Abstract
The metabolism of indole-3-acetic acid (IAA) was investigated in 14-d-old Arabidopsis plants grown in liquid culture. After ruling out metabolites formed as an effect of nonsterile conditions, high-level feeding, and spontaneous interconversions, a simple metabolic pattern emerged. Oxindole-3-acetic acid (OxIAA), OxIAA conjugated to a hexose moiety via the carboxyl group, and the conjugates indole-3-acetyl aspartic acid (IAAsp) and indole-3-acetyl glutamate (IAGlu) were identified by mass spectrometry as primary products of IAA fed to the plants. Refeeding experiments demonstrated that none of these conjugates could be hydrolyzed back to IAA to any measurable extent at this developmental stage. IAAsp was further oxidized, especially when high levels of IAA were fed into the system, yielding OxIAAsp and OH-IAAsp. This contrasted with the metabolic fate of IAGlu, since that conjugate was not further metabolized. At IAA concentrations below 0.5 μm, most of the supplied IAA was metabolized via the OxIAA pathway, whereas only a minor portion was conjugated. However, increasing the IAA concentrations to 5 μm drastically altered the metabolic pattern, with marked induction of conjugation to IAAsp and IAGlu. This investigation used concentrations for feeding experiments that were near endogenous levels, showing that the metabolic pathways controlling the IAA pool size in Arabidopsis are limited and, therefore, make good targets for mutant screens provided that precautions are taken to avoid inducing artificial metabolism.
The plant hormone IAA is an important signal molecule in the regulation of plant development. Its central role as a growth regulator makes it necessary for the plant to have mechanisms that strictly control its concentration. The hormone is believed to be active primarily as the free acid, and endogenous levels are controlled in vivo by processes such as synthesis, oxidation, and conjugation. IAA has been shown to form conjugates with sugars, amino acids, and small peptides. Conjugates are believed to be involved in IAA transport, in the storage of IAA for subsequent use, in the homeostatic control of the pool of the free hormone, and as a first step in the catabolic pathways (Cohen and Bandurski, 1978; Nowacki and Bandurski, 1980; Tuominen et al., 1994; Östin et al., 1995; Normanly, 1997). It is generally accepted that in some species conjugated IAA is the major source of free IAA during the initial stages of seed germination (Ueda and Bandurski, 1969; Sandberg et al., 1987; Bialek and Cohen, 1989), and there is also evidence that in some plants (but not all; see Bialek et al., 1992), the young seedling is entirely dependent on the release of free IAA from conjugated pools until the plant itself is capable of de novo synthesis (Epstein et al., 1980; Sandberg et al., 1987).
The function of conjugated IAA during vegetative growth is somewhat less clear. It has been shown that conjugated IAA constitutes as much as 90% of the total IAA in the plant during vegetative growth (Normanly, 1997). However, the role of the IAA conjugates at this stage of the plant's life cycle remains unknown. Analysis of endogenous IAA conjugates in vegetative tissues has revealed the presence of a variety of different compounds, including indole-3-acetyl-inositol, indole-3-acetyl-Ala, IAAsp, and IAGlu (Anderson and Sandberg, 1982; Cohen and Baldi, 1983; Chisnell, 1984; Cohen and Ernstsen, 1991; Östin et al., 1992). Studies of vegetative tissues have indicated that IAAsp, one of the major conjugates in many plants, is the first intermediate in an irreversible deactivation pathway (Tsurumi and Wada, 1986; Tuominen et al., 1994; Östin, 1995). Another mechanism that is believed to be involved in the homeostatic control of the IAA pool is catabolism by direct oxidation of IAA to OxIAA, which has been shown to occur in several plant species (Reinecke and Bandurski, 1983; Ernstsen et al., 1987).
One area in the study of IAA metabolism in which our knowledge is increasing is the analysis of the homeostatic controls of IAA levels in plants. It has been possible, for instance, to increase the levels of IAA in transgenic plants expressing iaaM and iaaH genes from Agrobacterium tumefaciens. Analysis of these transgenic plants has indicated that plants have several pathways that can compensate for the increased production of IAA (Klee et al., 1987; Sitbon, 1992). It is expected that future studies using now-available genes will provide further insight into IAA metabolism. For example, a gene in maize encoding IAA-Glc synthetase has been identified, and several genes (including ILR1, which may be involved in hydrolysis of the indole-3-acetyl-Leu conjugate) have been cloned from Arabidopsis (Szerszen et al., 1994; Bartel and Fink, 1995). Furthermore, Chou et al. (1996) identified a gene that hydrolyzes the conjugate IAAsp to free IAA in the bacterium Enterobacter aggloremans.
Because of its small genome size, rapid life cycle, and the ease of obtaining mutants, Arabidopsis is increasingly used as a genetic model system to investigate various aspects of plant growth and development. IAA signal transduction is also being investigated intensively in Arabidopsis in many laboratories (Leyser, 1997). Mutants with altered responses to externally added auxins or IAA conjugates have been identified in Arabidopsis. The identified mutants are either signal transduction mutants such as axr1-4 (Lincoln et al., 1990), or have mutations in genes involved in auxin uptake or transport, such as aux1 and pin1 (Okada et al., 1991; Bennett et al., 1996). A few mutants that are unable to regulate IAA levels or are unable to hydrolyze IAA conjugates, sur1-2 and ilr1, respectively, have also been identified (Bartel and Fink, 1995; Boerjan et al., 1995). To our knowledge, no mutant that is auxotrophic for IAA has been identified to date, which may reflect the redundancy in IAA biosynthetic pathways or the lethality of such mutants.
In spite of the work reported thus far, many aspects of the metabolism of IAA in Arabidopsis require further investigation, because few details of the processes involved in IAA regulation are known. This lack of knowledge puts severe constraints on genetic analysis of IAA metabolism in Arabidopsis. For example, it is essential to have prior knowledge of IAA metabolism to devise novel and relevant screens with which to identify mutants of IAA metabolism. We have sought to address this issue by identifying the metabolic pathways involved in catabolism and conjugation under conditions that minimally perturb physiological processes. In this investigation we studied the conjugation and catabolic pattern of IAA by supplying relatively low levels of labeled IAA and identifying the catabolites and conjugates by MS. Different feeding systems were tested to optimize the application of IAA and to avoid irregularities in metabolism attributable to culturing, feeding conditions, or microbial activity. It is well documented that IAA metabolism is altered according to the amount of exogenous auxin applied; therefore, we placed special emphasis on distinguishing between catabolic routes that occur at near-physiological concentrations and those that occur at the high auxin concentrations commonly used in mutant screens.
