Abstract
MicroRNAs (miRNAs) are small RNAs responsible for the post-transcriptional regulation of a variety of human genes. To date, their involvement in the regulation of CBR1 is unknown. This study reports for the first time the identification of microRNA-574-5p (hsa-miR-574-5p) and microRNA-921 (hsa-miR-921) as two miRNAs capable of interacting with the 3′-untranslated region (3′-UTR) of the CBR1 gene and downregulating CBR1 expression. Furthermore, we demonstrate that a common single-nucleotide polymorphism (SNP) in the CBR1 3′-UTR (rs9024, CBR1 1096G>A) differentially impacts the regulation of CBR1 by hsa-miR-574-5p and hsa-miR-921 dependent on genotype. First, four candidate miRNAs were selected based on bioinformatic analyses, and were tested in Chinese hamster ovary (CHO) cells transfected with CBR1 3′-UTR constructs harboring either the G or A allele for rs9024. We found that hsa-miR-574-5p and hsa-miR-921 significantly decreased luciferase activity in CHO cells transfected with the CBR1 3′-UTR construct carrying the major rs9024 G allele by 35% and 46%, respectively. The influence of these miRNAs was different in cells transfected with a CBR1 3′-UTR construct containing the minor rs9024 A allele in that only hsa-miR-574-5p had a demonstrable effect (i.e., 52% decrease in lucifersase activity). To further determine the functional effects of miRNA-mediated regulation of polymorphic CBR1, we assessed CBR1 protein expression and CBR1 enzymatic activity for the prototypical substrate menadione in human lymphoblastoid cell lines with distinct rs9024 genotypes. We found that hsa-miR-574-5p and hsa-miR-921 significantly decreased CBR1 protein (48% and 40%, respectively) and CBR1 menadione activity (54% and 18%, respectively) in lymphoblastoid cells homozygous for the major rs9024 G allele. In contrast, only hsa-miR-574-5p decreased CBR1 protein and CBR1 activity in cells homozygous for the minor rs9024 A allele, and did so by 49% and 56%, respectively. These results suggest that regulation of human CBR1 expression by hsa-miR-574-5p and hsa-miR-921 depends upon rs9024 genotype status.
Introduction
MicroRNAs (miRNAs) are a family of small (∼22 nt), highly conserved, non-protein coding RNAs that have been recognized as important gene expression regulators [1], [2], [3]. It is estimated that the human genome contains around 1000 miRNA genes, and that each miRNA gene harbors the flexibility to potentially interact with multiple genes [4]. MiRNAs have been linked to the post-transcriptional regulation of a diverse host of genes that participate in the majority of important biological functions [5], [6], [7]. This class of RNAs regulates gene expression through partial, complementary Watson-Crick base-pairing of target mRNAs in the 3′-untranslated region (3′-UTR) of the message.
Carbonyl reductase 1 (CBR1) is a ubiquitous, cytosolic, short-chain dehydrogenase that catalyzes the two-electron reduction of a broad range of endogenous and xenobiotic compounds [8]. Among its most relevant pharmacological substrates are the antipsychotic haloperidol and the chemotherapeutic anthracyclines doxorubicin and daunorubicin [9], [10]. Recently, we documented extensive variability in CBR1 expression in human liver [11] and heart [12] tissues. For example, normalized hepatic CBR1 mRNA expression was found to vary almost 50-fold while cardiac CBR1 mRNA expression levels varied 13-fold. Similarly, wide variability was found in CBR1 protein levels determined by quantitative immunoblotting (7-fold and 8-fold in liver and heart tissues, respectively). In line, and most therapeutically relevant, is the reported variability in CBR1 activity for the reduction of anthracycline substrates. For example, Gonzalez et al. and Kassner et al. found that hepatic CBR1 activity for the anthracycline substrate doxorubicin varied by approximately 22-fold [11], [13]. Functional single-nucleotide polymorphisms (SNPs) on the CBR1 gene may impact CBR1 expression and CBR1 activity. For instance, we reported that a relatively common SNP in the 3′-UTR of CBR1, CBR1 1096G>A (rs9024; q≈12.5% in Europeans and≈30% in Asians), impacts CBR1 phenotypes. In liver, homozygosis for the major G allele was associated with significantly higher CBR1 protein expression and CBR doxorubicin reductase activity [11]. We hypothesized that rs9024 might influence the binding of miRNAs to the 3′-UTR of CBR1 and represent another example of miRNA pharmacogenomics [14].
To date, there are no reports of functional miRNA interactions with human CBR1. Therefore, our primary objective was to determine if any miRNAs are capable of regulating CBR1. First, we set out to identify miRNAs that might bind to the 3′-UTR of CBR1 in silico. Next, we tested candidate miRNA binding with human CBR1 3′-UTR constructs in cultures of the Chinese hamster ovary-derived cell line (CHO). To confirm the potential of the miRNA candidates to regulate polymorphic CBR1, we assessed CBR1 protein levels and CBR enzymatic activity in human lymphoblastoid cell lines with distinct rs9024 genotype status. Finally, we assessed the co-expression of the miRNA candidates and CBR1 in human liver and heart tissue.
Materials and Methods
Ethics statement
The Health Sciences Institutional Review Board of the State University of New York at Buffalo has determined that this research project does not involve human subjects as defined under HHS regulations 45 CFR 46.102 (f).
Bioinformatics
The full 3′-UTR sequence of human CBR1 (GenBank sequence NM_001757.2) was retrieved using Entrez (http://www.ncbi.nlm.nih.gov/Entrez/). Potential miRNA binding sites along the CBR1 3′-UTR were found by comparing the results from multiple bioinformatic prediction databases including PITA [15], TargetScan [16], Microcosm Targets (formerly miRBase Targets) [17], [18], RNAhybrid [19], and PolymiRTS [20], [21].
