Abstract
This study demonstrated that foliar infection by Pseudomonas syringae pv tomato DC3000 induced malic acid (MA) transporter (ALUMINUM-ACTIVATED MALATE TRANSPORTER1 [ALMT1]) expression leading to increased MA titers in the rhizosphere of Arabidopsis (Arabidopsis thaliana). MA secretion in the rhizosphere increased beneficial rhizobacteria Bacillus subtilis FB17 (hereafter FB17) titers causing an induced systemic resistance response in plants against P. syringae pv tomato DC3000. Having shown that a live pathogen could induce an intraplant signal from shoot-to-root to recruit FB17 belowground, we hypothesized that pathogen-derived microbe-associated molecular patterns (MAMPs) may relay a similar response specific to FB17 recruitment. The involvement of MAMPs in triggering plant innate immune response is well studied in the plant’s response against foliar pathogens. In contrast, MAMPs-elicited plant responses on the roots and the belowground microbial community are not well understood. It is known that pathogen-derived MAMPs suppress the root immune responses, which may facilitate pathogenicity. Plants subjected to known MAMPs such as a flagellar peptide, flagellin22 (flg22), and a pathogen-derived phytotoxin, coronatine (COR), induced a shoot-to-root signal regulating ALMT1 for recruitment of FB17. Micrografts using either a COR-insensitive mutant (coi1) or a flagellin-insensitive mutant (fls2) as the scion and ALMT1pro:β-glucuronidase as the rootstock revealed that both COR and flg22 are required for a graft transmissible signal to recruit FB17 belowground. The data suggest that MAMPs-induced signaling to regulate ALMT1 is salicylic acid and JASMONIC ACID RESISTANT1 (JAR1)/JASMONATE INSENSITIVE1 (JIN1)/MYC2 independent. Interestingly, a cell culture filtrate of FB17 suppressed flg22-induced MAMPs-activated root defense responses, which are similar to suppression of COR-mediated MAMPs-activated root defense, revealing a diffusible bacterial component that may regulate plant immune responses. Further analysis showed that the biofilm formation in B. subtilis negates suppression of MAMPs-activated defense responses in roots. Moreover, B. subtilis suppression of MAMPs-activated root defense does require JAR1/JIN1/MYC2. The ability of FB17 to block the MAMPs-elicited signaling pathways related to antibiosis reflects a strategy adapted by FB17 for efficient root colonization. These experiments demonstrate a remarkable strategy adapted by beneficial rhizobacteria to suppress a host defense response, which may facilitate rhizobacterial colonization and host-mutualistic association.
Plant roots are the first organs that come in contact with diverse belowground microflora. Rhizospheric microbes, which utilize plant root exudates for growth and multiplication (Lugtenberg et al., 2001; Bais et al., 2006; Rudrappa et al., 2008b), are attracted to the rhizosphere, and may have either beneficial or deleterious effects on the plant. Classical examples are beneficial mycorrhizal fungi that provide the host with an enhanced root surface for absorbing water and mineral nutrients, notably phosphate (Harrison, 2005), and Rhizobium spp. that fix atmospheric nitrogen into ammonium that can be used by the plant for amino acid biosynthesis (Spaink, 2000). Several other types of beneficial soilborne microbes, such as plant growth-promoting rhizobacteria (PGPR) and fungi, can stimulate plant growth by suppressing plant diseases (van Loon et al., 1998) or insect herbivory (van Oosten et al., 2008). The biological control activity is exerted either directly through antagonism of soilborne pathogens or indirectly by eliciting a plant-mediated resistance response (van Loon et al., 1998; Pozo and Azcón-Aguilar, 2007).
The resistance responses mediated by PGPRs are either through systemic acquired resistance (SAR) or induced systemic resistance (ISR), both of which function systemically throughout the plant (Conrath et al., 2002). PGPRs activate ISR (van der Ent et al., 2009) whereas SAR is triggered by necrotizing pathogens (Conrath et al., 2002). SAR is controlled by the salicylic acid (SA)-dependent signaling pathway, and its onset involves local and systemic increases in endogenously synthesized SA, leading to activation of the regulatory protein NPR1 and the subsequent NPR1-dependent expression of genes encoding pathogenesis-related (PR) proteins, including PR1, PR2, and PR5 (van Loon and van Strien, 1999). Nonpathogenic PGPRs regulate ISR by jasmonic acid (JA)- and ethylene (ET)-dependent signaling pathways and are associated with the downstream regulation of plant defensin1.2 (PDF1.2; van Oosten et al., 2008). Pieterse et al. (1998) reported that ISR triggered by Pseudomonas fluorescens WCS417r signals resistance responses through JA- and ET-dependent pathways. Rhizobacteria-mediated ISR has been demonstrated in a variety of plants including bean (Phaseolus vulgaris), carnation (Dianthus caryophyllus), cucumber (Cucumis sativus), radish (Raphanus sativus), tobacco (Nicotiana tabacum), tomato (Solanum lycopersicum), and the model plant Arabidopsis (Arabidopsis thaliana; van Loon et al., 1998). Beneficial rhizobacteria trigger ISR by priming the plant for potentiated activation of various cellular defense responses, which are subsequently induced upon pathogen attack (Conrath et al., 2006). The potentiated responses include oxidative burst (Iriti and Faoro, 2003), cell wall reinforcements (Benhamou et al., 1996), accumulation of defense-related materials and enzymes (Chen et al., 2000), secondary metabolite production (Ongena et al., 2000), and impediment of infection processes of pathogens such as inhibition of sporangia and zoospore germination (Yan et al., 2002). Lipopolysaccharides (LPS), siderophores, or SA from rhizobacteria also are indispensable for successful disease protection (De Meyer et al., 1999; for review, see Ramamoorthy et al., 2001). In connection with ISR and PGPR, Niu et al. (2011), reported that Bacillus cereus AR156 induces ISR in Arabidopsis by simultaneously activating SA- and JA/ET-dependent signaling pathways.