MATERIALS AND METHODS
Chemicals and Isotopically Labeled Substrates
[1′-14C]IAA, with a specific activity of 55 mCi/mmol, was purchased from American Radiolabeled Chemicals (St. Louis, MO). [13C6]IAA was from Cambridge Isotope Laboratories (Andover, MA). All other chemicals were from Sigma if not stated otherwise. Labeled IAAsp, IAAGlu, and OxIAA were synthesized from [1′-14C]IAA according to the methods of Tuominen et al. (1995) and Ilić et al. (1997). To obtain metabolic profiles, a solution of 50% Murashige-Skoog medium (Duchefa, Haarlem, The Netherlands) with 5 μm radiolabeled [1′-14C]IAA metabolite was used. For the identification of metabolites, a 10 μm 1:1 mixture of nonlabeled IAA:[13C6]IAA spiked with 1 μm [1′-14C]IAA as a tracer in one-half-strength Murashige-Skoog medium was used. For feeds with low concentrations of IAA, 0.1 and 0.5 m solutions of [5-3H]IAA were used (specific activity of 20 Ci/mmol, American Radiolabeled Chemicals).
Plant Material
Seeds of Arabidopsis ecotype Columbia were surface sterilized in 5% calcium hypochlorite plus 0.02% (v/v) Triton X-100 for 30 min, and then washed three times with sterile water. They were then dried overnight and stored at 4°C until use. Seeds grown in soil were planted without any surface sterilization.
Feeding Conditions
Plantlets were potted and grown under short-day conditions until they had about 20 leaves. They were then picked from the soil, the roots were rinsed with distilled water, and the root system was removed under water approximately 5 mm from the lowest leaf. The shoots were transferred to a tissue-culture dish with 200 μL of medium containing labeled IAA in each well. After the labeling solution was taken up by the transpiration stream, excess 50% Murashige-Skoog medium was added to provide the plant with enough liquid for the rest of the incubation period. Plants were also grown in Petri dishes containing Murashige-Skoog medium, 0.3% agar, and 3% Suc. After 14 d in short-day conditions, the plantlets were carefully rinsed with sterile water, transferred to 24-well plates (Costar 24, Cambridge, MA), and fed with 5 μm radiolabeled [1′-14C]IAA in full-strength Murashige-Skoog medium, pH 5.6, in the dark. Finally, 20 surface-sterilized seeds were transferred to a 250-mL conical flask containing 50 mL of Murashige-Skoog medium, pH 5.6, with 3% Suc. Germination and further growth took place in long-day conditions for 14 d, during which time the medium was replaced once. The plants were then fed with radiolabeled [1′-14C]IAA, [1′-14C]IAAsp, [1′-14C]IAGlu, or [1′-14C]OxIAA (at approximately 5 μm) in the dark. In separate experiments, 0.5 and 0.1 μm radiolabeled [5-3H]IAA were fed to replicate plants in the dark.
To assess the effects of OxIAA, IAA, IAGlu, and IAAsp, Arabidopsis seeds were germinated on Murashige-Skoog medium with 3% Suc or on Murashige-Skoog medium with 3% Suc supplemented with these compounds at a concentration of 50 μm. After 4°C treatment of the seeds in the dark for 2 d, the plates were placed in continuous light. The effect of the compounds on root growth was assessed 10 d after germination.
Extraction and Purification
The plant material was homogenized in liquid nitrogen using a cold mortar and pestle. The homogenized material was then extracted with methanol containing 0.02% (w/v) diethylcarbamatic acid as an antioxidant for 5 h. Before reduction to the aqueous phase in a rotary evaporator, 10 mL of distilled water was added to the filtered extraction mixture. The aqueous phase was then adjusted to pH 2.7, and the sample was passed through a C18 solid-phase extraction column (Varian, Harbor City, CA) and eluted with 80% methanol. The aqueous phase of the samples used for metabolite identification was partitioned three times against ethyl acetate, after adjustment to pH 2.7, and then partitioned three times against water-saturated butanol. Each of the three phases, ethyl acetate, butanol, and aqueous, was dried and redissolved in methanol, diluted with 1% acetic acid, and applied to C18 solid-phase extraction columns as described above. The eluates from the solid-phase extraction were reduced in volume and centrifuged in 1.7-mL microcentrifuge tubes at 20,800g for 5 min before HPLC.
HPLC
Metabolic profiles were obtained, and the metabolites were purified by HPLC before MS analysis. The HPLC system consisted of a controller and pump (type 600, Waters, Millipore), and samples were introduced by an autosampler onto a 3.9- × 150-mm, 5-μm ODS symmetry column (Waters, Millipore). The mobile phase was delivered at a flow rate of 0.8 mL/min with an initial mixture of 5% methanol in water (with 1% acetic acid in both solvents) for 5 min, followed by a 45-min linear gradient to 60% methanol, and finally, a linear gradient to 90% methanol over 5 min. For analysis of 14C-labeled samples, the eluate was passed through a radioactivity monitor (model 9701, Reeve Analytical Ltd., Glasgow, Scotland) with a heterogeneous flow cell packed with cerium-activated lithium glass scintillate. For analysis of 3H samples, the heterogenous flow cell was replaced with a 500-μL flow cell and operated in homogenous mode. The scintillant was Permablend III (Packard Instrument Co., Meriden, CT) used in a 4:1 (v/v) scintillant:eluate ratio. One-minute fractions were collected (fraction collector, ISCO, Lincoln, NE) for further MS analysis. Overall control, data sampling, and data handling were performed by the Millennium data system (Millipore). Aliquots of the extracts were mixed with [14C]OxIAA, [14C]IAAsp, or [14C]IAGlu before injection into the HPLC system to identify peaks comigrating with these potential metabolites.
Hydrolysis Experiments
Aliquots from HPLC-purified metabolites were stirred in 1 m NaOH for 1 h at room temperature (25°C). Under these conditions, ester (but not amide) conjugates of IAA are hydrolyzed (Bandurski and Schulze, 1974). At the end of the incubation period, samples were neutralized with glacial acetic acid and purified on C18 solid-phase extraction columns, then analyzed by HPLC as described above. Metabolites were also treated with 7 m NaOH at 100°C for 3 h, under a stream of water-saturated nitrogen gas, which results in the hydrolysis of amide IAA conjugates (Bandurski and Schulze, 1974). After strong alkaline hydrolysis, samples were cooled on ice for 15 min, adjusted to pH 3.0 with phosphoric acid, and processed in the same way as the samples from weak alkaline hydrolysis.
Derivatization
To improve chromatography and sensitivity in the HPLC-MS analysis (Östin, 1995), free carboxylic acids were methylated in HPLC-purified fractions according to the method of Shlenk and Gellerman (1960). After methylation, the samples were reseparated by HPLC as described above. Radioactive fractions were then analyzed by HPLC-MS. Aliquots of the methylated radioactive fractions were purified a second time by HPLC, and were then dried, dissolved in 25 μL of acetonitrile, and silylated (before further analysis by GC-MS) by adding 25 μL of bis(trimethylsilyl)-trifluoroacetamide with 1% (v/v) trimethylchlorosilane (Pierce) and heating in sealed vials at 70°C for 15 min. Finally, they were reduced to dryness, redissolved in acetonitrile, and analyzed by GC-MS.