Cell culture and reagents
CHO-K1 cells (Chinese hamster ovary-derived cell line, CCL-61) were obtained from the American Type Culture Collection (Manassas, VA). Common cell culture reagents were purchased from Invitrogen (Carlsbad, CA). CHO-K1 cells were routinely cultured in 10 cm2 tissue culture dishes using RPMI 1640 medium supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Sigma-Aldrich, St. Louis, MO), 100 U/ml penicillin, and 100 µg/ml streptomycin. Cultures were grown and maintained at low passage numbers (n<12) using standard incubator conditions of 37°C, 5% CO2, and 95% relative humidity.
Human lymphoblastoid cell lines (Coriell Institute, Camden, NJ) were routinely cultured in 75 cm tissue culture flasks using RPMI 1640 medium supplemented with 15% (v/v) heat-inactivated fetal bovine serum (Sigma-Aldrich, St. Louis, MO), 100 U/ml penicillin, and 100 µg/ml streptomycin. Suspension cultures were maintained in standard incubator conditions of 37°C, 5% CO2, and 95% relative humidity. These particular cell lines originate from the International HapMap project, a collaborative effort to create a haplotype map of the human genome, and were chosen for their utility in identifying genetic determinants of variability in gene expression [22], [23].
Constructs
MicroRNA mimics and hairpin inhibitors for microRNA-574-5p (hsa-miR-574-5p), microRNA-656 (hsa-miR-656), microRNA-877-5p (hsa-miR-877-5p), and microRNA-921 (hsa-miR-921), as well as mimic and inhibitor negative controls, were obtained from Dharmacon (Chicago, IL). Full-length CBR1 3′-UTR constructs, one containing the G allele of CBR1 1096G>A (pCBR1-1096G) and the other containing the A allele (pCBR1-1096A), were synthesized by OriGene (Rockville, MD). The 3′-UTR fragments were cloned into a pMirTarget vector (OriGene) downstream of the firefly luciferase gene. All inserts were verified by direct DNA sequencing.
Transfection
CHO-K1 cells were plated in 48-well plates 24 to 48 hours prior to transfection. CHO-K1 cells were co-transfected with CBR1 3′-UTR luciferase reporter constructs (500 ng) plus internal control plasmid pRL-TK (50 ng) and 15 nM miRNA mimic (hsa-miR-574-5p, hsa-miR-656, hsa-miR-877-5p, or hsa-miR-921) in the presence or absence of 5 nM specific miRNA inhibitor using DharmaFECT Duo transfection reagent (Dharmacon). Identical concentrations of miRNA mimic and inhibitor negative controls were also used. 24 hours post-transfection, cultures were washed once with phosphate-buffered saline solution and cells were lysed in freshly diluted passive lysis buffer (100 µl/well; Promega, Madison, WI) by incubating the plates at room temperature on a shaker at 200 rpm for 60 minutes. Luciferase reporter gene activities were determined with the Dual-Luciferase Reporter Assay System (Promega) per the manufacturer's instructions. Light intensity was measured in a Synergy HT luminometer equipped with proprietary software for data analysis (BioTek, Winooski, VT). Light intensity values from cell cultures transfected with CBR1 3′-UTR luciferase reporter constructs and Renilla luciferase control plasmids were used to correct for background. Corrected firefly luciferase activities were normalized to Renilla luciferase activities and expressed as fold increases with respect to the values obtained from control transfections with miRNA mimic negative control. Three independent experiments were performed in triplicate to evaluate reproducibility.
Lymphoblastoid cells were plated in 48-well plates 24 hours prior to transfection. Cells were co-transfected with 15 nM miRNA mimic (hsa-miR-574-5p, hsa-miR-656, hsa-miR-877, or hsa-miR-921) and miRNA mimic negative control in the presence or absence of 5 nM of specific miRNA inhibitor using the Neon transfection system (Invitrogen). 48 hours post-transfection, cells were pelleted for cytosolic protein extraction.
Genotyping of lymphoblastoid cells for CBR1 1096G>A genotype status
The CBR1 1096G>A polymorphism was investigated by allelic discrimination with fluorescent probes and real-time PCR (Assays-by-Design; Applied Biosystems, Carlsbad, CA). Genotyping reactions were run as described [11], [12]. DNA samples of known rs9024 genotype status were used as positive controls.
Quantitative immunoblotting of CBR1 in lymphoblastoid cells
To assess the influence of microRNA binding on CBR1 protein expression, lymphoblastoid cells (≈1×106 cells) were incubated for 24 hours with 15 nM miRNA mimic in the presence or absence of 5 nM specific hairpin inhibitor. Cytosolic protein was isolated using the Minute Cytosolic and Nuclear Extraction kit (Invent Biotechnologies, Eden Prairie, MN) according to the manufacturer's instructions. CBR1 protein levels were quantitated essentially as described [24]. In brief, lymphoblast cytosols (35 µg) were loaded into 12% precast polyacrylamide gels (Invitrogen, Carlsbad, CA) and separated by electrophoresis. Protein blots were probed with a specific polyclonal anti-human CBR1 antibody (1∶1,000; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or anti-human β-actin antibody (1∶1,000; Santa Cruz Biotechnology, Inc.) and a secondary goat anti-rabbit IgG antibody conjugated with horseradish peroxidase (1∶10,000; Sigma-Aldrich). Immunoreactive bands were visualized with the ECL plus western blotting detection system (GE Healthcare, Chalfont St. Giles, UK). CBR1 band intensity values (pixels/mm2) were quantified with a ChemiDoc XRS gel documentation system (Bio-Rad Laboratories). Cytosolic CBR1 levels were calculated relative to the band density of cells transfected with miRNA mimic negative control.
