Abstract
Mitochondria from diverse organisms are capable of transporting large amounts of Ca2+ via a ruthenium-red-sensitive, membrane-potential-dependent mechanism called the uniporter1–4. Although the uniporter’s biophysical properties have been studied extensively, its molecular composition remains elusive. We recently used comparative proteomics to identify MICU1 (also known as CBARA1), an EF-hand-containing protein that serves as a putative regulator of the uniporter5. Here, we use whole-genome phylogenetic profiling, genome-wide RNA co-expression analysis and organelle-wide protein coexpression analysis to predict proteins functionally related to MICU1. All three methods converge on a novel predicted transmembrane protein, CCDC109A, that we now call ‘mitochondrial calcium uniporter’ (MCU). MCU forms oligomers in the mitochondrial inner membrane, physically interacts with MICU1, and resides within a large molecular weight complex. Silencing MCU in cultured cells or in vivo in mouse liver severely abrogates mitochondrial Ca2+ uptake, whereas mitochondrial respiration and membrane potential remain fully intact. MCU has two predicted transmembrane helices, which are separated by a highly conserved linker facing the intermembrane space. Acidic residues in this linker are required for its full activity. However, an S259A point mutation retains function but confers resistance to Ru360, the most potent inhibitor of the uniporter. Our genomic, physiological, biochemical and pharmacological data firmly establish MCU as an essential component of the mitochondrial Ca2+ uniporter.
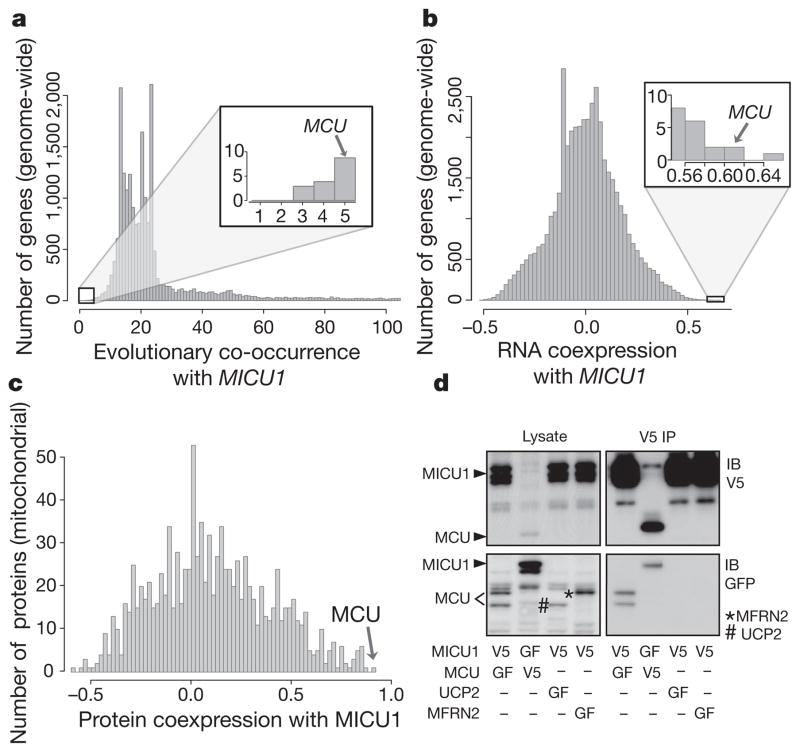
To predict proteins that are functionally related to MICU1 (ref. 5) and essential for mitochondrial calcium (Ca2+) uptake, we performed three systematic computational analyses. First, we ranked all ~20,000 mammalian genes on the basis of the similarity of their phylogenetic profile to MICU1, where the phylogenetic profile of a gene is defined as the binary vector of presence or absence of its homologues across 500 evolutionarily diverse organisms6,7. Second, we scored all ~20,000 mammalian genes for their expression similarity to MICU1 across 81 mouse cell types and tissues using a genome-wide RNA expression atlas8. Third, we scored protein expression similarity between MICU1 and all ~1,100 mitochondrial proteins, based on their pattern of peptide abundance across 14 different mouse tissues6. All three computational methods (Fig. 1a–c) spotlight an unstudied protein (previously called CCDC109A, accession number NM_138357.1) that we now call ‘mitochondrial calcium uniporter’ (MCU). MCU, which has two predicted transmembrane domains, was first discovered in our proteomic analysis as a mitochondrial protein detected in 12 different mouse tissues6. MCU, which bears no sequence similarity to MICU1, is tied as the eighth closest phylogenetic neighbour genome-wide (Fig. 1a), being either co-present or co-absent with MICU1 in 495 of 500 organisms evaluated (Hamming distance = 5). MCU is also the second highest scoring gene in the genome-wide mRNA co-expression analysis (Fig. 1b) and is the top scoring protein amongst all ~1,100 mitochondrial proteins for protein coexpression with MICU1 across 14 mouse tissues (Fig. 1c).