Analysis by GC-MS and HPLC-MS
GC-MS was performed using a gas chromatograph (model 5890, Hewlett-Packard) linked to a mass spectrometer (model JMS-SX-102, JEOL). Samples were injected, splitless, at 280°C onto a 15-m × 0.25-mm fused-silica column with low-bleed MS film (Sil-8 CB, ChromPack, Middelburg, The Netherlands), 0.25 μm thick. The carrier gas was helium. The column temperature was programmed to increase 20°C/min from 70°C to 280°C, and was held at final temperature until elution of the sample. Ions were generated with 70 eV at an ionization current of 300 μA. The acceleration voltage was 10 kV. Positive-ion mass spectra were obtained at a rate of 1 s per scan for a mass range of m/z 25 to 800. The capillary HPLC-frit-FAB-MS system used for the identification of the metabolites has been described in detail elsewhere (Östin et al., 1992). For the initial HPLC separation, a 300- × 0.32-mm capillary column packed with 5-μm ODS symmetry packing material was used (LC Packings, Amsterdam, The Netherlands). The ion source temperature was 50°C, and ions were generated with a beam of 5 to 6 kV xenon atoms at an emission current of 20 mA. The acceleration voltage was 10 kV. Positive-ion mass spectra were obtained at a rate of 5 s per scan for a mass range of m/z 50 to 800, and the spectra were background subtracted. All MS data were processed by a data system (MD7000, JEOL).
RESULTS
Incubation Systems
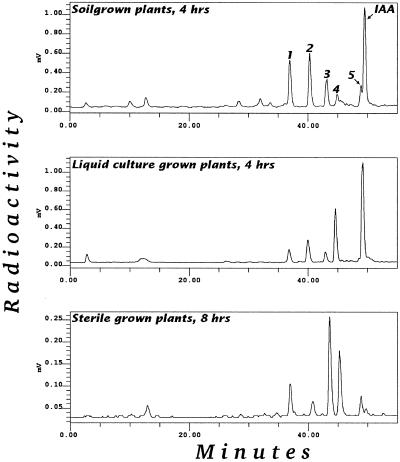
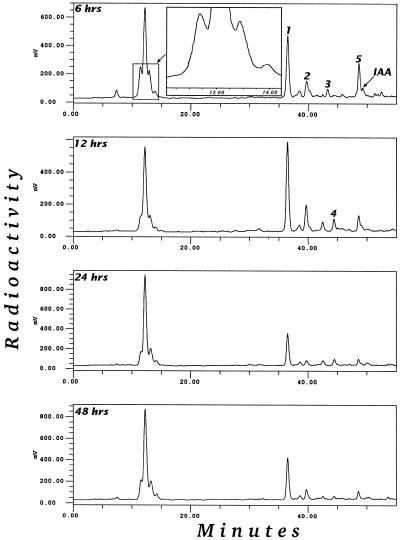
IAA metabolism was investigated in three different systems: soil-grown, tissue-cultured, and liquid-cultured Arabidopsis plants. Figure 1 shows the metabolic profile after feeding with 5 μm [1′-14C]IAA. In all three systems, five major metabolites, numbered 1 through 5, were always found. In the soil-grown plants and in plants fed with high levels of IAA, additional polar metabolites could be detected in some experiments. Some of these additional peaks were attributed to the nonsterile conditions and some to spontaneous conversions. In this investigation we chose to perform the incubation in darkness to minimize light-induced oxidation of IAA. Plants grown in sterile conditions were chosen for the rest of the investigations, partly to avoid complications arising from metabolism by contaminating microorganisms and partly to allow rapid and reproducible uptake of the label. The system with plants grown in liquid culture proved to be efficient for producing large amounts of metabolites, whereas incubation of single, sterile-grown plantlets in wells allowed specific labeling via the Arabidopsis transpiration stream.
Figure 1.
Gradient-elution reversed-phase HPLC-radioactivity chromatogram of partially purified extracts of Arabidopsis plants fed [1-14C]IAA. All incubations were done in darkness.
Identification of IAA Metabolites
To facilitate identification of the unknown metabolites, Arabidopsis plants grown in the dark in liquid culture were fed a 1:1 mixture of nonlabeled IAA and [13C6]IAA spiked with [1′-14C]IAA. The total IAA concentration was 5 μm. Using an equal amount of unlabeled and labeled ([13C6]IAA) gives characteristic fragments with two ions of equal size separated by six mass units. The metabolites denoted 1 through 5, plus four additional polar metabolites denoted 6 through 9, were purified as described in Methods. Fractions were collected after HPLC purification and processed for MS analysis as described below. The use of ion-suppression reversed-phase separation gives the possibility of calculating retention times relative to IAA, thus giving an indirect estimate of relative polarity at the pH of the analysis.
Metabolite 1
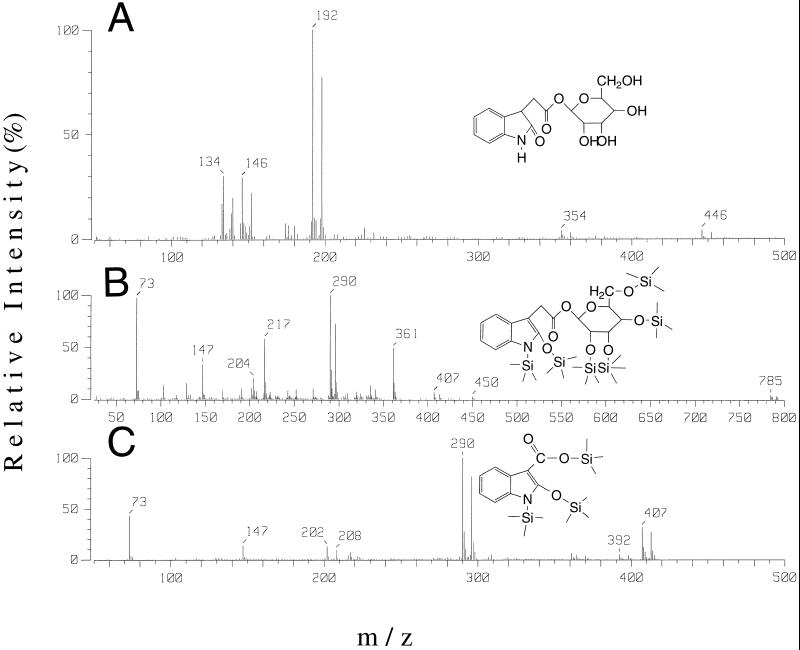
Metabolite 1 had a retention time relative to IAA of 0.74. When subjected to either mild or strong hydrolysis, only decomposition occurred, with no release of IAA, indicating that the compound was not an IAA conjugate. No co-elution with known standards occurred when aliquots of the extract were spiked with labeled OxIAA, IAGlu, or IAAsp. Methylation with diazomethane altered some of this compound to a species with an identical retention time to that of metabolite 5. The underivatized metabolite was subjected to capillary HPLC-frit-FAB-MS analysis, producing the spectrum shown in Figure 2A, and GC/MS analysis of the silylated compound gave a spectrum as in Figure 2B. The HPLC-MS spectrum, corresponding to that of OxIAA linked to a hexose sugar, consists of a protonated molecular ion [MH]+ at m/z 354/360. Loss of the sugar moiety gives OxIAA, as indicated by the fragment at m/z 192/198.