RNA degradation analysis
48 hours post-transfection with microRNA mimics and/or inhibitors, lymphoblastoid cells were treated with 10 µM actinomycin D (Enzo Life Sciences, Farmingdale, NY) to block de novo RNA synthesis. Cells were collected after 0, 2, 4, and 8 hours of actinomycin D incubation. Treatments were conducted in triplicate. CBR1 mRNA levels were quantitated as described [11]. Briefly, total RNA (75 ng) from lymphoblastoid cells was reverse-transcribed and amplified using a one-step QuantiTect SYBR Green RT-PCR kit (Qiagen, Valencia, CA) with the following primers: 5′-TCAAGCTGAAGTGACGATGA-3′ (forward) and 5′-GGTGCACTCCCTTCTTTGTA-3′ (reverse). CBR1 mRNA levels were determined by the comparative quantitation method. Individual β-actin mRNA levels were used for normalization. Experimental samples and standards for calibration curves were analyzed in quadruplicate. The relative amount of CBR1 mRNA in each sample was automatically calculated with a comparative quantitation algorithm (iQ5 Optical System Software version 2.0, Bio-Rad, Hercules, CA). CBR1 mRNA values were expressed relative to the normalized CBR1 mRNA content of negative control-transfected cells.
Detection of miRNA expression in human liver and heart tissue
The Institutional Review Board of the State University of New York at Buffalo approved this research. Human heart (left ventricle) and liver tissue samples from organ donors were procured from the National Disease Research Interchange (Philadelphia, PA) and the Liver Tissue Procurement and Distribution System (National Institutes of Health Contract N01-DK-9-2310), respectively. Cardiac and hepatic total RNA were extracted as previously described [11], [12]. To determine the gene expression of our candidate miRNAs in human tissues, we conducted real-time RT-PCR in triplicate in heart and liver total RNA (15 ng) using miScript Primer Assays (Qiagen). MiRNA expression levels in both tissue types were normalized to individual U6 gene levels and then expressed relative to corresponding amounts of hsa-miR-15a expression.
Kinetic analysis
Maximal CBR1 enzymatic activities for the prototypical substrate menadione were measured in lymphoblastoid cell cytosols in a Synergy HT Multi-Detection Microplate Reader (Bio-Tek, Winooski, VT) [25], [26], [27]. Briefly, CBR1 enzymatic activity was assessed by measuring the oxidation rate of the CBR1 co-factor NADPH at 340 nm (molar absorption coefficient = 6220 M−1 cm−1). Reaction mixtures (0.25 ml) contained potassium phosphate buffer (0.1 M; pH 7.4), NADPH (200 µM; Sigma-Aldrich), and menadione (250 µM; Sigma-Aldrich) and were incubated at 37°C for 5 minutes. Kinetic reactions were started by the addition of cytosols (40 µl, total protein concentration: 1.1±0.4 mg/ml). Enzymatic velocities were automatically calculated by linear regression of the Δabsorbance/Δtime points with Bio-Tek's proprietary software for enzyme kinetic analysis.
Results
Bioinformatic analyses
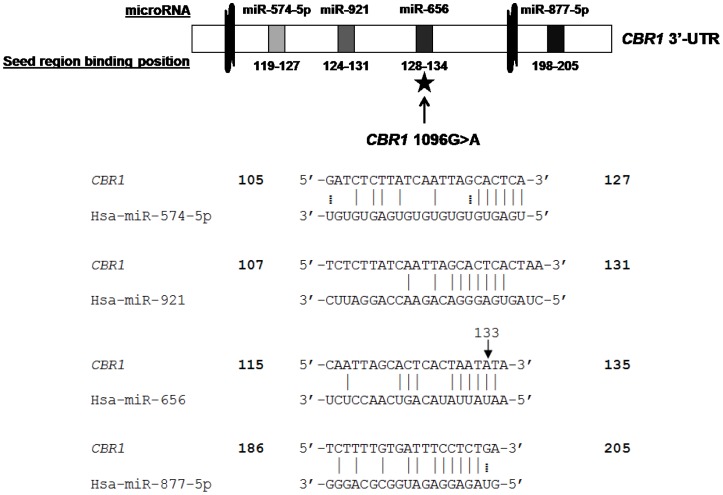
PITA, TargetScan, Microcosm Targets, RNAhybrid, and PolymiRTS were used in our computational screening for human microRNA antisense matches of the CBR1 3′-UTR. Each algorithm produced several potential candidate interactions, with the exception of PolymiRTS which is a specialized algorithm that matches microRNA sequences to known 3′-UTR SNPs. Interestingly, PolymiRTS identified a single microRNA, hsa-miR-656, which might bind to the 3′-UTR of CBR1 only when the minor rs9024 A allele is present. To prioritize these candidate microRNAs, we first identified miRNAs that were predicted by more than one algorithm. Candidates were then sorted by the free energy criteria ΔGopen, the free energy lost by opening the target site, ΔGduplex, the free binding energy of a miRNA to the target, and ΔΔG, the difference between ΔGopen and ΔGduplex (Table 1). These free energy values are critical in determining which microRNA/mRNA interactions are most energetically favorable. The algorithms we used pinpointed hsa-miR-574-5p, hsa-miR-656, hsa-miR-877-5p, and hsa-miR-921 as the most viable candidates for interacting with the 3′-UTR of CBR1 (Figure 1).
Table 1. Potential human miRNAs that target the CBR1 3′-UTR (G allele/A allele).
| Free energy predictions from the PITA algorithm | ||||||
| microRNA | ▵Gduplex | ▵Gopen | ▵▵G | |||
| G | A | G | A | G | A | |
| hsa-miR-574-5p | −24.49 | −24.49 | −6.54 | −5.41 | −17.94 | −19.07 |
| hsa-miR-877-5p | −19.4 | −19.4 | −12.75 | −13.33 | −6.64 | −6.06 |
| hsa-miR-921 | −11.6 | −5.8 | −5.92 | −8.04 | −5.67 | 2.24 |
| hsa-miR-656 | N/A | −10 | N/A | −5.05 | N/A | −4.94 |
▵G values expressed in kcal/mol
Figure 1. Schematic diagram of the CBR1 3′-UTR.