Figure 1. Integrative genomics predicts MCU to be functionally related to MICU1.
a–c, Phylogenetic profile neighbours (a), RNA co-expression neighbours (b) and protein co-expression neighbours (c) of MICU1. Hamming distances between phylogenetic profiles were computed genome-wide for all 20,000 mammalian genes across 500 fully sequenced organisms. Genes co-expressed with MICU1 were computed genome-wide by Pearson correlation using a mouse atlas of 81 tissues. Protein expression correlation with MICU1 was analysed for all mitochondrial proteins across 14 mouse tissues. d, Coimmunoprecipitation of MICU1 and MCU. HEK-293 cells stably expressing MICU1–V5 or MCU–V5 were transfected with MICU1–GFP, MCU–GFP, MFRN2–GFP or UCP2–GFP. Cell lysates were incubated with anti-V5 antibody, immunoprecipitates were resolved on SDS–PAGE, and input lysates and immunoprecipitates were blotted with anti-V5 (top) or anti-GFP (bottom) antibodies. Data are representative of three independent experiments. IP, immunoprecipitation; GF, green fluorescent protein; IB, immunoblot.
Our prediction of a functional relationship between MICU1 and MCU was further corroborated by evidence of a physical interaction. By transfecting green-fluorescent-protein-tagged MCU (MCU–GFP) into cells stably expressing V5-tagged MICU1 (MICU1–V5) and vice versa, we were able to recover both GFP-tagged proteins following immunoprecipitation with an anti-V5 antibody (Fig. 1d). The interaction was specific as MICU1–V5 was incapable of pulling down two different GFP-tagged inner membrane proteins (UCP2 and MFRN2, also known as SLC25A28). Similar results were also obtained by immunoprecipitating Flag-tagged MCU and probing for endogenous MICU1 (Supplementary Fig. 1).
Collectively, our three complementary genomic analyses combined with our biochemical data (Fig. 1) predict that MCU is functionally related to MICU1 and that it, too, may participate in mitochondrial Ca2+ uptake.
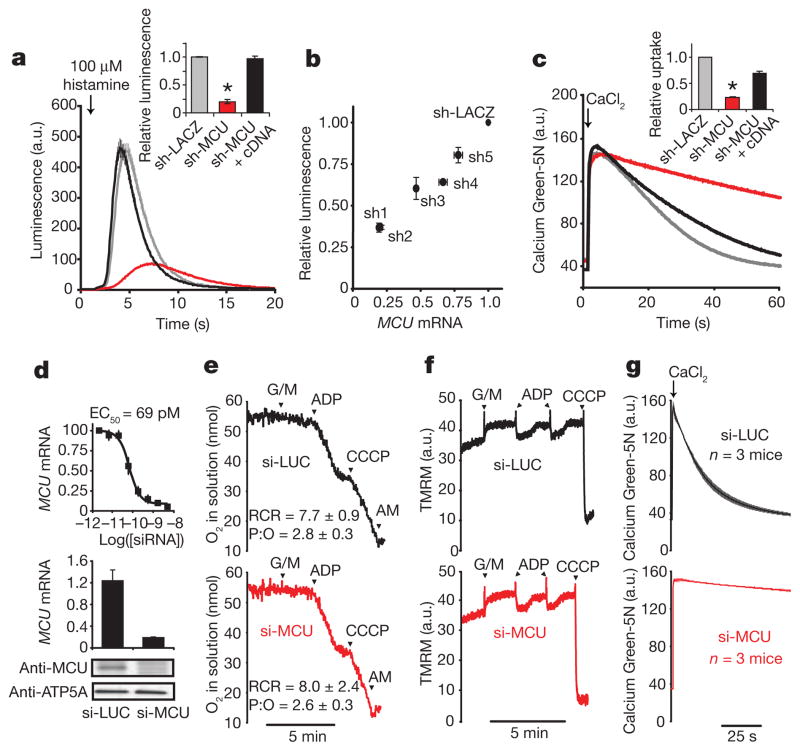
We evaluated the impact of silencing MCU on mitochondrial Ca2+ uptake in intact and permeabilized cells using RNA interference (RNAi). Silencing MCU in a HeLa cell line expressing a mitochondria-targeted aequorin (mt-Aeq) reporter9 attenuates mitochondrial Ca2+ uptake (Fig. 2a) proportionate to the strength of knockdown (Fig. 2b). The RNAi-induced phenotype is not off-target because coexpression of a full-length MCU cDNA together with a short hairpin RNA (shRNA) that targets the MCU 3′ untranslated region (3′UTR) fully rescues Ca2+ uptake (Fig. 2a). Moreover, the RNAi effect is not a trivial consequence of interrupting upstream signalling, because histamine mobilization of cytosolic Ca2+ remains intact (Supplementary Fig. 2a), and because we obtain similar results when measuring clearance of exogenously added Ca2+ by mitochondria in permeabilized HEK-293 (Fig. 2c) and HeLa cells (Supplementary Fig. 3). In HeLa cells, basal and uncoupled respiration were intact (Supplementary Fig. 2b), mitochondrial membrane potential (ψm) was not depolarized (Supplementary Fig. 2c), and mitochondrial morphology remained grossly unchanged (data not shown) after silencing of MCU. Basal