor protein required for axon outgrowth and guidance.
Discussion
We demonstrate that Caskin mediates a novel Lar RPTP signaling cascade during axonogenesis. Analysis of a panel of ckn LOF alleles indicates that ckn is necessary for motor axon pathfinding as homozygous mutants display classic bypass defects in the ISNb motor nerve. This phenotype is identical with that displayed by Lar mutants, and genetic and biochemical interaction data demonstrate that Ckn is a Lar-interacting protein. These studies position Caskin to be a core member of a Lar-associated signaling complex that mediates its function during axonogenesis.
Drosophila caskin encodes a conserved neuron-specific adaptor protein
Vertebrate Caskin was identified as a binding partner of the synaptic adaptor protein CASK and competes for binding to the CaM kinase domain of CASK with the PDZ (postsynaptic density-95/Discs large/zona occludens-1) protein Mint1 (Tabuchi et al., 2002). The CASK-binding site on Caskin maps to an N-terminal region not conserved in Drosophila, suggesting that fly Caskin does not bind CASK. Consistent with this finding, Drosophila Caskin and CASK do not interact in a yeast interaction assay (data not shown). However, both mouse and fly Caskin homologs bind LAR family members and Nck/Dock, in support of considerable shared functions (Fawcett et al., 2007; Balázs et al., 2009). Furthermore, overexpression of the Lar-binding domain of mouse Caskin in Drosophila neurons yields a pathfinding phenotype like that of Lar and ckn LOF, suggesting that mouse Caskin competes with fly Ckn for binding to the Lar receptor to function as a dominant negative. These biochemical studies indicate that, whereas Caskins may have species-specific binding partners, Ckn function in Lar signal transduction is conserved. Drosophila Lar also physically interacts with the Abl tyrosine kinase and its substrate the cytoskeletal regulator Ena (Wills et al., 1999). We are unable to detect physical interactions between Ckn and Abl or Ena, suggesting they bind Lar independently (data not shown). This raises the possibility that Caskin and Abl/Ena constitute parallel pathways downstream of the Lar receptor.
Our allelic series enabled us to analyze the in vitro and in vivo activities of four Caskin mutant proteins: Ckn-A, Ckn-C, Ckn-K, and Ckn-Y. Ckn-A and Ckn-K contain alterations in the first SAM domain and block the interaction of Ckn with Lar, pointing to the importance of this domain for Lar/Ckn complex formation. The in vivo analysis of Ckn-A and Ckn-K is in strong agreement with the in vitro data as motor axon phenotypes are not associated with their overexpression, suggesting they do not interfere with Lar signaling in vivo. The behavior of Ckn-A and Ckn-K contrasts that of Ckn-C, which contains a C-terminal deletion. Ckn-C interacts with Lar, and its neuronal overexpression yields dominant-negative-like effects. In fact, the penetrance of ISNb bypass associated with Ckn-C overexpression is comparable with that observed in embryos lacking both maternal and zygotic Lar (Fox and Zinn, 2005), suggesting that it effectively interferes with Lar activity. Although Ckn-C binds Lar, cknC homozygous LOF mutants display a “Lar-like” ISNb phenotype, indicating that Lar signaling is blocked downstream of receptor binding. Ckn-C does not interact with Dock, but this interaction is insufficient to explain the cknC mutant phenotype since ISNb bypass is not associated with dock LOF. The pathfinding phenotype observed in cknC embryos argues that the allele also disrupts the interaction between Caskin and another downstream protein(s) essential for Lar signaling.
Ckn and Dock function redundantly in axon outgrowth
Dock/Nck are SH2/SH3-containing adaptor proteins that couple phosphotyrosines on activated receptors to downstream signaling molecules via SH2 and SH3 domain interactions, respectively (Buday et al., 2002). Dock also engages in a ligand-regulated SH3 domain interaction with the Robo receptor (Fan et al., 2003), demonstrating that it is involved in diverse interactions downstream of guidance receptors. In this work, we demonstrate that Caskin interacts with the second SH3 domain of Dock (SH3–2). This domain has also been shown to interact with the cytoskeletal effector Pak (Hing et al., 1999), raising the issue of the relationship between Caskin and Pak. It will be informative to determine whether Dock forms alternative complexes with Caskin and Pak, or whether Dock binds Caskin and Pak simultaneously.