Potential miRNA binding sites on the 3′-UTR of CBR1 and their relative locations are indicated by shaded regions. The location of the CBR1 1096G>A polymorphism is designated with a star in the diagram and by an arrow in the sequence alignment.
Hsa-miR-574-5p and hsa-miR-921 bind differentially to CBR1 3′-UTR constructs
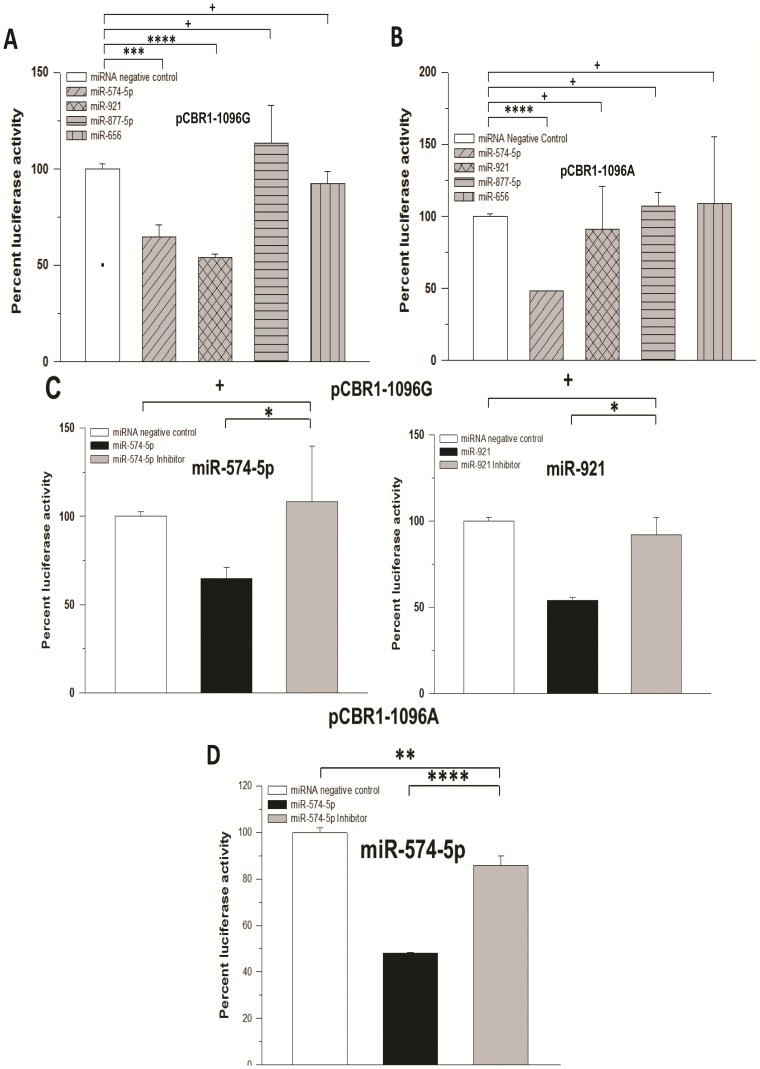
We employed dual-luciferase assays to evaluate the binding of miRNA candidates to two full-length CBR1 3′-UTR constructs identical except for the rs9024 polymorphic site (pCBR1-1096G and pCBR1-1096A). We found that CHO cells co-transfected with pCBR1-1096G and hsa-miR-574-5p or hsa-miR-921 exhibited decreased luciferase activity compared to a miRNA negative control mimic, 35% and 46%, respectively, which suggested a significant interaction between these miRNAs and the 3′-UTR of CBR1. Meanwhile, transfection of hsa-miR-656 or hsa-miR-877-5p resulted in no change in luciferase activity. Additionally, hsa-miR-656 or hsa-miR-877-5p failed to significantly alter luciferase activity when paired with the pCBR1-1096A construct (Figure 2A). On the other hand, co-transfection of hsa-miR-574-5p and hsa-miR-921 with pCBR1-1096A resulted in differential regulation of luciferase activity. That is, co-transfection of hsa-miR-921 with the pCBR1-1096A construct failed to produce a significant decrease in luciferase activity while hsa-miR-574-5p decreased luciferase activity by 52% (p<0.0001), representing a≈1.5-fold stronger binding interaction compared to its activity with pCBR1-1096G (Figure 2B). Co-transfection of specific hairpin inhibitors for hsa-miR-574-5p was sufficient to significantly rescue the luciferase activities of the pCBR1-1096G and pCBR1-1096A constructs (Figure 2C and 2D). Similar experiments with specific hairpin inhibitors for hsa-miR-921 showed rescue of luciferase activity for the pCBR1-1096G construct (Figure 2C).
Figure 2. Luciferase activity in CHO cells transfected with various microRNA mimics.
Luciferase activities in pCBR1-1096G (A) and pCBR1-1096A (B) constructs transfected with miRNA mimics in the presence (C) or absence (D) of specific miRNA inhibitors were measured as light intensity with a dual-luciferase assay. For each construct, normalized luciferase activities were expressed relative to the values from control transfections (miRNA negative control mimic). Data represent the mean±S.D. from three independent experiments performed in triplicate. *: p<0.05, **: p<0.01; ***: p<0.001; ****: p<0.0001; +: p>0.05 (Student's t-test).