cytosolic Ca2+ levels were 53.9 nM ± 16.3 (n = 30) in MCU knockdown HeLa cells and 70.9 nM ± 11.7 (n = 14) in control sh-LACZ cells. Although mitochondrial Ca2+ buffering is known to shape cytosolic Ca2+ transients in many cell types, its inhibition through silencing of MCU did not show a significant impact on cytosolic Ca2+ clearance after histamine stimulation (Supplementary Fig. 2a). However, this result could reflect an incomplete silencing of MCU-mediated mitochondrial Ca2+ uptake in HeLa cells (Fig. 2a, b). Mitochondrial Ca2+ uptake has also previously been shown to stimulate ATP production by allosterically activating three tricarboxylic acid (TCA) cycle dehydrogenases10. Using a previously reported protocol11, we found that permeabilized sh-MCU cells exhibit attenuated NAD(P)H elevation in response to exogenously added Ca2+ (Supplementary Fig. 2d), demonstrating that silencing MCU, like MICU1 (ref. 5), attenuates Ca2+ activation of the TCA cycle.
Figure 2. MCU is required for mitochondrial Ca2+ uptake in cultured cells and in purified mouse liver mitochondria.
a, Representative luminescence measurements of a mitochondrial aequorin Ca2+ reporter after histamine stimulation in HeLa cells expressing sh-LACZ, sh-MCU, or a combination of sh-MCU and an RNAi-resistant cDNA for MCU (mean ± s.e.m., n = 10 traces). Inset shows statistical analysis of the maximal luminescence (mean ± s.d., n= 10 traces, *P<0.001). a.u., arbitrary units. b, Relationship between MCU mRNA expression and histamine-induced mitochondrial Ca2+ uptake (maximal aequorin luminescence) recorded from five independent shRNAs targeting MCU and normalized to sh-LACZ (mean ± s.d., n= 3). c, Representative traces of Ca2+ uptake in digitonin-permeabilized sh-MCU HEK-293 cells or sh-LACZ control cells after addition of 50 μM final concentration of CaCl2. Inset reports linear fits of uptake kinetics between 15 and 20 s, normalized to sh-LACZ (mean ± s.d., n = 3, *P <0.001). Ca2+ was measured with Calcium Green-5N. d, In vitro dose–response of a selected siRNA duplex targeting mouse MCU. Relative expression of MCU mRNA in livers following weekly injections of si-MCU or si-LUC for 3 weeks, normalized to expression in PBS-treated mice. Immunoblot analysis of liver mitochondria isolated from mice treated with si-MCU or control si-LUC using antibodies against MCU and ATP5A1 as a loading control. e, Oxygen consumption measurements of isolated mitochondria in a well-stirred cuvette. Glutamate and malate (G/M), ADP, uncoupler (carbonyl cyanide m-chlorophenyl hydrazone, CCCP), antimycin (AM) were added at indicated time points. Respiratory control ratio (RCR) and ADP:O ratio (P:O) were computed from three separate mice for each group. f, Mitochondrial membrane potential (ψm) measured by tetramethyl rhodamine methyl ester (TMRM) in isolated liver mitochondria. g, Ca2+ uptake kinetics in energized liver mitochondria following the addition of 50 μM final CaCl2. Extra-mitochondrial Ca2+ was measured with Calcium Green-5N (mean ± s.e.m., n = 3 mice). Traces depicted in e and f are representative of measurements made from three independent mouse experiments performed on separate days.