The contrast between the ckn and dock single- and double-mutant phenotypes demonstrates that the adaptors have mostly redundant functions. Single-mutant analyses indicate that ckn plays a nonredundant role in Lar signaling, whereas dock has a unique role in synaptogenesis of the RP3 motorneuron. However, the outgrowth defects observed in dock ckn double mutants argue that these adaptors have overlapping roles in a number of signaling events. These data caution against drawing conclusions of cellular function based solely on single mutant analysis, as this obviously uncovers only the nonredundant functions of a protein. The issue of genetic redundancy may be particularly acute in signaling systems involving multi-subunit complexes with many opportunities for parallel functions. It will be important to identify additional binding partners of dock and ckn to determine whether they have a common set of interactors, or whether they impinge on the cytoskeleton via distinct, yet redundant, paths.
A Lar–Ckn pathway for axon pathfinding
The Lar receptor is a member of the type IIA subfamily of RPTPs, comprising Lar and PTP69D in flies (Johnson and Van Vactor, 2003). The single-mutant phenotypes of Lar and PTP69D indicate they have nonredundant functions in motor axon guidance, NMJ growth, and photoreceptor axon targeting (Desai et al., 1997; Clandinin et al., 2001; Maurel-Zaffran et al., 2001; Kaufmann et al., 2002; Hofmeyer and Treisman, 2009). Several observations hint that the unique functions implied by the divergent phenotypes of Lar and PTP69D stem in part from distinct ligand-binding activities. Fox and Zinn (2005) observed that Lar and PTP69D alkaline phosphatase fusion proteins have distinct embryonic staining patterns suggesting the presence of unique ligands. Furthermore, overexpression of a chimeric receptor composed of the Lar extracellular domain fused to the PTP69D intracellular domain rescues the LOF photoreceptor defect of Lar, whereas a PTP69D extracellular domain fusion to the Lar intracellular domain does not (Maurel-Zaffran et al., 2001)—arguing that Lar and PTP69D have overlapping intracellular partners and (at least partially) nonoverlapping extracellular ones. However, more recent data open the door for functional differences between the intracellular pathways activated by Lar and PTP69D. R7 photoreceptor axon targeting is independent of Lar phosphatase activity, but dependent on PTP69D phosphatase activity, suggesting that the receptors have distinct binding partners (Hofmeyer and Treisman, 2009; Prakash et al., 2009). These findings are consistent with the work presented here. Both fly and vertebrate Caskins interact with subsets of LAR family receptors, raising the possibility that the intracellular signaling cascade(s) organized by Ckn contributes to the functional differences between Lar and PTP69D.
We investigated the physical relationship between Lar, Ckn, and Liprin-α and do not detect a ternary complex. These binding data support mapping studies indicating that Ckn and Liprin-α both interact with the D2 phosphatase domain of Lar via SAM domain-mediated interactions. They further suggest sequential/competitive binding of Ckn and Liprin-α to the Lar receptor and raise the possibility of distinct neuronal functions. It is conceivable that Ckn and Liprin-α both act downstream of Lar to mediate its activity during axon outgrowth/pathfinding and synaptogenesis, respectively. To determine whether Ckn function is specific for Lar signaling during axonogenesis, it will be informative to test whether ckn LOF mutants exhibit defects in the assembly/localization of presynaptic components similar to that observed in Lar mutants (Kaufmann et al., 2002; Miller et al., 2005). Alternatively, the function of Liprin-α in Lar signaling may be primarily to localize or maintain Lar at the presynaptic terminal, whereas Ckn functions in downstream signal transduction. This hypothesis is supported by evidence for a conserved function for Liprin-α in synaptic protein targeting or anchoring (Serra-Pagès et al., 1998; Kaufmann et al., 2002; Ackley et al., 2005; Miller et al., 2005). A role for Liprin-α in trafficking is further bolstered by conserved physical interactions between Liprin-α and Kinesin (Shin et al., 2003; Miller et al., 2005), suggesting it is an adaptor protein for anterograde transport of synaptic proteins. In this scenario, it is notable that Liprin-α function is not required for pathfinding (Hofmeyer et al., 2006), arguing either that another protein serves to localize Lar during guidance or that Lar activity in this process does not require its tight localization to the axon terminal. This model is consistent with the broad axonal localization of Lar during embryogenesis (Krueger et al., 2003).