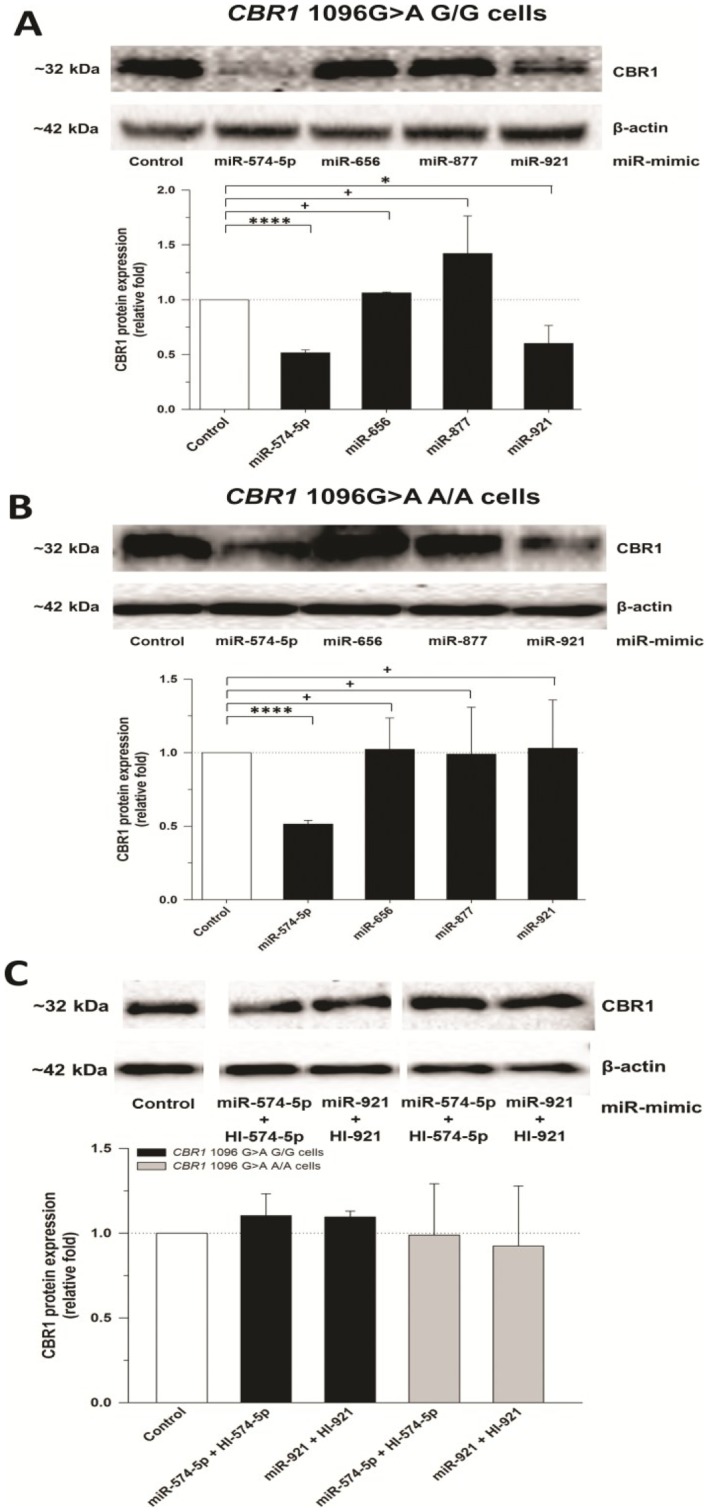
Hsa-miR-574-5p and hsa-miR-921 regulate CBR1 protein expression in human lymphoblastoid cell lines
The results in CHO cells showed that hsa-miR-574-5p interacts with CBR1 3′-UTR constructs carrying the G or the A allele for rs9024 while hsa-miR-921 selectively binds only to the CBR1 3′-UTR construct carrying the G allele. To test the effect of these microRNAs in human cells we chose to utilize lymphoblastoid cell lines from the HapMap Project with known rs9024 (CBR1 1096G>A) genotype status. We have shown that CBR1 1096G>A genotype status impacts CBR1 mRNA and CBR1 activity in lymphoblastoid cell lines [11]. Therefore, these cell lines represent a useful model to further investigate the effect of rs9024 genotype status on miRNA/CBR1 interactions. Here, we utilized lymphoblastoid cells from two from donors with homozygosis for the major G allele (GM 16688 and GM 10857) and two from individuals with homozygosis for the minor A allele (GM 10853 and GM 17240). Lymphoblastoid cells homozygous for the major rs9024 G allele treated with hsa-miR-574-5p or hsa-miR-921 showed a 48% (p<0.0001) and 40% (p<0.05) reduction in CBR1 protein expression, respectively (Figure 3A). Of note, in lymphoblastoid cells homozygous for the minor rs9024 A allele, treatment with miR-574-5p reduced CBR1 protein expression by 49% (p<0.0001), while miR-921 incubation had no significant effect (Figure 3B). Consistent with their lack of interaction with CBR1 3′-UTR reporters in CHO cells, miR-656 and miR-877 failed to induce significant changes in CBR1 protein levels in lymphoblastoid cells homozygous for either rs9024 G or A alleles (Figure 3A and 3B). In both cell lines, specific miRNA inhibitors were capable of completely rescuing CBR1 protein expression (Figure 3C).
Figure 3. CBR1 protein expression in human lymphoblastoid cells transfected with microRNAs.
β-actin-normalized CBR1 levels in lymphoblastoid cell lines with homozygosis for the CBR1 1096G>A SNP G allele (A) and the A allele (B) were interpolated from calibration curves of recombinant CBR1. Experiments were repeated in the presence of specific miRNA inhibitors (C). Shown first are representative western blots of CBR1 and β-actin protein. Data represent the mean±S.D. from three independent experiments performed in duplicate. ****: p<0.0001; +: p>0.05 (Student's t-test).