Figure 2.
Frit-FAB-MS spectra of metabolite 1 (A); and GC-MS spectrum of silylated metabolite 1 (B) and of the silylated hydrolysis product of metabolite 1 (C).
The OxIAA moiety is then further fragmented to m/z 174/180 and m/z 146/152. Silylation of the catabolite before GC-MS caused significant hydrolysis to free OxIAA, identified by analysis of its trimethylsilyl derivative (Fig. 2C). The silylated derivative of metabolite 1 yielded at least two isomers, with spectra showing a molecular ion at m/z 785/791, corresponding to the incorporation of six trimethylsilyl groups into an OxIAA-hexose conjugate. The m/z 450, 361, 217, 204, 191, 147, and 73 fragmentation pattern has been demonstrated previously using 1-O-(indole-3-acetyl)-β-d-glucopyranose (Ehman, 1974) to represent a silylated sugar moiety. Furthermore, the ions m/z 290 and 407 represent a shift of +88 mass units, corresponding to a silylated oxygen group in the number 2 position of an oxindole. These peaks also show the diagnostic double labeling from [13C6]IAA (plus 6 mass units), with fragments at m/z 296 and 413. We thus conclude that metabolite 1 is OxIAA linked to a hexose sugar via the carboxyl group. We did not proceed with a detailed characterization of the sugar because this was not the main focus of the study.
Metabolite 2
This metabolite had a retention time of 0.81 relative to IAA. Both mild and strong hydrolysis caused breakdown of the metabolite without any release of IAA. Spiking experiments demonstrated co-elution with [14C]OxIAA (Fig. 3). Methylation with diazomethane changed the retention time to the same as that of metabolite 5. Analysis with capillary HPLC-frit-FAB-MS of the methylated derivative revealed a spectrum corresponding to an OxIAA methyl ester (Table I), and GC-MS of the methylated and silylated derivative revealed a spectrum corresponding to that of a di-(trimethylsilyl)-OxIAA methyl ester (Table II). We concluded, therefore, that metabolite 2 is OxIAA.
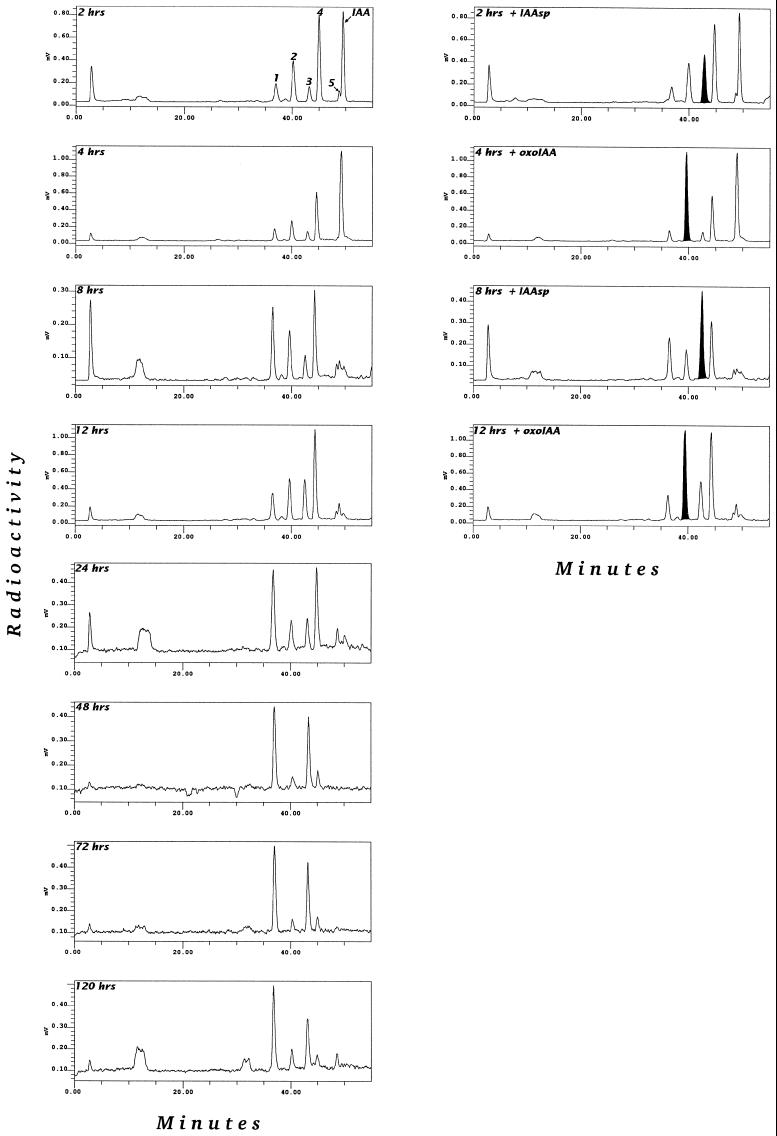
Figure 3.
Time-course study of a [1-14C]IAA feed to Arabidopsis plants grown in liquid culture for 2 to 120 h and of partially purified extracts spiked with [14C]OxIAA and [14C]IAAsp.
Table I.
Tabulated FAB spectra of methylated IAA metabolites
| Compound | [MH]+ | Other Ions |
|---|---|---|
| Metabolite 1 | 354/360 (5) | 192/198 (Bp), 146/152 (33), 134/140 (31) |
| Metabolite 2 | 206/212 (Bp) | 174/180 (10), 146/152 (32) |
| Metabolite 3 | 319/325 (55) | 162 (25), 130/136 (Bp) |
| Metabolite 4 | 333/339 (81) | 205 (35), 176 (23), 130/136 (Bp), 116 (9) |
| Metabolite 5 | 206/212 (Bp) | 174/180 (5), 146/152 (30) |
| Metabolite 6 | 335/341 (Bp) | 174/180 (14), 162 (30), 146/152 (37), 130/136 (25), 130/132 (21), 102 (21) |
| Metabolite 7 | 335/341 (Bp) | 174/180 (14), 162 (24), 146/152 (36), 130/136 (17), 130/132 (19), 102 (15) |
| Metabolite 8 | 335/341 (Bp) | 204 (23), 174/180 (14), 162 (46), 146/152 (42), 102 (43) |
| Metabolite 9 | 335/341 (Bp) | 204 (26), 174/180 (8), 162 (44), 146/152 (38), 102 (36) |
The intensity of nonlabeled peaks are shown in parentheses, the intensity of the 13C6 label from the IAA feed is of approximately equal intensity as that of nonlabeled ions. Bp, Base peak with relative intensity 100%. All spectra also contain an adduct ion [M + glycerol + H]+ with an intensity of 2% to 10%; these are not tabulated.