Extracellularly, LAR family members interact with HSPGs and CSPGs. In Drosophila, mutations in the HSPG syndecan (sdc) interact with Lar in motor axon guidance, but homozygous LOF sdc embryos do not display appreciable bypass phenotypes (Fox and Zinn, 2005), arguing that other ligands are involved. Once these ligands are identified, it will be critical to determine whether ligand binding influences the association of intracellular adaptors such as Liprin-α and Caskin with Lar. Recently, vertebrate LAR family members have moved into the spotlight in the field of axon regeneration, as PTPσ has been shown to be a receptor for CSPGs, which are dramatically upregulated at the lesion site and are strongly inhibitory to axon growth (Shen et al., 2009; Fry et al., 2010). Strikingly, axons in PTPσ mutant mice have a greatly enhanced ability for long-distance regeneration relative to wild-type mice. These studies suggest that blocking PTPσ signaling in injured axons might enhance recovery after spinal cord injury. Hence, the truncated forms of fly and vertebrate Caskins that interfere with Lar signaling are particularly interesting. The identification of such dominant-negative reagents allowing the blockade of Lar signal transduction in vivo may have clinical implications in neuronal regeneration.
Footnotes
This work was supported by National Institutes of Health Grants NS055245 (H.T.B.) and DA020812 (A.D.). We are grateful to Jim Skeath in whose laboratory this project was initiated. We thank Scott Portman and Jonathan Paek for excellent technical assistance. Greg Bashaw and Susan Lindquist generously supplied fly strains and reagents. We thank members of the Broihier Laboratory, and Kate O'Connor-Giles, Jessica Treisman, and Kirsten Hofmeyer for critical comments on this manuscript.
References
- Ackley BD, Harrington RJ, Hudson ML, Williams L, Kenyon CJ, Chisholm AD, Jin Y. The two isoforms of the Caenorhabditis elegans leukocyte-common antigen related receptor tyrosine phosphatase PTP-3 function independently in axon guidance and synapse formation. J Neurosci. 2005;25:7517–7528. doi: 10.1523/JNEUROSCI.2010-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Gitler AD, Lindquist S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24:913–919. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aricescu AR, McKinnell IW, Halfter W, Stoker AW. Heparan sulfate proteoglycans are ligands for receptor protein tyrosine phosphatase sigma. Mol Cell Biol. 2002;22:1881–1892. doi: 10.1128/MCB.22.6.1881-1892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balázs A, Csizmok V, Buday L, Rakács M, Kiss R, Bokor M, Udupa R, Tompa K, Tompa P. High levels of structural disorder in scaffold proteins as exemplified by a novel neuronal protein, CASK-interactive protein1. FEBS J. 2009;276:3744–3756. doi: 10.1111/j.1742-4658.2009.07090.x. [DOI] [PubMed] [Google Scholar]
- Bateman J, Shu H, Van Vactor D. The guanine nucleotide exchange factor Trio mediates axonal development in the Drosophila embryo. Neuron. 2000;26:93–106. doi: 10.1016/s0896-6273(00)81141-1. [DOI] [PubMed] [Google Scholar]
- Broihier HT, Skeath JB. Drosophila homeodomain protein dHb9 directs neuronal fate via crossrepressive and cell-nonautonomous mechanisms. Neuron. 2002;35:39–50. doi: 10.1016/s0896-6273(02)00743-2. [DOI] [PubMed] [Google Scholar]
- Buday L, Wunderlich L, Tamas P. The Nck family of adapter proteins: regulators of actin cytoskeleton. Cell Signal. 2002;14:723–731. doi: 10.1016/s0898-6568(02)00027-x. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, Lee CH, Herman T, Lee RC, Yang AY, Ovasapyan S, Zipursky SL. Drosophila LAR regulates R1-R6 and R7 target specificity in the visual system. Neuron. 2001;32:237–248. doi: 10.1016/s0896-6273(01)00474-3. [DOI] [PubMed] [Google Scholar]
- Desai CJ, Gindhart JG, Jr, Goldstein LS, Zinn K. Receptor tyrosine phosphatases are required for motor axon guidance in the Drosophila embryo. Cell. 1996;84:599–609. doi: 10.1016/s0092-8674(00)81035-1. [DOI] [PubMed] [Google Scholar]