To complement these cell-based studies, we analysed mitochondrial calcium uptake in mouse liver mitochondria upon in vivo silencing of MCU. A key advantage of this experimental system is that Ca2+ uptake phenotypes are directly attributable to mitochondria, and additional bioenergetic parameters, notably respiratory flux and ψm, can be carefully measured under classic respiratory state transitions12. As respiratory state transitions are dependent on TCA cycle metabolism, electron transport, H+ pumping, ATP/ADP exchange, and transport of phosphate, such studies allow us to evaluate the specificity of MCU’s role in mitochondrial Ca2+ transport. Using previously described siRNA design and delivery technology13, we screened 46 distinct siRNA duplexes. We selected a duplex characterized by a half-maximal inhibitory concentration of 69 pM that achieved >90% knockdown at a concentration of 5 nM in cultured mouse liver cells (Fig. 2d). We performed a large-scale synthesis of this siRNA duplex and encapsulated it into a lipid-based formulation optimized for liver-specific delivery14,15. As a negative control we used a siRNA duplex specific for the firefly luciferase gene (si-LUC). We performed weekly tail vein injections of the siRNA duplexes over a 3 week period and achieved 84 ± 2% (n = 3) in vivo mRNA knockdown of MCU in the liver, which was corroborated by immunoblot analysis (Fig. 2d). Both groups of mice showed normal weight gain (5.6 ± 1.3% of body weight for si-LUC and 5.0 ± 3.0% of body weight for si-MCU, n = 3) and the gross appearance of the livers did not differ. Mitochondria isolated from these livers were physiologically intact, capable of undergoing robust respiratory state transitions (Fig. 2e, f). The respiratory control ratio (RCR) was comparable in si-LUC (7.7 ± 0.9, n = 3) and si-MCU animals (8.0 ± 2.4, n = 3). The ADP:O ratios were 2.77 ± 0.3 (n = 3) and 2.61 ± 0.3 (n = 3) in si-LUC and si-MCU mitochondria, respectively, indicating intact respiratory chain physiology. However, mitochondria from si-MCU mice show a profound and near complete loss in Ca2+ uptake in response to extramitochondrial pulses of Ca2+ (Fig. 2g and Supplementary Fig. 4).
Collectively, our physiology studies, comprising three different experimental systems (intact cells, permeabilized cells and mitochondria purified from mouse organs), three different cell types (HeLa, HEK-293 and mouse liver), and two different silencing modalities (in vitro lentiviral shRNA and in vivo siRNA), firmly establish that MCU is essential for high capacity Ca2+ transport into mitochondria, and that the phenotype is not a secondary consequence of alterations in cytosolic Ca2+ signalling or of impaired ion or metabolite transport across the mitochondrial inner membrane. To our knowledge, this is the first time that mitochondrial Ca2+ uptake has been silenced in vivo in an animal.