Kinetic analysis
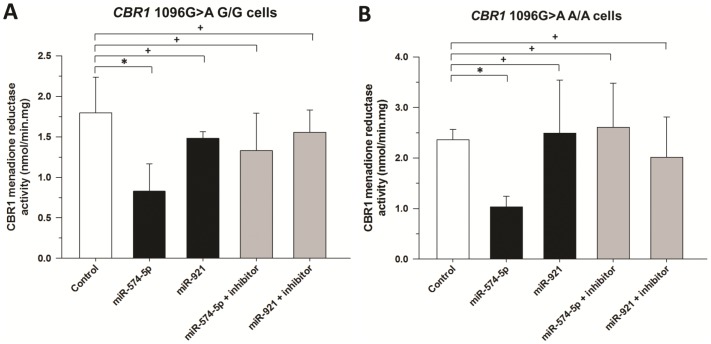
Cytosols from lymphoblastoid cells transfected with miRNA mimics or miRNA mimics plus inhibitors were incubated with 250 µM menadione, a prototypical CBR1 substrate. CBR1 activity was significantly decreased in lymphoblastoid cells transfected with hsa-miR-574-5p (0.83±0.34 nmol/min.mg; 54% decrease; p = 0.04) and moderately reduced by hsa-miR-921 (1.48±0.08 nmol/min.mg; 18% decrease; p = 0.29), whereas co-transfection of hsa-miR-574-5p (1.33±0.46 nmol/min.mg) and hsa-miR-921 (1.55±0.27 nmol/min.mg) inhibitors rescued CBR1 menadione activity (Figure 4A). In agreement with the pattern of inhibition of CBR1 protein expression in lymphoblastoid lines homozygous for the minor rs9024 A allele, miR-574-5p diminished CBR1 activity by 56% (p = 0.001) while miR-921 exerted no significant impact on CBR1 activity (p = 0.84) (Figure 4B).
Figure 4. CBR1 activity in lymphoblastoid cells for the substrate menadione.
Maximal CBR1 activities for the substrate menadione were determined in human lymphoblastoid cells with homozygosis for the CBR1 1096G>A SNP G allele (A) and the A allele (B). Enzymatic activity was assayed in triplicate. *: p<0.05; +: p>0.05.
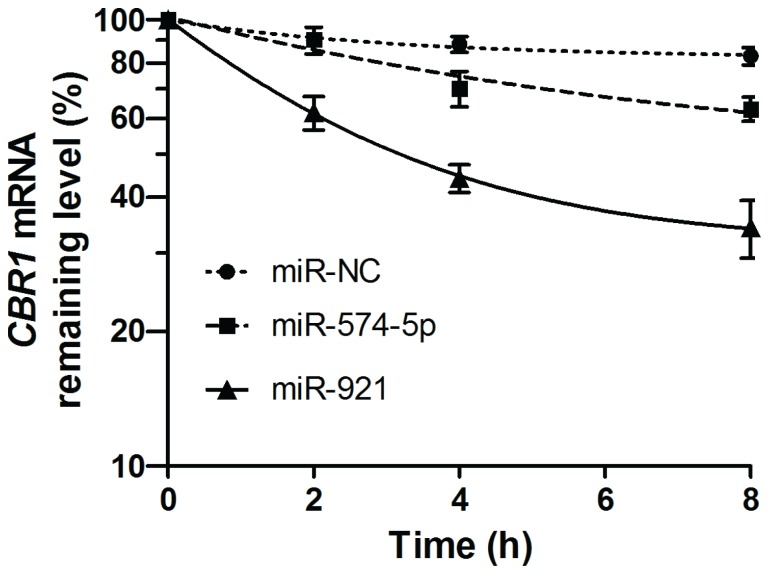
Hsa-miR-574-5p- and hsa-miR-921-mediated regulation of CBR1 involves mRNA degradation
To elucidate the role of mRNA degradation in hsa-miR-574-5p- and hsa-miR-921-mediated post-transcriptional regulation of CBR1, we conducted RNA degradation experiments. The lymphoblastoid cell line GM 10857 was co-transfected with hsa-miR-574-5p and hsa-miR-921 for 48 hours and then treated with actinomycin D, a potent de novo transcription inhibitor. Inspection of apparent RNA half-lives demonstrated that both hsa-miR-574-5p (CBR1 mRNA t1/2>8 hours, but<<CBR1 mRNA t1/2 of miRNA negative control) and hsa-miR-921 (CBR1 mRNA t1/2≈3.5 hours) accelerated decay of CBR1 mRNA compared to the miRNA mimic negative control (Figure 5).
Figure 5. CBR1 RNA degradation.
Relative CBR1 mRNA levels were determined with a comparative quantitation method in cells transfected with miRNA negative control (hsa-miR-NC), hsa-miR-574-5p, or hsa-miR-921. Individual β-actin mRNA levels were used as normalizers. Results shown are mean±SD from triplicate exposures.
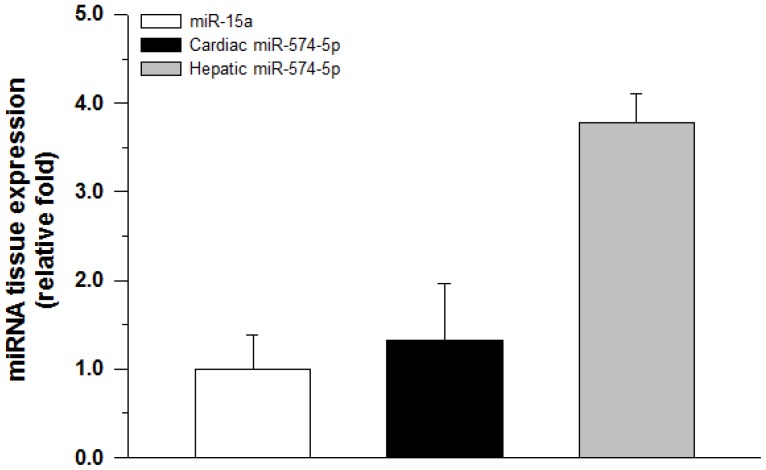
Hsa-miR-574-5p is expressed in human liver and heart tissues
Direct sequencing of real-time PCR products confirmed the specific amplification of our positive control hsa-miR-15a as well as hsa-miR-574-5p. However, commercial primers for hsa-miR-921 failed to amplify the correct mature miRNA sequence. Quantitative RT-PCR gene expression detection of hsa-miR-15a and hsa-miR-574-5p in total RNA isolated from 4 human liver and 4 heart samples showed that both miRNAs were present in all samples (Figure 6). U6-normalized expression levels of hsa-miR-574-5p were favorable compared to individual hsa-miR-15a expression levels in liver (3.8±0.3 relative fold) and heart (1.3±0.7 relative fold).