- Desai CJ, Krueger NX, Saito H, Zinn K. Competition and cooperation among receptor tyrosine phosphatases control motoneuron growth cone guidance in Drosophila. Development. 1997;124:1941–1952. doi: 10.1242/dev.124.10.1941. [DOI] [PubMed] [Google Scholar]
- Desai CJ, Garrity PA, Keshishian H, Zipursky SL, Zinn K. The Drosophila SH2-SH3 adapter protein Dock is expressed in embryonic axons and facilitates synapse formation by the RP3 motoneuron. Development. 1999;126:1527–1535. doi: 10.1242/dev.126.7.1527. [DOI] [PubMed] [Google Scholar]
- Ensslen-Craig SE, Brady-Kalnay SM. Receptor protein tyrosine phosphatases regulate neural development and axon guidance. Dev Biol. 2004;275:12–22. doi: 10.1016/j.ydbio.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Fan X, Labrador JP, Hing H, Bashaw GJ. Slit stimulation recruits Dock and Pak to the roundabout receptor and increases Rac activity to regulate axon repulsion at the CNS midline. Neuron. 2003;40:113–127. doi: 10.1016/s0896-6273(03)00591-9. [DOI] [PubMed] [Google Scholar]
- Fawcett JP, Georgiou J, Ruston J, Bladt F, Sherman A, Warner N, Saab BJ, Scott R, Roder JC, Pawson T. Nck adaptor proteins control the organization of neuronal circuits important for walking. Proc Natl Acad Sci U S A. 2007;104:20973–20978. doi: 10.1073/pnas.0710316105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AN, Zinn K. The heparan sulfate proteoglycan syndecan is an in vivo ligand for the Drosophila LAR receptor tyrosine phosphatase. Curr Biol. 2005;15:1701–1711. doi: 10.1016/j.cub.2005.08.035. [DOI] [PubMed] [Google Scholar]
- Fry EJ, Chagnon MJ, López-Vales R, Tremblay ML, David S. Corticospinal tract regeneration after spinal cord injury in receptor protein tyrosine phosphatase sigma deficient mice. Glia. 2010;58:423–433. doi: 10.1002/glia.20934. [DOI] [PubMed] [Google Scholar]
- Garrity PA, Rao Y, Salecker I, McGlade J, Pawson T, Zipursky SL. Drosophila photoreceptor axon guidance and targeting requires the dreadlocks SH2/SH3 adapter protein. Cell. 1996;85:639–650. doi: 10.1016/s0092-8674(00)81231-3. [DOI] [PubMed] [Google Scholar]
- Hing H, Xiao J, Harden N, Lim L, Zipursky SL. Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell. 1999;97:853–863. doi: 10.1016/s0092-8674(00)80798-9. [DOI] [PubMed] [Google Scholar]
- Hofmeyer K, Treisman JE. The receptor protein tyrosine phosphatase LAR promotes R7 photoreceptor axon targeting by a phosphatase-independent signaling mechanism. Proc Natl Acad Sci U S A. 2009;106:19399–19404. doi: 10.1073/pnas.0903961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeyer K, Maurel-Zaffran C, Sink H, Treisman JE. Liprin-α has LAR-independent functions in R7 photoreceptor axon targeting. Proc Natl Acad Sci U S A. 2006;103:11595–11600. doi: 10.1073/pnas.0604766103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KG, Van Vactor D. Receptor protein tyrosine phosphatases in nervous system development. Physiol Rev. 2003;83:1–24. doi: 10.1152/physrev.00016.2002. [DOI] [PubMed] [Google Scholar]
- Johnson KG, Tenney AP, Ghose A, Duckworth AM, Higashi ME, Parfitt K, Marcu O, Heslip TR, Marsh JL, Schwarz TL, Flanagan JG, Van Vactor D. The HSPGs Syndecan and Dallylike bind the receptor phosphatase LAR and exert distinct effects on synaptic development. Neuron. 2006;49:517–531. doi: 10.1016/j.neuron.2006.01.026. [DOI] [PubMed] [Google Scholar]
- Kaufmann N, DeProto J, Ranjan R, Wan H, Van Vactor D. Drosophila liprin-alpha and the receptor phosphatase Dlar control synapse morphogenesis. Neuron. 2002;34:27–38. doi: 10.1016/s0896-6273(02)00643-8. [DOI] [PubMed] [Google Scholar]
- Krueger NX, Van Vactor D, Wan HI, Gelbart WM, Goodman CS, Saito H. The transmembrane tyrosine phosphatase DLAR controls motor axon guidance in Drosophila. Cell. 1996;84:611–622. doi: 10.1016/s0092-8674(00)81036-3. [DOI] [PubMed] [Google Scholar]
- Krueger NX, Reddy RS, Johnson K, Bateman J, Kaufmann N, Scalice D, Van Vactor D, Saito H. Functions of the ectodomain and cytoplasmic tyrosine phosphatase domains of receptor protein tyrosine phosphatase Dlar in vivo. Mol Cell Biol. 2003;23:6909–6921. doi: 10.1128/MCB.23.19.6909-6921.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Fan J, Woodley DT. Nck/Dock: an adapter between cell surface receptors and the actin cytoskeleton. Oncogene. 2001;20:6403–6417. doi: 10.1038/sj.onc.1204782. [DOI] [PubMed] [Google Scholar]