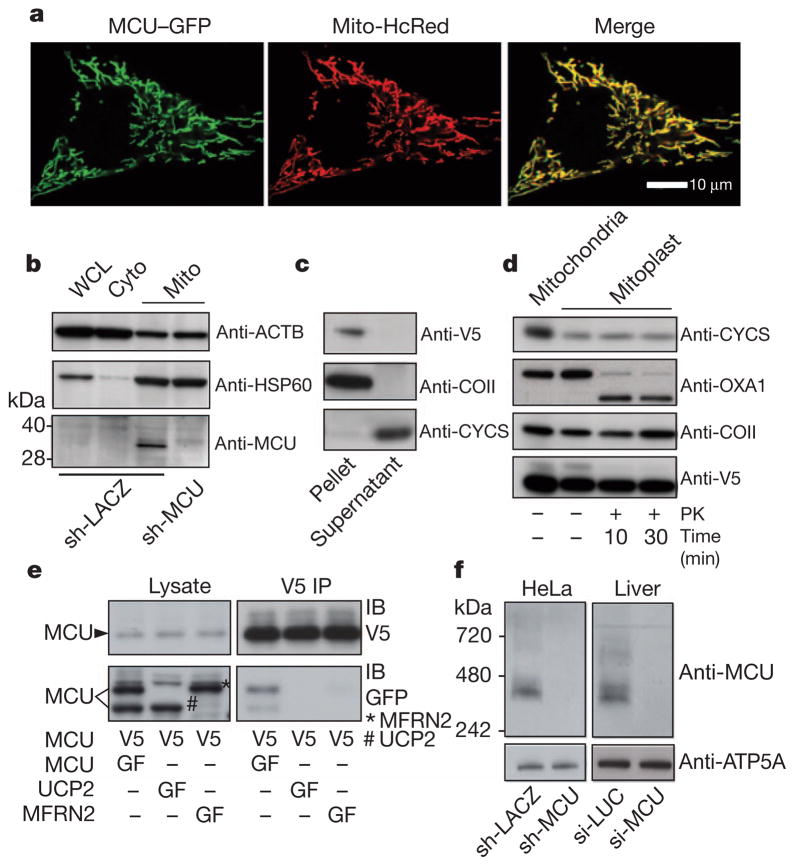
We next established MCU’s precise sub-organellar localization and topology. MCU was first identified as a mitochondrial protein in our previous proteomic survey6, where it was detected as a low abundance protein in 12 out of 14 mouse tissues, being missed only in heart and kidney. Confirmatory immunoblot analysis suggests it is also present in the heart and the kidney (data not shown), indicating it is universally expressed. As expected, a carboxy terminus GFP-tagged version of MCU localizes exclusively to mitochondria in HeLa cells (Fig. 3a) as well as in HEK-293 cells (data not shown). To biochemically validate MCU’s localization, we fractionated HeLa cell lysate and observed significant enrichment of the approximately 35 kDa native MCU in the mitochondrial fraction (Fig. 3b). The observed molecular weight of MCU is lower than the predicted 40 kDa and is consistent with a predicted cleavable amino-terminal mitochondrial targeting sequence16. Multiple algorithms predict that MCU contains two transmembrane domains17,18. To experimentally confirm mitochondrial inner-membrane integration and to discriminate between transmembrane and peripheral membrane configurations, we performed alkaline carbonate extraction of proteins from intact mitochondria isolated from HEK-293 cells expressing MCU-V5. We find that MCU remains in the insoluble fraction, proving that it is a bona fide transmembrane protein (Fig. 3c). To establish the topology of MCU across the inner membrane, we disrupted the mitochondrial outer membrane by hypotonic swelling and treated the resulting mitoplasts with proteinase K. Proteins such as OXA1 that are exposed to the mitochondrial inter-membrane space are digested by proteinase K, whereas COII (an integral inner membrane protein, also known as MT-CO2) and MCU are completely protected (Fig. 3d), despite being substrates for proteinase K (Supplementary Fig. 5). Our data therefore indicate that MCU is localized to the inner membrane with both its N- and C-termini facing the matrix space.
Figure 3. MCU is oligomeric and resides in the mitochondrial inner membrane as a larger complex.
a, Confocal imaging of MCU–GFP co-expressed with mitochondria-targeted HcRed (Mito-HcRed) in HeLa cells. b, Immunoblot analysis of HeLa whole-cell lysate (WCL), cytosol (Cyto) or crude mitochondrial fractions (Mito), using antibodies against MCU, HSP60 (matrix protein, also known as HSPD1), or ACTB (cytosol). c, Immunoblot analysis of soluble (supernatant) and insoluble (pellet) fractions following alkaline carbonate extraction of mitochondrial fractions from HEK-293 cells expressing MCU–V5. Immunoblot analysis was performed using antibodies against V5, COII (integral inner membrane protein) and CYCS (soluble intermembrane space protein). d, Immunoblot analysis after proteinase K (PK) treatment of MCU–V5-expressing HEK-293 mitoplasts for indicated times. e, Anti-V5 immunoprecipitations performed as in Fig. 1d using lysates from HEK-293 cells stably expressing MCU–V5 and transiently transfected with MCU–GFP, UCP2–GFP, or MFRN2–GFP. f, Blue native PAGE analysis of mitochondrial fractions from HeLa cells (stably expressing sh-LACZ or sh-MCU, left panel) or from livers of mice (si-LUC or si-MCU, right panel) and immunoblotted for MCU. ATP5A1 is used as a loading control.