Figure 6. miR-574-5p expression in human tissues.
Relative miR-574-5p miRNA levels were determined with a comparative quantitation method in total RNA extracted from human heart and human liver samples. Individual U6 expression levels were used as normalizers. Results shown represent the mean±SD expression levels of triplicate experiments from 4 tissue samples of each tissue type.
Discussion
Previously, we reported that hepatic and cardiac CBR1 expression is highly variable [11], [12]. We also pinpointed a SNP in the 3′-UTR of CBR1, rs9024 (CBR1 1096G>A), that has a functional impact on CBR1 mRNA and CBR1 protein expression as well as CBR activity [11]. The physiologic origin of this genotype/phenotype association has not yet been elucidated. Here, we posited that rs9024 may affect microRNA regulation of CBR1 due to its proximity to several bioinformatically-predicted miRNA binding sites on the CBR1 3′-UTR. This interplay could play a significant role in the observed variability of CBR1 expression.
The current study was designed to identify potential miRNA interactions with the 3′-UTR of the CBR1 gene and assess the influence of rs9024 on miRNA-mediated post-transcriptional regulation of CBR1 (Table 2). Bioinformatic predictions led us to examine hsa-miR-574-5p, hsa-miR-656, hsa-miR-877-5p, and hsa-miR-921 as prime CBR1 binding candidates. We demonstrated that miR-574-5p and hsa-miR-921 both influence CBR1 protein expression in human cell lines with homozygosis for the major CBR1 1096G>A G allele (Figure 3A and Table 2). On the other hand, in cell lines homozygous for the minor A allele only hsa-miR-574-5p produced significant decreases in CBR1 protein levels (Figure 3B and Table 2). These reductions in CBR1 protein levels corresponded to substantially diminished maximal CBR1 menadione activity. Of note, a priori it was anticipated that hsa-miR-656 would demonstrate no binding capacity for the rs9024 G allele, but a significant ability to interact with the CBR1 3′-UTR in its rs9024 polymorphic state (A allele) due to the establishment of a novel binding site by the single base-pair substitution. Contrary to these predictions, hsa-miR-656 exhibited no effect on CBR1 expression or activity. It appears that this seemingly surprising result can be explained by the energetically unfavorable free energy conditions that would accompany the binding of hsa-miR-656 to CBR1 (Table 1).
Table 2. Comprehensive CBR1/miRNA results summary.
| G/G/A/A | ||||
| Bioinformatic predictions | miR-574-5p | miR-877-5p | miR-921 | miR-656 |
| CHO cells | 35% ↓/52% ↓ | NC/NC | 46% ↓/NC | N/A/NC |
| Lymphoblastoid protein | 48% ↓/49% ↓ | NC/NC | 40% ↓/NC | N/A/NC |
| Lymphoblastoid activity | 54% ↓/56% ↓ | NC/NC | 18% ↓/NC | N/A/NC |
Although the SNP rs9024 does not directly alter the 3′-UTR target binding sites of hsa-miR-574-5p (position 120) or hsa-miR-921 (position 125) seed regions, it is notable that both binding sites are proximal to the polymorphic site at position 133 (Figure 1). The juxtaposition of the SNP and these target sites may alter the binding of these miRNAs by affecting factors such as ΔGopen, the free energy lost by opening the target site and ΔGduplex, the free binding energy of a miRNA to the target. Indeed, Mishra et al. found that a SNP in the 3′-UTR of the dihydrofolatereductase gene 14 bp downstream of the hsa-miR-24 binding site led to a phenotypic change in methotrexate resistance [28]. Hu and Bruno conducted a genome-wide study of SNPs in genes known to be targeted by miRNAs and discovered that SNPs in seed target regions as well as the entire 3′-UTR of miRNA target genes were under significantly greater negative selection compared to non-miRNA target genes. Moreover, they found that local RNA sequences ∼67 nucleotides symmetrically surrounding miRNA target sites were under moderate strong selection [29]. It is possible that the proximity of hsa-miR-574-5p and hsa-miR-921 binding sites, 13 bp and 8 bp, respectively, upstream of the polymorphic site, is the underlying cause of the changes observed in free energy predictions between the two rs9024 variants of the CBR1 3′-UTR. The accessibility of the CBR1 message to miRNA binding partners may impart differential miRNA regulation of the CBR1 gene leading to downstream impacts on CBR1 protein expression and activity. We hypothesize that this phenomenon contributes to interindividual variability in CBR1 mRNA, CBR1 protein expression and CBR activity which could have major implications in the metabolism of a host of endogenous and xenobiotic substrates.
Importantly, hsa-miR-574-5p and hsa-miR-921 were co-expressed with CBR1 in human liver and heart tissue. Previous studies have reported that CBR1 is the primary anthracycline reductase in liver [11], [13] and participates in the formation of cardiotoxic alcohol metabolites in the heart [12], [30], [31]. MicroRNAs may play a significant role in the regulation of CBR1, although it is important to recognize that they represent one part of a complex array of contributing regulatory factors [12], [24], [32]. Our pilot work detecting the presence of miR-574-5p in liver and heart tissues suggests that there is significant interindividual variability in the expression of this miRNA in these tissues (Figure 6). Future work will attempt to characterize the extent of interindividual variability in the expression of hsa-miR-574-5p and hsa-miR-921 in human tissues in tandem with CBR1 1096G>A genotype and assess their combined impact on CBR1 phenotypic variability.
