Nothing can happen in biology unless something binds to something else. Although much of the effort in bioinformatics to date has focused on the detection of homology or the deduction of structure and/or function from sequence, in the long run, for bioinformatics to make a real contribution to drug design and cell biology, it will be necessary to be able to predict what other molecules a given gene product will bind to and how tight that binding will be. At present, no one knows how to do this routinely. The collection of large data sets for molecular interactions will help discover predictable features of molecular interactions.
Methods to measure molecular interactions are increasingly able to determine interactions on a large scale. For example, the creation of protein arrays allows probing of proteins with lipids, kinases and DNA recognition sequences to determine molecular interactions and ultimately why one interaction occurs and another does not. These collections of data not only contribute to a better understanding of the biology of the molecular interactions, but can form data sets to aid the development of prediction methods. For example, over the last 10 years two strategies to generate experimental information about protein-protein interactions have appeared. These methods have been generating data that can inform computer models to predict protein-protein interactions and to create networks or perform network analysis. The first method is the yeast two-hybrid that uses the S. cerevisiae GAL4 transcriptional activator to determine if two proteins interact. Transcription factors have two domains- a binding domain and an activation domain. They are quite modular and can be split in two without affecting the function of either domain. Fields and Song recognized the modularity of transcription factors could make them a useful tool to determine if two proteins interact (Fields and Song, 1989). By fusing one protein to the binding domain and a second protein to the activation domain, if the two proteins interact they will activate transcription. Fields created a powerful assay with this method to screen protein-protein interactions in Escherichia coli bacteriophage T7 (Bartel et al., 1996) and then S. cerevisiae (Uetz et al., 2000). These methods have also been applied to large scale analyses of human protein-protein interactions (Rual et al., 2005). Data sets of protein-protein interactions present unique opportunities for algorithm development to analyze the data (Schwikowski et al., 2000) and Units 8.8 and 8.13 discuss software to visualize large networks such as those created by the 2-hybrid method.
Protein complexes can be enriched by immunoprecipitation or epitope tagging of a member of the complex (AP-MS). Precipitated complexes are then analyzed by tandem mass spectrometry to identify and potentially quantitate members of the complex. Protein-protein interactions are revealed by this method. Several large-scale studies have revealed protein-protein interactions in S. cerevisiae using affinity purification and mass spectrometry (AP-MS) (Gavin et al., 2002; Ho et al., 2002). Hazbun et al employed a variety of methods including the yeast 2-hybrid genetic method, AP-MS and cytofluorescence to determine the functions of 100 essential hypothetical open reading frames (Hazbun et al., 2003). They were able to assign function to almost all of the proteins. These large data sets provide a wealth of information that requires sophisticated software algorithms to extract and organize the information and to reconstruct networks of interactions. As more large-scale experiments are performed increasingly sophisticated tools will be needed for analysis of the data. Gomez et al described methods to predict protein-protein interaction networks in Unit 8.2 and the 8.8 and 8.13 Units mentioned above will also be useful for studies of protein networks. Unit 8.8 describes the use of VisANT a tool to develop and display molecular networks based on functional and physical relations from the Predictome database. Unit 8.13 describes the use of a Cytoscape to determine and analyze the interconnectivity of genes or proteins.