If MCU is an integral component of the uniporter, we might expect that it oligomerizes and operates within a larger complex. To test for MCU oligomerization, we performed pull-down experiments and found that V5-tagged MCU is capable of immunoprecipitating GFP-tagged MCU (Fig. 3e), indicating that MCU can form multimers. To control for nonspecific interactions we used two inner mitochondrial membrane spanning transporters as controls, neither of which immunoprecipitated with MCU–V5 (Fig. 3e). We performed blue native gel separation of digitonin-solubilized mitochondria purified from HeLa cells and found that MCU migrates at an apparent molecular weight of ~450 kDa. This larger complex disappears following silencing with sh-MCU (Fig. 3f, left panel). We obtained similar results using mitochondria from livers of si-LUC and si-MCU mice (Fig. 3f, right panel). Collectively, these studies indicate that MCU oligomerizes in the mitochondrial inner membrane as part of a larger molecular weight complex.
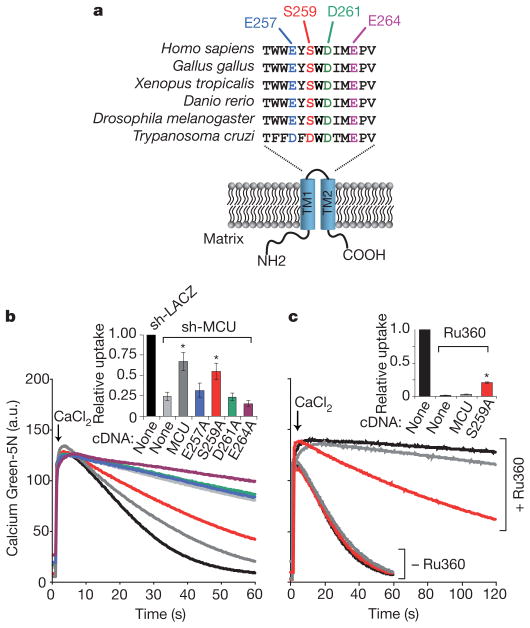
Our topology analysis (Fig. 3c, d) combined with computational predictions of membrane spanning domains indicate that MCU’s two transmembrane helices, TM1 and TM2, are linked by a short stretch of amino acids that face the intermembrane space and contain what we now term a ‘DIME’ motif (Fig. 4a). We sought to determine whether conserved amino acids near and within the DIME motif are required for MCU-mediated Ca2+ uptake. We created a series of four alanine mutants at conserved residues (E257A, S259A, D261A, E264A) and expressed them on an sh-MCU background to evaluate their ability to rescue Ca2+ transport (Fig. 4b). One mutant, S259A, was capable of providing significant rescue of the sh-MCU phenotype (Fig. 4b). The remaining three mutants (E257A, D261A, E264A), however, failed to restore mitochondrial calcium uptake despite comparable expression to the S259A mutant (Supplementary Fig. 6a), signifying that these highly conserved acidic residues are critical for Ca2+ transport.
Figure 4. Impact of point mutations on MCU activity and its sensitivity to Ru360.
a, Schematic of MCU topology across the mitochondrial inner membrane and a multiple sequence alignment of the linker sequence containing a DIME motif. TM1 and TM2 are two transmembrane domains. b, Ca2+ uptake in permeabilized sh-MCU HEK-293 cells transiently expressing MCU mutants. Inset reports linear fits of uptake kinetics between 15 and 25 s, normalized to sh-LACZ (mean ± s.d., n = 3, *P <0.01) c, Ca2+ uptake in HEK-293 cells transiently expressing wild-type MCU or the S259A mutant, in the presence or absence of 0.5 μM Ru360. Inset reports linear fits of uptake kinetics between 30 and 60 s for Ru360-treated cells, and between 15 and 25 s for untreated cells. Uptake rates are normalized to untreated HEK-293 cells (mean ± s.d., n = 3, *P <0.01).
Ruthenium red and a related compound, Ru360, are the most potent inhibitors of the mitochondrial Ca2+ uniporter, yet their target remains unknown19,20. These cell-impermeant inhibitors have been useful pharmacological tools in studies of isolated mitochondria21,22 and are believed to act on the outer face of the inner membrane. We next sought to determine whether the functional S259A mutant might influence Ru360 sensitivity. Ru360 strongly abolishes mitochondrial Ca2+ uptake in permeabilized HEK-293 cells (Fig. 4c). Transient over-expression of MCU leads to a mild resistance to Ru360 compared to control (Fig. 4c). However, transient expression of the S259A mutant confers marked resistance to Ru360 in both HEK-293 and HeLa cells (Fig. 4c, Supplementary Fig. 6b, c, d, e). These experiments clearly indicate a functional role for the linker region in both activity and Ru360 sensitivity of MCU.