Funding Statement
This work was supported by National Institutes of Health/National Institute of General Medical Sciences grant GM73646. James L. Kalabus is a Daiichi Sankyo fellow. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ambros V (2004) The functions of animal microRNAs. Nature 431: 350–355. [DOI] [PubMed] [Google Scholar]
- 2. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 3. He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531. [DOI] [PubMed] [Google Scholar]
- 4. Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ambros V (2003) MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113: 673–676. [DOI] [PubMed] [Google Scholar]
- 6. Chang TC, Mendell JT (2007) microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet 8: 215–239. [DOI] [PubMed] [Google Scholar]
- 7. Kloosterman WP, Plasterk RH (2006) The diverse functions of microRNAs in animal development and disease. Dev Cell 11: 441–450. [DOI] [PubMed] [Google Scholar]
- 8. Forrest GL, Gonzalez B (2000) Carbonyl reductase. Chem Biol Interact 129: 21–40. [DOI] [PubMed] [Google Scholar]
- 9. Forrest GL, Gonzalez B, Tseng W, Li X, Mann J (2000) Human carbonyl reductase overexpression in the heart advances the development of doxorubicin-induced cardiotoxicity in transgenic mice. Cancer Res 60: 5158–5164. [PubMed] [Google Scholar]
- 10. Hoffmann F, Maser E (2007) Carbonyl reductases and pluripotent hydroxysteroid dehydrogenases of the short-chain dehydrogenase/reductase superfamily. Drug Metab Rev 39: 87–144. [DOI] [PubMed] [Google Scholar]
- 11. Gonzalez-Covarrubias V, Zhang J, Kalabus JL, Relling MV, Blanco JG (2009) Pharmacogenetics of human carbonyl reductase 1 (CBR1) in livers from black and white donors. Drug Metab Dispos 37: 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kalabus JL, Sanborn CC, Jamil RG, Cheng Q, Blanco JG (2010) Expression of the anthracycline-metabolizing enzyme carbonyl reductase 1 in hearts from donors with Down syndrome. Drug Metab Dispos 38: 2096–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kassner N, Huse K, Martin HJ, Godtel-Armbrust U, Metzger A, et al. (2008) Carbonyl reductase 1 is a predominant doxorubicin reductase in the human liver. Drug Metab Dispos 36: 2113–2120. [DOI] [PubMed] [Google Scholar]
- 14. Rukov JL, Shomron N (2011) MicroRNA pharmacogenomics: post-transcriptional regulation of drug response. Trends Mol Med 17: 412–423. [DOI] [PubMed] [Google Scholar]
- 15. Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39: 1278–1284. [DOI] [PubMed] [Google Scholar]
- 16. Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 17. Griffiths-Jones S (2006) miRBase: the microRNA sequence database. Methods Mol Biol 342: 129–138. [DOI] [PubMed] [Google Scholar]
- 18. Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kruger J, Rehmsmeier M (2006) RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34: W451–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bao L, Zhou M, Wu L, Lu L, Goldowitz D, et al. (2007) PolymiRTS Database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Res 35: D51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ziebarth JD, Bhattacharya A, Chen A, Cui Y (2012) PolymiRTS Database 2.0: linking polymorphisms in microRNA target sites with human diseases and complex traits. Nucleic Acids Res 40: D216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, et al. (2005) Mapping determinants of human gene expression by regional and genome-wide association. Nature 437: 1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, et al. (2007) Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet 39: 226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kalabus JL, Cheng Q, Jamil RG, Schuetz EG, Blanco JG (2012) Induction of carbonyl reductase 1 (CBR1) expression in human lung tissues and lung cancer cells by the cigarette smoke constituent benzo[a]pyrene. Toxicol Lett 211: 266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bohren KM, von Wartburg JP, Wermuth B (1987) Kinetics of carbonyl reductase from human brain. Biochem J 244: 165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gonzalez-Covarrubias V, Kalabus JL, Blanco JG (2008) Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid 7-monohydroxyethyl rutoside (monoHER). Pharm Res 25: 1730–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lakhman SS, Ghosh D, Blanco JG (2005) Functional significance of a natural allelic variant of human carbonyl reductase 3 (CBR3). Drug Metab Dispos 33: 254–257. [DOI] [PubMed] [Google Scholar]
- 28. Mishra PJ, Humeniuk R, Longo-Sorbello GS, Banerjee D, Bertino JR (2007) A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci U S A 104: 13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu Z, Bruno AE (2011) The Influence of 3′UTRs on MicroRNA Function Inferred from Human SNP Data. Comp Funct Genomics 2011: 910769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Menna P, Recalcati S, Cairo G, Minotti G (2007) An introduction to the metabolic determinants of anthracycline cardiotoxicity. Cardiovasc Toxicol 7: 80–85. [DOI] [PubMed] [Google Scholar]
- 31. Mordente A, Meucci E, Silvestrini A, Martorana GE, Giardina B (2009) New developments in anthracycline-induced cardiotoxicity. Curr Med Chem 16: 1656–1672. [DOI] [PubMed] [Google Scholar]
- 32. Lakhman SS, Chen X, Gonzalez-Covarrubias V, Schuetz EG, Blanco JG (2007) Functional characterization of the promoter of human carbonyl reductase 1 (CBR1). Role of XRE elements in mediating the induction of CBR1 by ligands of the aryl hydrocarbon receptor. Mol Pharmacol 72: 734–743. [DOI] [PubMed] [Google Scholar]