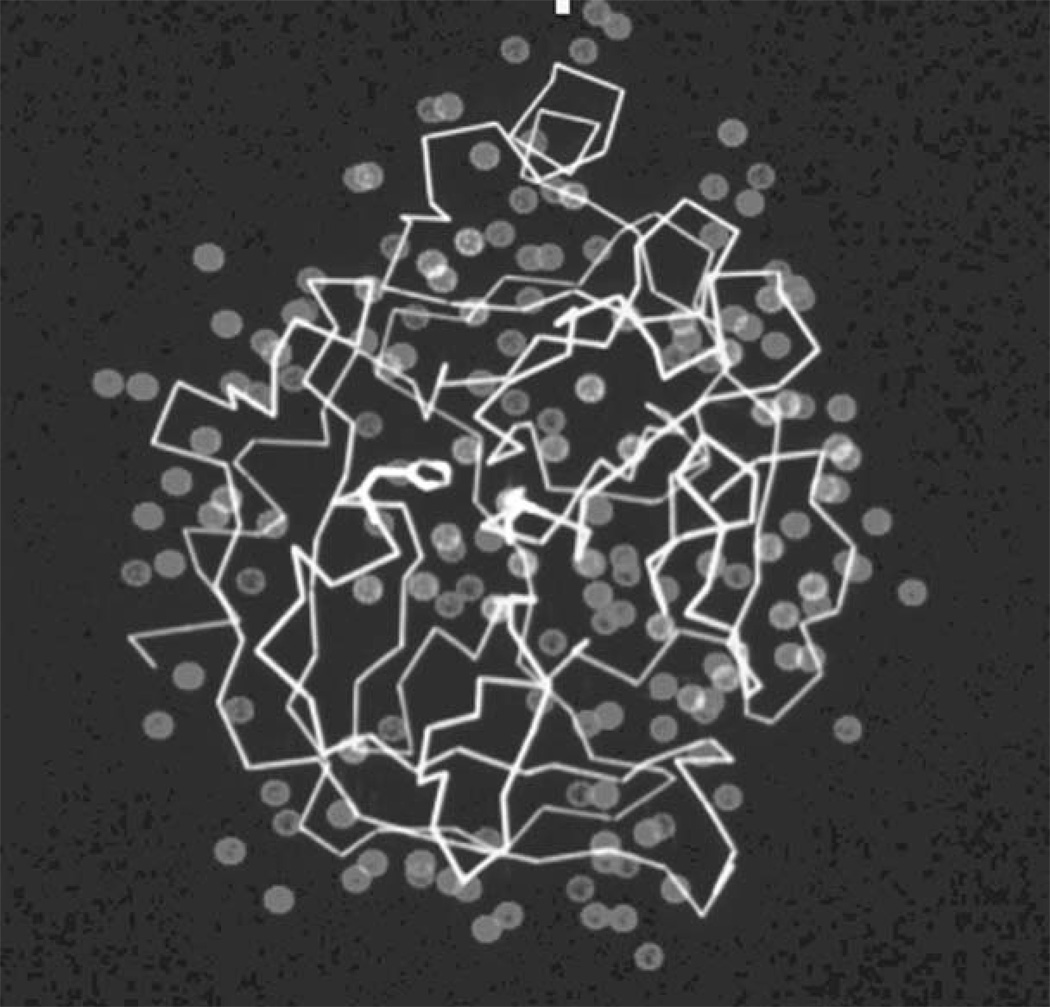
Fundamental to any treatment of molecular interactions is recognition of the fact that, when anything tries to bind to the surface of a protein, it does so in the presence of a 55 M concentration of a competing ligand: water. The surfaces of protein molecules are coated with a layer of bound solvent about 1 to 2 molecules deep (Fig. 8.1.1). A typical protein will have at least 2 to 3 bound waters per amino acid, numerically, and although most will be on the surface, a few will be buried in cavities or at the interfaces between subunits. Displacement of bound solvent from a potential binding site can be easy or difficult; it seems reasonable to assume that the degree of difficulty must relate in some fashion to how tightly any ligand can bind to that site or whether the site is accessible to ligands at all. Yet, most computational approaches to analyzing or predicting ligand binding sites and affinities have either ignored the role of bound water or treated it in a very general way. There is experimental evidence that a general treatment may be as bad as neglecting solvent altogether. Crystallographic analyses of protein structures in different solvents, or in the same solvent but in different crystal lattices, have suggested the existence of at least three different classes of water molecules on a protein surface. Tightly bound solvent molecules are observed under all conditions; disordered solvent molecules are either never observed or are found in only one or two structures of the same protein, indicating very weak binding. A third, intermediate class of waters appears in many but not all structures and has positions that vary somewhat from structure to structure, suggesting binding sites of intermediate strength. Methods to analyze these classes of water molecules offer the intriguing suggestion that ligand binding sites primarily involve displacement of the intermediate waters rather than the other two classes (Mattos, 2002). It may be possible to rationalize this striking fact by considering solvent entropy and the contributions it makes to binding. Tightly bound waters are simply too strongly associated with the protein surface to be displaced; they basically occlude the sites they are on. Disordered solvent molecules can be displaced easily, but no entropy gain occurs when they are—they are already conformationally unrestricted. Intermediate waters, on the other hand, are not held so tightly that they cannot be displaced by a ligand, yet are held tightly enough that freeing them up to go into the solvent will produce an entropy gain that can help drive ligand binding thermodynamically. If this analysis is correct, it suggests that computational approaches to finding water sites of intermediate affinity could provide a means for identifying ligand binding sites on the surface of a protein, even when nothing is known about what binds there. Since it has also been shown that the locations of bound waters trace the conformation of bound ligands in the binding site, reliable methods for predicting solvent positions could also provide a first-pass outline for the design of drugs.