Table II.
Tabulated EI spectra of methylated/silylated IAA metabolites
| Compound | [MH]+ | Other Ions |
|---|---|---|
| Metabolite 1 | 785/791 (5) | 450 (4), 407/413 (8), 361 (50), 290/296 (Bp), 217 (61), 204 (20), 191 (10), 147 (36), 73 (98) |
| Metabolite 2 | 349/355 (47) | 290/296 (Bp), 244/250 (14), 216/222 (22), 202/208 (34), 89 (8), 73 (99) |
| Metabolite 3 | 390/396 (20) | 202/208 (Bp), 73 (30) |
| Metabolite 4 | 404/410 (20) | 202/208 (Bp), 73 (27) |
The intensity of the nonlabeled peaks are shown in parentheses, the intensity of the 13C6 label from the IAA feed is of approximately equal intensity as that of nonlabeled ions. Bp, Base peak with a relative intensity of 100%.
Metabolite 3
Metabolite 3 had a retention time of 0.87 relative to IAA. Hydrolysis experiments resulted in the release of IAA with strong but not with mild hydrolysis, indicating that the compound is an amide conjugate of IAA. Spiking experiments resulted in co-elution with [14C]IAAsp (Fig. 3). Methylation with diazomethane changed the retention time of metabolite 3 to a relative retention of 1.06, corresponding to the methylation of the two carboxylic acids in the Asp moiety (as demonstrated by standard IAAsp having the same shift in retention time after methylation). The structure of IAAsp was verified by comparing the HPLC-MS spectrum of its methylated derivative and the GC-MS spectrum of its methylated/silylated derivative with appropriate standards, as shown in Tables I and II.
Metabolite 4
Metabolite 4 had a retention time of 0.90 relative to IAA. Hydrolysis experiments indicated that the compound was an amide conjugate of IAA. Spiking experiments resulted in co-elution with [14C]IAGlu (data not shown). Methylation with diazomethane changed the retention time of metabolite 4 to a relative retention of 1.23, corresponding to the methylation of the two carboxylic acids in the Glu moiety. The structure of IAGlu was verified by analyzing the HPLC-MS spectrum of its methylated derivative and the GC-MS spectrum of its methylated/silylated derivative (Tables I and II).
Metabolite 5
In samples from all feeding systems, a metabolite with more or less the same retention time as IAA (0.98 relative to IAA) on ion-suppression chromatography was found. This compound was also formed from metabolite 1 during sample preparation and methylation with diazomethane. Methylated metabolite 2 has the same retention time in HPLC. After derivatization and subsequent GC-MS analysis, this peak was identified as a di-(trimethylsilyl)-OxIAA methyl ester. We believe that this metabolite is a result of spontaneous methylation of metabolite 1, a phenomenon we have earlier observed for IAA-1-O-β-d-Glc during storage in acid methanol. Therefore, we excluded this metabolite from the following discussion of physiologically relevant metabolites.
Metabolites 6 to 9
In soil-grown plants and in sterile-grown plants given high dosages of IAA, additional oxidative IAAsp metabolites were detected as their methylated derivatives by HPLC-MS. Metabolites 6 to 9 have molecular ions that correspond to an extra oxygen incorporated into IAAsp ([M + H]+ m/z 335/341 with [13C6] label). These metabolites had a relative retention to IAA of 0.21, 0.31, 0.41, and 0.47, respectively, on a 3.9- × 150-mm, 5-μm ODS column (Nova Pac, Waters, Millipore), using the same solvent system as described above. Metabolites 8 and 9 were major constituents that showed the same fragmentation pattern as the standard OxIAAsp methyl ester and the same retention time as the two spontaneously formed stereoisomers (Table I). Compared with methylated OxIAAsp, metabolites 6 and 7 lacked the fragment m/z 204 (which corresponds to loss of the entire methylated side chain) and have in addition two pairs of ions, m/z 130/136 and 132/138, which probably result from the loss of oxygen from the m/z 146/152 quinolonium fragment. These spectra are characteristic of hydroxylated forms of IAAsp (Table I). Furthermore, some other minor metabolites with the characteristic 12C6/13C6 label were detected from plants given high dosages of IAA, but these were not fully characterized because of their questionable physiological relevance. OxIAAsp is most likely metabolized to a metabolite with a molecular ion m/z 592/598. Two compounds with the expected 12C6/13C6 ratio in their molecular ion (m/z 365/371 and 379/385) were detected (data not shown). An unknown ester-bound conjugate of IAA with a molecular ion of m/z 372/378 was also detected (data not shown). Methylated OxIAAsp can be partially oxidized to a hydroxylated OxIAAsp during isolation or when stored in solution for longer times (data not shown).
Time-Course Studies
In a pulse-chase experiment 5 μm [1-14C]IAA was fed to liquid-culture-grown plants that were harvested at various times from 2 to 120 h after treatment. The resulting metabolic profiles are illustrated in Figure 3. OxIAA is formed early and is subsequently conjugated to form an OxIAA hexose. Amide conjugation is seen initially by measuring the formation of IAGlu and, at a later stage, IAAsp. IAGlu accumulated over time and was not further metabolized, whereas IAAsp accumulated as long as there was free IAA available. IAAsp was further metabolized to the polar catabolites described above. It is also worth noting that accumulation of more stable forms occurs mainly through the formation of OxIAA hexose in older plants and IAAsp in younger plants (data not shown).