- Maurel-Zaffran C, Suzuki T, Gahmon G, Treisman JE, Dickson BJ. Cell-autonomous and -nonautonomous functions of LAR in R7 photoreceptor axon targeting. Neuron. 2001;32:225–235. doi: 10.1016/s0896-6273(01)00471-8. [DOI] [PubMed] [Google Scholar]
- Miller CM, Page-McCaw A, Broihier HT. Matrix metalloproteinases promote motor axon fasciculation in the Drosophila embryo. Development. 2008;135:95–109. doi: 10.1242/dev.011072. [DOI] [PubMed] [Google Scholar]
- Miller KE, DeProto J, Kaufmann N, Patel BN, Duckworth A, Van Vactor D. Direct observation demonstrates that Liprin-alpha is required for trafficking of synaptic vesicles. Curr Biol. 2005;15:684–689. doi: 10.1016/j.cub.2005.02.061. [DOI] [PubMed] [Google Scholar]
- Prakash S, McLendon HM, Dubreuil CI, Ghose A, Hwa J, Dennehy KA, Tomalty KM, Clark KL, Van Vactor D, Clandinin TR. Complex interactions amongst N-cadherin, DLAR, and Liprin-alpha regulate Drosophila photoreceptor axon targeting. Dev Biol. 2009;336:10–19. doi: 10.1016/j.ydbio.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero-Lezcano OM, Marcilla A, Sameshima JH, Robbins KC. Wiskott-Aldrich syndrome protein physically associates with Nck through Src homology 3 domains. Mol Cell Biol. 1995;15:5725–5731. doi: 10.1128/mcb.15.10.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol P, Collier S, Bush M, Shaw P, Doonan JH. Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J Cell Sci. 2007;120:3678–3687. doi: 10.1242/jcs.004119. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Serra-Pagès C, Kedersha NL, Fazikas L, Medley Q, Debant A, Streuli M. The LAR transmembrane protein tyrosine phosphatase and a coiled-coil LAR-interacting protein co-localize at focal adhesions. EMBO J. 1995;14:2827–2838. doi: 10.1002/j.1460-2075.1995.tb07282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Pagès C, Medley QG, Tang M, Hart A, Streuli M. Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J Biol Chem. 1998;273:15611–15620. doi: 10.1074/jbc.273.25.15611. [DOI] [PubMed] [Google Scholar]
- Shen Y, Tenney AP, Busch SA, Horn KP, Cuascut FX, Liu K, He Z, Silver J, Flanagan JG. PTPsigma is a receptor for chondroitin sulfate proteoglycan, an inhibitor of neural regeneration. Science. 2009;326:592–596. doi: 10.1126/science.1178310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Wyszynski M, Huh KH, Valtschanoff JG, Lee JR, Ko J, Streuli M, Weinberg RJ, Sheng M, Kim E. Association of the kinesin motor KIF1A with the multimodular protein liprin-alpha. J Biol Chem. 2003;278:11393–11401. doi: 10.1074/jbc.M211874200. [DOI] [PubMed] [Google Scholar]
- Song J, Wu L, Chen Z, Kohanski RA, Pick L. Axons guided by insulin receptor in Drosophila visual system. Science. 2003;300:502–505. doi: 10.1126/science.1081203. [DOI] [PubMed] [Google Scholar]
- Tabuchi K, Biederer T, Butz S, Sudhof TC. CASK participates in alternative tripartite complexes in which Mint 1 competes for binding with caskin 1, a novel CASK-binding protein. J Neurosci. 2002;22:4264–4273. doi: 10.1523/JNEUROSCI.22-11-04264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vactor D, Sink H, Fambrough D, Tsoo R, Goodman CS. Genes that control neuromuscular specificity in Drosophila. Cell. 1993;73:1137–1153. doi: 10.1016/0092-8674(93)90643-5. [DOI] [PubMed] [Google Scholar]
- Viquez NM, Li CR, Wairkar YP, DiAntonio A. The B′ protein phosphatase 2A regulatory subunit well-rounded regulates synaptic growth and cytoskeletal stability at the Drosophila neuromuscular junction. J Neurosci. 2006;26:9293–9303. doi: 10.1523/JNEUROSCI.1740-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills Z, Bateman J, Korey CA, Comer A, Van Vactor D. The tyrosine kinase Abl and its substrate enabled collaborate with the receptor phosphatase Dlar to control motor axon guidance. Neuron. 1999;22:301–312. doi: 10.1016/s0896-6273(00)81091-0. [DOI] [PubMed] [Google Scholar]