Our genomic, physiological, biochemical and pharmacological data firmly establish that MCU is an oligomeric, mitochondrial inner membrane protein that is essential for calcium uniporter activity. In our current studies, overexpression of MCU alone did not give rise to a marked gain of Ca2+ uptake in HeLa cells (Fig. 2a), indicating that additional components or chaperones may be limiting in vivo. Although its precise mechanism remains to be elucidated, MCU fulfils multiple criteria for being an integral component of the uniporter, where it could serve as the pore-forming subunit or as a closely associated partner protein. First, loss of MCU leads to a profound defect in mitochondrial Ca2+ uptake in three distinct systems: intact cells, per-meabilized cells and mouse liver (Fig. 2). Second, it has two bona fide transmembrane domains and oligomerizes to form a larger molecular weight complex in the mitochondrial inner membrane (Fig. 3e, f). Third, mutations at evolutionarily conserved acidic residues attenuate its activity (Fig. 4b). Fourth, a point mutation in MCU confers resistance against Ru360, the most potent inhibitor of the uniporter (Fig. 4c and Supplementary Fig. 6). The current study further supports our previously reported phylogenomic signature of the mitochondrial Ca2+ uniporter5, and together these studies identify MICU1 and MCU as an evolutionarily conserved duo that are critical for mitochondrial Ca2+ uptake. Whereas MICU1 may serve as a regulator, MCU is likely to represent a core component of the uniporter. Future studies will be necessary to establish the exact mechanism by which these proteins function together to orchestrate the mitochondrial Ca2+ cycle.
METHODS SUMMARY
Candidate human genes required for mitochondrial Ca2+ uptake were prioritized on the basis of the co-occurrence of their homologues with MICU1 across 500 evolutionarily diverse organisms6,7 and their RNA and protein co-expression across mouse cell types and tissues6,8. Stable knockdown of MCU in HeLa and HEK-293 cells was achieved using lentiviral shRNA-expressing constructs available from the Broad Institute RNAi Consortium23. In vivo silencing of MCU in mouse liver was achieved via weekly tail vein injections of selected siRNAs in lipid-based formulations13–15,24. cDNA rescue studies in MCU knockdown cells were carried out by overexpression of wild-type or mutant versions of full-length MCU cDNAs together with a shRNA that targets the MCU 3′UTR. Agonist-induced rises in mitochondrial Ca2+ in intact HeLa cells were measured by luminescence of a mitochondria-targeted aequorin reporter9. Extra-mitochondrial calcium in per-meabilized HeLa cells, HEK-293 and isolated liver mitochondria was measured using Calcium Green-5N (ref. 5). Single-cell studies of cytosolic calcium were performed as previously described25. Mitochondrial respiration, membrane potential and NAD(P)H were measured via established protocols11,26. Crude mitochondria and mitoplasts were prepared from cultured HEK-293 cells expressing a C terminus V5-tagged version of MCU5. Protein topology was assessed by alkaline carbonate extraction from crude mitochondria and proteinase K digestion of mitoplasts27. Protein interaction studies were performed by immunoprecipitation with anti-V5 and anti-Flag antibodies. Native gel electrophoresis was performed as previously described28. Unless otherwise indicated, data are summarized as mean ± standard deviation (s.d.), and P-values were computed from t-tests.
Supplementary Material
Acknowledgments
We thank R. Nilsson, J. Engreitz and S. Calvo for bioinformatics assistance; D. Root and S. Silver for assistance in lentiviral RNAi; B. R. Bettencourt, K. Charisse, S. Kuchimanchi and L. Speciner for siRNA design, synthesis and formulation; M. Blower, J. Avruch and R. Ward for advice; and members of the Mootha laboratory for valuable feedback. J.M.B. and L.S. were supported by graduate student fellowships from the National Science Foundation. This work was supported by grants from the National Institutes of Health (GM0077465, DK080261) awarded to V.K.M.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Author Contributions J.M.B., F.P. and V.K.M. conceived of the project and its design. J.M.B., F.P., H.S.G., M.P., O.G., L.S., C.A.B.-T., X.R.B., Y.S. and R.L.B. performed experiments and data analysis. V.K. aided in experimental design. V.K.M., J.M.B., F.P. and M.P. wrote the manuscript.