Metabolism of OxIAA, IAAsp, and IAGlu
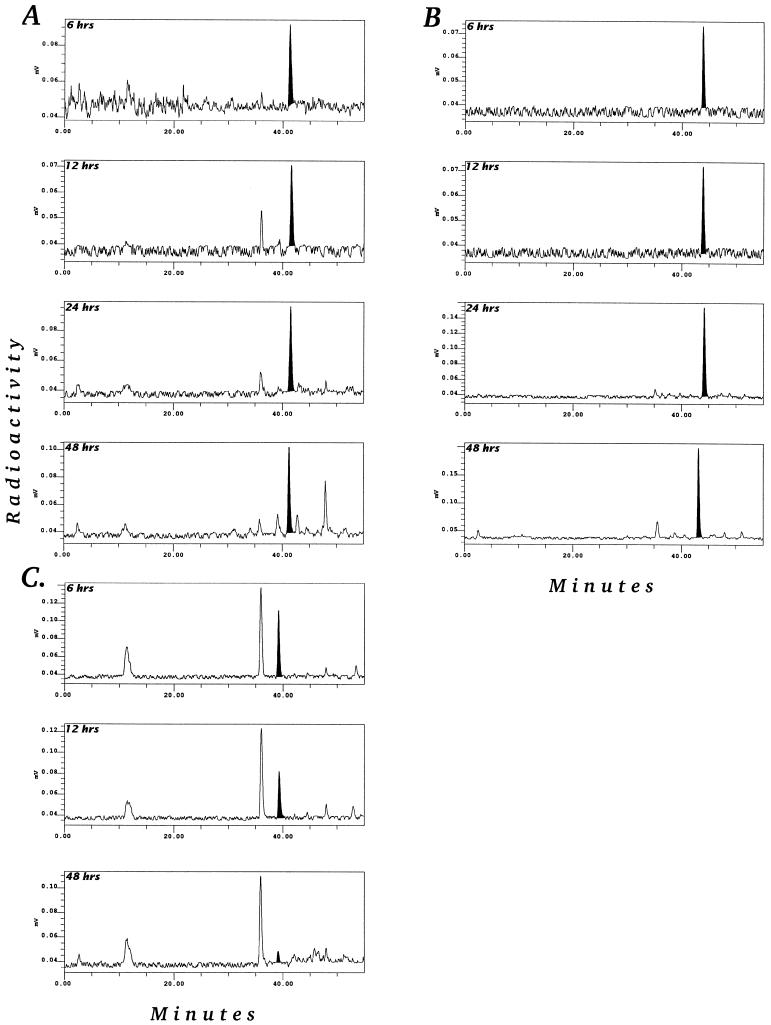
An important question is whether the major metabolites formed after feeding plants with labeled IAA are strictly catabolites or if they can be reused (i.e. converted back to IAA). This was investigated in two separate experiments. [14C]OxIAA, [14C]IAAsp, and [14C]IAGlu were separately fed to liquid-culture-grown Arabidopsis plants. Feeding with IAAsp indicated that this conjugate was stable in Arabidopsis tissues, with almost no metabolism occurring during the first 24 h. However, after 48 h there was significant metabolism, with both a minor level of hydrolysis to IAA and formation of additional catabolites of IAAsp (Fig. 4A). The results of feeding with labeled IAGlu are presented in Figure 4B. IAGlu was stable over time, with only minor levels of metabolism occurring. This is consistent with the time-course studies of IAA metabolism shown in Figure 3, which show that IAGlu accumulated over time and did not appear to be metabolized further. Finally, Figure 4C shows that OxIAA was, as expected, not converted back to IAA, and that the major product of supplied OxIAA was OxIAA hexose (peak 1). There was also a polar peak that co-eluted with DiOxIAAsp (metabolite 10) and was probably formed via OxIAAsp.
Figure 4.
Time-course study of [1-14C]IAAsp (A), [1-14C]IAGlu (B), and [1-14C]OxIAA (C) feeds to Arabidopsis plants grown in liquid culture for 6, 12, and 48 h.
In a second experiment the sensitivity of Arabidopsis seedlings to the major catabolites/conjugates OxIAA, IAAsp, and IAGlu was tested using a protocol described earlier for IAAsp (Bartel and Fink, 1995). OxIAA, IAAsp, and IAGlu had no effect at all on the phenotype after the feedings, even at very high concentrations (data not shown). This, together with the data from the feeding experiments, clearly shows that these three catabolites/conjugates cannot be reconverted back to IAA at measurable rates in Arabidopsis at this developmental stage.
Physiological Relevance of the Metabolic Studies
Many studies involving metabolic analysis of IAA, especially studies using Arabidopsis, have used relatively high concentrations of IAA during the feeding of plants. To determine the consequence of feeding level, we investigated the effect of the concentration of IAA fed to the Arabidopsis on the metabolites detected. Metabolic profiles were obtained from plants fed with 5, 0.5, and 0.1 μm IAA. Figure 5 shows a time-course study with 0.5 μm IAA fed to the plants. This experiment demonstrates that different metabolic profiles were obtained by feeding with 0.5 and 5 μm IAA. Low-level feeding with IAA resulted in the detection of mostly OxIAA and OxIAA hexose, with a minor level of conjugation to IAAsp and IAGlu. A set of polar catabolites was also detected, which most likely represented polar products of OxIAA and IAAsp. The 3H profiles presented in Figure 5 were analyzed using an on-line radioactivity monitor operated in the homogenous mode. The signal-to-noise ratio is significantly improved using this detection method compared with the use of a heterogeneous detection system. This is demonstrated by the resolution of the clustered peaks at approximately 13 min in Figure 5 (homogenous detection) compared with the same unresolved peaks in Figure 3 (heterogeneous detection). Another interesting observation was that the [5-3H]IAA low-level feed indicated a more rapid turnover of IAA compared with the [1′-14C]IAA feed. When 0.1 μm IAA was fed to the plants, the same general metabolism as with a 0.5 μm concentration (Fig. 5) was found (data not shown).
Figure 5.
Gradient-elution reversed-phase HPLC-radioactivity chromatogram of aliquots of partially purified extracts of sterile-grown plants fed [5-3H]IAA in liquid culture in darkness for 6 to 48 h.
DISCUSSION
We analyzed the metabolism of IAA by Arabidopsis using three different experimental systems. Irrespective of the feeding system, four metabolites of IAA were consistently detected. The metabolites were identified as the conjugates IAAsp and IAGlu, the oxidative catabolite OxIAA, and an OxIAA-hexose conjugate. Some additional metabolites, such as oxidative products of IAAsp, were found in profiles from older soil-grown material and from plants that were fed very high levels of auxin. Metabolic studies with synthetically made [14C]OxIAA, [14C]IAGlu, or [14C]IAAsp indicated that free IAA was not released from these conjugates in measurable amounts by Arabidopsis at this developmental stage. Irreversible oxidation occurred mainly via OxIAA, but the possibility that IAAsp can also function as an initial catabolic intermediate is indicated by its further metabolism to OxIAAsp and OH-IAAsp. In the soil-grown plants, relatively more oxidative catabolites were formed, with OxIAA hexose accumulating over time, whereas in the sterile-grown, young plant material, IAAsp and IAGlu were the major end products.
Earlier results based on TLC separation indicated that IAA applied to Arabidopsis was metabolized to compounds with an RF similar to that of IAAsp and/or IAAGlu, IAGlc, and indole-3-acetyl-Leu and/or indole-3-acetyl-Ile (Sztein et al., 1995). Our results differ from the earlier observations made by Sztein et al. (1995) in that we did not find any peaks that comigrated with indole-3-acetyl-Leu, indole-3-acetyl-Ile, or IAGlc. Because IAGlc might be spontaneously converted to IAA methyl ester during methanolic extraction, we also attempted extraction with 80% acetone. This extraction resulted in reduced spontaneous conversion of OxIAA hexose to OxIAA methyl ester, but also in increased spontaneous degradation of OxIAA and reduced recovery of the amide. No IAGlc was detected in either case. Based on our results (see below), we believe that IAGlc is a product that is detectable only when high levels of IAA are fed to plant tissues. The overall function of conjugation to Glc needs to be investigated further. For example, Catala et al. (1992) observed that the only metabolite of IAA in tomato in which formation was not inhibited by cyclohexamide was IAGlc. This, together with our data, indicates that the induction of this pathway is complex.