Figure 8.1.1.
The crystal structure of the bacterial serine protease subtilisin with the bound water molecules observed crystallographically indicated as blue spheres. Figure courtesy of Dagmar Ringe, Brandeis University.
Once ligands can be modeled into protein binding sites—a task that sounds straightforward but is actually extremely difficult—the next step is determining affinity. Once it was thought that computing affinities to within an order or magnitude or two would be satisfactory, but current methods aim to do much better than that—it is not unreasonable to expect an accuracy to within a few kilojoules or less in favorable situations. Coping with the effects of protein conformational changes and nonstandard binding modes is still the challenge for such calculations. Better methods for predicting these situations in advance are sorely needed. Reliable tools for calculating electrostatic interaction energies are equally necessary. Of all the forces that exist between molecules, the electrostatic term is the hardest to compute accurately. One reason is that the charges, and partial charges, on the ionizable groups and polar groups involved are simply not known with certainty. Another reason is that the dielectric constant term in the familiar Coulomb potential term is almost impossible to estimate. The dielectric constant is really a property of bulk solvent, and whatever else one may say about the environment of a ligand binding site, it seems certain that in most respects it does not resemble bulk solvent. There is no universally agreed upon method for calculating the screening effect of solvent and protein atoms in the microenvironment of a protein binding pocket. These approaches offer hope of a forthcoming solution to this most thorny problem of estimating interaction energies. Although the fields of drug design and metabolic biochemistry are mostly concerned with the interactions between proteins and small molecules, cell biology is more often interested in macromolecular interactions, usually those of proteins with one another. Bioinformatics is only beginning to tackle this most challenging problem: given the structure (or ultimately, the sequence) of a gene product, how does one predict, from first principles or from comparative analysis, what other gene’s product it will associate with, and at what sites and with what consequences (Al-Lazikani et al., 2001)? Cesarini and Gomez describe databases and tools that provide the first tentative steps in answering these questions (UNIT 8.2).
In the end, bioinformatics will need to address even more difficult questions than these in regard to interactions. Many protein complexes in the cell are dynamic; how are we to predict the lifetime of a complex? Can computationally determined affinities be related to on- and off-rates in the absence of experimental data? What about protein turnover rates: can they be predicted from sequence and/or structural information? Many proteins interact with one another only in the vicinity of the membrane: what are the effects of membranes on protein conformation and binding properties? Can we ever predict how a protein’s structure and dynamics will change in response to ligand binding? These and other challenges for the future of bioinformatics will most likely require completely new methods for analyzing intermolecular interactions.
Contributor Information
Gregory A. Petsko, Brandeis University, Waltham, Massachusetts
John R. Yates, III, The Scripps Research Institute, LaJolla, CA.
References
- Al-Lazikani B, Sheinerman FB, Honig B. Combining multiple structure and sequence alignments to improve sequence detection and alignment: application to the SH2 domains of Janus kinases. Proc Natl Acad Sci U S A. 2001;98:14796–14801. doi: 10.1073/pnas.011577898. 64938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel PL, Roecklein JA, SenGupta D, Fields S. A protein linkage map of Escherichia coli bacteriophage T7. Nat Genet. 1996;12:72–77. doi: 10.1038/ng0196-72. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Hazbun TR, Malmstrom L, Anderson S, Graczyk BJ, Fox B, Riffle M, Sundin BA, Aranda JD, McDonald WH, Chiu CH, et al. Assigning function to yeast proteins by integration of technologies. Mol Cell. 2003;12:1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Mattos C. Protein-water interactions in a dynamic world. Trends Biochem Sci. 2002;27:203–208. doi: 10.1016/s0968-0004(02)02067-4. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Schwikowski B, Uetz P, Fields S. A network of protein-protein interactions in yeast. Nat Biotechnol. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]