References
- 1.DeLuca HF, Engstrom GW. Calcium uptake by rat kidney mitochondria. Proc Natl Acad Sci USA. 1961;47:1744–1750. doi: 10.1073/pnas.47.11.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasington FD, Murphy JV. Ca++ ion uptake by rat kidney mitochondria and its dependence on respiration and phosphorylation. J Biol Chem. 1962;237:2670–2677. [PubMed] [Google Scholar]
- 3.Carafoli E, Lehninger AL. A survey of the interaction of calcium ions with mitochondria from different tissues and species. Biochem J. 1971;122:681–690. doi: 10.1042/bj1220681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunter K, Gunter TE. Transport of calcium by mitochondria. J Bioenerg Biomembr. 1994;26:471–485. doi: 10.1007/BF00762732. [DOI] [PubMed] [Google Scholar]
- 5.Perocchi F, et al. MICU1 encodes a mitochondrial EF hand protein required for Ca2+ uptake. Nature. 2010;467:291–296. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lattin JE, et al. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizzuto R, Simpson AW, Brini M, Pozzan T. Rapid changes of mitochondrial Ca2+ revealed by specifically targeted recombinant aequorin. Nature. 1992;358:325–327. doi: 10.1038/358325a0. [DOI] [PubMed] [Google Scholar]
- 10.Denton RM, McCormack JG. The role of calcium in the regulation of mitochondrial metabolism. Biochem Soc Trans. 1980;8:266–268. doi: 10.1042/bst0080266. [DOI] [PubMed] [Google Scholar]
- 11.Territo PR, Mootha VK, French SA, Balaban RS. Ca2+ activation of heart mitochondrial oxidative phosphorylation: role of the F0/F1-ATPase. Am J Physiol Cell Physiol. 2000;278:C423–C435. doi: 10.1152/ajpcell.2000.278.2.C423. [DOI] [PubMed] [Google Scholar]
- 12.Chance B, Williams GR. Respiratory enzymes in oxidative phosphorylation I Kinetics of oxygen utilization. J Biol Chem. 1955;217:383–393. [PubMed] [Google Scholar]
- 13.Musunuru K, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akinc A, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nature Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akinc A, et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 17.Bernsel A, Viklund H, Hennerdal A, Elofsson A. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 2009;37:W465–W468. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 19.Moore CL. Specific inhibition of mitochondrial Ca++ transport by ruthenium red. Biochem Biophys Res Commun. 1971;42:298–305. doi: 10.1016/0006-291x(71)90102-1. [DOI] [PubMed] [Google Scholar]
- 20.Ying WL, Emerson J, Clarke MJ, Sanadi DR. Inhibition of mitochondrial calcium ion transport by an oxo-bridged dinuclear ruthenium ammine complex. Biochemistry. 1991;30:4949–4952. doi: 10.1021/bi00234a016. [DOI] [PubMed] [Google Scholar]
- 21.Bernardi P. Mitochondrial transport of cations: channels, exchangers, and permeability transition. Physiol Rev. 1999;79:1127–1155. doi: 10.1152/physrev.1999.79.4.1127. [DOI] [PubMed] [Google Scholar]
- 22.Kirichok Y, Krapivinsky G, Clapham DE. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427:360–364. doi: 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- 23.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 24.Semple SC, et al. Rational design of cationic lipids for siRNA delivery. Nature Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 25.Rebres RA, et al. Synergistic Ca2+ responses by Gαi- and Gαq-coupled G-protein-coupled receptors require a single PLCβ isoform that is sensitive to both Gβγ and Gαq. J Biol Chem. 2011;286:942–951. doi: 10.1074/jbc.M110.198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gohil VM, et al. Nutrient-sensitized screening for drugs that shift energy metabolism from mitochondrial respiration to glycolysis. Nature Biotechnol. 2010;28:249–255. doi: 10.1038/nbt.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan MT, Voos W, Pfanner N. Assaying protein import into mitochondria. Methods Cell Biol. 2001;65:189–215. doi: 10.1016/s0091-679x(01)65012-x. [DOI] [PubMed] [Google Scholar]
- 28.Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.