Our strategy in this study was to use [1′-14C]IAA, first to establish the metabolic profiles, and second, to produce larger amounts of catabolites for identification. The results from [1′-14C]IAA feeding were then complemented by results from [5-3H]IAA feeding, which were used to support the physiological relevance of the identified metabolites. The average values for endogenous concentration of IAA in the Arabidopsis plants used for these studies varied between 10 and 25 ng/g fresh weight, depending on the growth conditions and developmental stage. In a separate investigation, we found by GC-MS microanalysis that the concentration range within individual Arabidopsis plants ranges from 5 to more than 900 ng/g (K. Ljung, R. Bhalerao, and G. Sandberg, unpublished data), the higher values being found in actively growing parts of the plant.
The concentration of IAA conjugates in the Arabidopsis plants was very high, with average values for the amide conjugates reaching micrograms per gram fresh weight levels. We used an average of 5 μm IAA solution, added in 5-fold excess (v/w), in our initial feeding experiments and this would be expected to cause changes in the endogenous IAA pool. However, because the values in different organs ranged from 5 to 900 ng/g, the endogenous pool in the old, nonexpanding leaves was increased a maximum of 175-fold, whereas the pool in the most rapidly expanding leaves could be increased less than 2-fold by such an application. The metabolic profiles were therefore reanalyzed using [5-3H]IAA at a concentration of 0.5 μm. In this case the dilution of the average pool could be expected to increase 5-fold, whereas the dilution of the pool in the nonactively growing parts would be 17.5-fold, and that in actively dividing parts, less than 4%. The concentration used may alter the endogenous pools markedly in some of the plant compartments, but not in others. We decided, therefore, to further reduce the concentration of IAA fed to the plants. A new feed was performed with 0.1 μm [5-3H]IAA. In this case, the dilutions of the IAA pools were 1-fold when calculated at the whole-plant level, 3-fold in the old leaves, and less than 1% in the actively growing parts. These estimates are based on the fact that no active accumulation occurred, so at most a 5-fold increase is theoretically possible.
As shown in Figure 5, OxIAA and OxIAA hexose are the major catabolites when low levels of IAA are fed to the plant. Thus, at IAA concentrations less than 0.5 μm, the majority of the metabolism is via OxIAA, and only a minor portion of IAA is conjugated. However, at 5 μm concentrations of IAA, the metabolic pattern is altered (Figs. 1 and 3), with a marked induction of conjugation to IAAsp and IAGlu. The conjugative pathways to IAAsp and IAGlu may be relevant to normal physiological processes, because the actively growing parts of the Arabidopsis plant do contain high levels of IAA. Thus, the addition of 5 μm exogenous IAA may not cause totally unphysiological conditions in these specific tissues. This aspect of tissue-specific effects has to be evaluated in future experiments in which the metabolism in each tissue is related to the actual endogenous concentration.
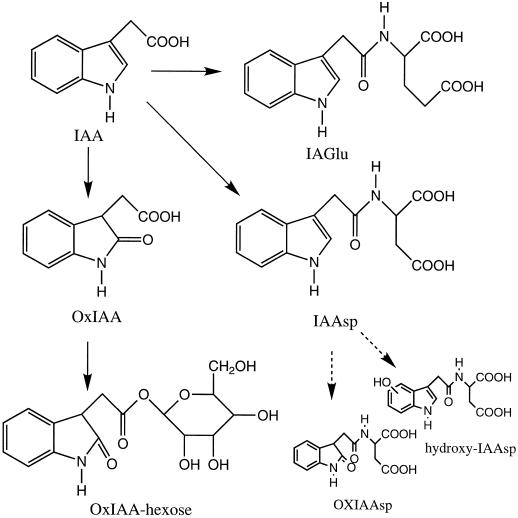
We can now conclude that the metabolism of IAA in Arabidopsis plants at the developmental stage studied follows the pathway presented in Figure 6. These results also demonstrate that nonsterile conditions or feeding high concentrations of IAA will induce alternative metabolic pathways. Additionally, the complexities of such analysis are increased by the inherent instability of the indolic compounds, since they are both light sensitive and easily oxidized. Therefore, data obtained using high IAA levels and nonsterile conditions should be treated with caution, because they are unlikely to reflect any normal physiological process in the plant. Until this report, extremely high auxin dosages have been used in experiments with Arabidopsis aimed at analyzing metabolism of IAA and isolating mutants. One exception is the study of IAA biosynthesis in Arabidopsis by Normanly et al. (1993), in which low levels of IAA precursors were fed. In our investigation, one of the major metabolites identified was OxIAA hexose, which, during isolation, can be spontaneously methylated to form OxIAA methyl ester, a compound that comigrates almost precisely with IAA. Therefore, if identification and analysis are not done carefully, this metabolite could easily be mistaken for free IAA released from a conjugated form upon hydrolysis. Feeds with OxIAA and hydroxylated forms of IAAsp also show that there are a number of polar catabolites of both compounds that can also be formed by spontaneous oxidation and that should not be confused with genuine in planta catabolites. Thus, the results presented here are of consequence not only for determining the metabolites of IAA produced by Arabidopsis, but also for providing important information necessary for future designs of screens aimed at the isolation of mutants in metabolic pathways for IAA.
Figure 6.
Proposed metabolic pathway for IAA in Arabidopsis plants. The solid arrows represent the steps that have been demonstrated by in planta conversions under physiologically relevant conditions.
Footnotes
This work was financed by the Swedish Foundation for Strategic Research and the European Commission DG XII biotechnology program.
Abbrevations: FAB, fast-atom bombardment; IAAsp, indole-3-acetyl Asp; IAGlc, indole-3-acetyl Glc; IAGlu, indole-3-acetyl Glu; OxIAA, oxindole-3-acetic acid.
LITERATURE CITED
- Andersson B, Sandberg G. Identification of endogenous N-(3-indoleacetyl)aspartic acid in Scots pine (Pinus sylvestris L.) by combined gas chromatography-mass spectrometry, using high-performance liquid chromatography for quantification. J Chromatogr. 1982;238:151–156. [Google Scholar]
- Bandurski RS, Schulze A. Concentrations of indole-3-acetic acid and its esters in Avena and Zea. Plant Physiol. 1974;54:257–262. doi: 10.1104/pp.54.3.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Fink GR. ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science. 1995;268:1745–1748. doi: 10.1126/science.7792599. [DOI] [PubMed] [Google Scholar]
- Bennett JM, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schultz B, Feldmann KA. Arabidopsis AUX 1 gene, a permease-like regulator of root gravitropism. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- Bialek K, Cohen JD. Free and conjugated indole-3-acetic acid in developing bean seeds. Plant Physiol. 1989;91:775–779. doi: 10.1104/pp.91.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek K, Michalczuk L, Cohen JD. Auxin biosynthesis during seed germination in Phaseolus vulgaris. Plant Physiol. 1992;100:509–517. doi: 10.1104/pp.100.1.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Cervera M-T, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Onckelen HV, Montagu MV, Dirk I. Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell. 1995;7:1405–1419. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala C, Östin A, Chamarro J, Sandberg G, Crozier A. Metabolism of indole-3-acetic acid by pericarp discs from immature and mature tomato (Lycopersicon esculentum Mill.) Plant Physiol. 1994;100:1457–1463. doi: 10.1104/pp.100.3.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisnell JR. Myo-inositol esters of indole-3-acetic acid are endogenous components of Zea mays L. shoot tissue. Plant Physiol. 1984;74:278–283. doi: 10.1104/pp.74.2.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou JC, Kuleck GA, Cohen JD, Mulbry WW. Partial purification and characterization of an inducible indole-3-acetyl-l-aspartic acid hydrolase from Enterobacter aggloremans. Plant Physiol. 1996;112:1281–1287. doi: 10.1104/pp.112.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JD, Baldi B. Studies of endogenous indole-3-acetyl-L-aspartate during germination of soybeans. Proceedings of the Plant Growth Regulator Society of America. 1983;10:117–122. [Google Scholar]
- Cohen JD, Bandurski RS. The bound auxin: protection of indole-3-acetic acid from peroxidase-catalyzed oxidation. Planta. 1978;139:203–208. doi: 10.1007/BF00388631. [DOI] [PubMed] [Google Scholar]
- Cohen JD, Ernsten A. Indole-3-acetic acid and indole-acetylaspartic acid isolated from seeds of Heraculum laciniatum Horn. Plant Growth Regul. 1991;10:95–101. [Google Scholar]
- Ehman A. Identification of 2-O-(indole-3-acetyl)-D-glucopyranose, 4-O-(indole-3-acetyl)-D-glucopyranose and 6-O-(indole-3-acetyl)-D-glucopyranose from kernels of Zea mays. Carbohydr Res. 1974;34:99–114. doi: 10.1016/s0008-6215(00)80374-2. [DOI] [PubMed] [Google Scholar]
- Epstein E, Cohen JD, Bandurski RS. Concentration and metabolic turnover of indoles in germinating kernels of Zea mays L. Plant Physiol. 1980;65:415–421. doi: 10.1104/pp.65.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernstsen A, Sandberg G, Lundström K. Identification of oxindole-3-acetic acid, and metabolic conversion of indole-3-acetic acid to oxindole-3-acetic acid in seeds of Pinus sylvestris. Planta. 1987;172:47–52. doi: 10.1007/BF00403027. [DOI] [PubMed] [Google Scholar]
- Ilić N, Magnus V, Östin A, Sandberg G. Stable-isotope labeled metabolites of the phytohormone, indole-3-acetic acid. J Labelled Compd Radiopharm. 1997;39:433–440. [Google Scholar]
- Klee HJ, Horsch RB, Hinchee MA, Hein MB, Hoffman NL. The effects of overproduction of two Agrobacterium tumefaciens auxin biosynthetic genes in transgenic petunia plants. Genes Dev. 1987;1:86–96. [Google Scholar]
- Leyser O. Auxin: a lesson from a mutant weed. Physiol Plant. 1997;100:407–414. [Google Scholar]
- Lincoln C, Britton JH, Estelle M. Growth and development of axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J. Auxin metabolism. Physiol Plant. 1997;100:431–442. [Google Scholar]
- Normanly J, Cohen JD, Fink GR. Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc Natl Acad Sci USA. 1993;90:10355–10359. doi: 10.1073/pnas.90.21.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki J, Bandurski RS. Myo-inositol esters of indole-3-acetic acid as seed auxin precursors of Zea mays L. Plant Physiol. 1980;65:422–427. doi: 10.1104/pp.65.3.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki M, Bell C, Shimura Y. Requirement of the polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell. 1991;3:677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östin A (1995) Metabolism of indole-3-acetic acid in plants with emphasis on non-decarboxylative catabolism. Dissertation. Swedish University of Agricultural Sciences, Umeå
- Östin A, Catala C, Chamarro J, Sandberg G. Identification of glucopyranosyl-β-1,4-glucopyranosyl-β-1-N-oxindole-3-acetyl-N-aspartic acid, a new IAA-catabolite with liquid chromatography tandem mass spectrometry. J Mass Spectrom. 1995;30:1007–1017. [Google Scholar]
- Östin A, Moritz T, Sandberg G. Liquid chromatography-mass spectrometry of conjugates and oxidative metabolites of indole-3-acetic acid. Biol Mass Spectrom. 1992;21:292–298. [Google Scholar]
- Reinecke DM, Bandurski RS. Oxindole-3-acetic acid, an indole-3-acetic acid catabolite in Zea mays. Plant Physiol. 1983;86:868–872. doi: 10.1104/pp.71.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg G, Ernstsen A, Hamnede M. Dynamics of indole-3-acetic acid and indole-3-ethanol during development and germination of Pinus sylvestris seeds. Physiol Plant. 1987;71:411–418. [Google Scholar]
- Shlenk H, Gellerman JL. Esterification of fatty acids with diazomethane on a small scale. Anal Chem. 1960;32:1412–1414. [Google Scholar]
- Sitbon F (1992) Transgenic plants overproducing IAA, a model system to study regulation of IAA metabolism. Dissertation. Swedish University of Agricultural Sciences, Umeå
- Szerszen JB, Szczyglowski K, Bandurski RS. iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science. 1994;265:1699–1701. doi: 10.1126/science.8085154. [DOI] [PubMed] [Google Scholar]
- Sztein AE, Cohen JD, Slovin JP, Cooke TC. Auxin metabolism in representative land plants. Am J Bot. 1995;82:1514–1521. [Google Scholar]
- Tsurumi S, Wada S. Dioxindole-3-acetic acid conjugates formation from indole-3-acetylaspartic acid in Vicia seedlings. Plant Cell Physiol. 1986;27:1513–1522. [Google Scholar]
- Tuominen H, Östin A, Sandberg G, Sundberg B. A novel metabolic pathway for the indole-3-acetic acid in apical shoots of Populus tremula (L.) × Populus tremuloides (Michx.) Plant Physiol. 1994;106:1511–1520. doi: 10.1104/pp.106.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Bandurski RS. A quantitative estimation of alkali-labile indole-3-acetic acid compounds in dormant and germinating maize kernels. Plant Physiol. 1969;44:1175–1181. doi: 10.1104/pp.44.8.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]