Despite progress toward understanding the microbe-mediated plant responses in plant-microbe interactions, little headway has been made in identifying the genetic and biochemical changes responsible for the attraction of beneficial symbiotic rhizospheric microbes. Under herbivory, wounding of plant tissues by insect feeding triggers the release of volatile signals that attract natural enemies of insects (Kessler and Baldwin, 2001). Upon insect infestation, plants release compounds such as hormones, exogenous volatile organic compounds, and secondary metabolites as long-distance root-to-shoot signals (for review, see Erb et al., 2009). Whitefly infestation of pepper (Capsicum annuum) plants elicits SA-dependent signaling in leaves whereas roots showed increased colonization of Gram-positive bacterial populations (Yang et al., 2011), suggesting a signaling event between aboveground and belowground plant parts. Whereas considerable data exist on the occurrence of aboveground/belowground communication in the case of plant herbivory, evidence of similar phenomena in plant-pathogenic bacteria interactions is lacking. Some of the recent reports indicate that the hormonal activation of ISR by ET/JA or SAR by SA showed no significant effect on the density and structure of the rhizosphere bacterial community (Doornbos et al., 2011). On the contrary, a recent report suggests that ATP-binding cassette transporters involved in root secretions may structure the rhizospheric microbiome (Badri et al., 2008). Carbon enrichment of the rhizosphere especially carboxylate excretion and acidification at the root surface might have a strong general impact on structuring rhizospheric microbial communities (Marschner et al., 2002). Tricarboxylic acids such as malic acid (MA) and citrate are suitable carbon sources for many microorganisms (López-Bucio et al., 2000; for review, see Pineda et al., 2010). A recent study from our group showed that inoculation of Arabidopsis leaves with the foliar pathogen Pseudomonas syringae pv tomato (Pst) DC3000 induced MA excretion in roots (Rudrappa et al., 2008b). The study revealed that Pst DC3000-infected shoots relay chemical signal(s) underground through root MA secretion, resulting in specific chemotaxis to recruit rhizobacteria Bacillus subtilis strain FB17. The authors further demonstrated that infection of Arabidopsis leaves up-regulated root ALUMINUM-ACTIVATED MALATE TRANSPORTER1 (ALMT1). These findings were further validated by Chen et al. (2012), wherein exudates of tomato roots strongly stimulated B. subtilis biofilm formation ex planta and that an abundant small molecule in the exudates, l-MA, was able to stimulate biofilm formation at high concentrations in a manner that was dependent on the KinD CACHE domain.
When pathogenic bacteria infect, plants recognize molecules common to many classes of microbes called microbe-associated molecular patterns (MAMPs), such as bacterial flagellin (flg22; Felix et al., 1999) and bacterial elongation factor Tu (Kunze et al., 2004). Other MAMPs include chitin, a major component of the fungal cell wall (Wan et al., 2008), LPS (Zeidler et al., 2004), and peptidoglycans (PGNs; Gust et al., 2007). MAMPs in leaves including flg22 induce overlapping genes (Zipfel et al., 2004). MAMPs in leaves trigger an oxidative burst, ET, and nitric oxide leading to activation of defense response genes (for review, see Zhang and Zhou, 2010). First, purified flagellin from Pseudomonas putida WCS358, as well as LPS from P. fluorescens WCS417r and P. putida WCS358, were shown to trigger ISR against Pst DC3000 in Arabidopsis (Meziane et al., 2005). Further studies showed that flg22-induced gene expression and regulation is SA independent and SA dependent in early and late phases after flg22 administration (Vlot et al., 2009). Second, Rhizobium Nod factors, which are structurally related to chitin, are recognized by LysM receptor kinases in legume roots (Limpens et al., 2003). From the abovementioned studies, it is clear that MAMPs-based interactions modulate plant innate immunity to create a “primed” state. However, many pathogens have evolved strategies to counteract the plant immune response, including the involvement of direct injection of virulence effectors through the type III secretion system (Block et al., 2008). Many Pst DC3000 pathovars secrete coronatine (COR), a low Mr phytotoxin that functions in leaves as a mimic of JA-Ile (Kunkel and Brooks, 2002). By activating the JA pathway, COR triggers a mutually antagonistic interaction between the SA and JA signaling pathways and suppresses SA signaling, a key component in basal resistance against Pst DC3000. In addition, COR suppresses the flg22-elicited activation of the Arabidopsis gene NHO1, which is important for resistance against Pst DC3000 infection (Li et al., 2005). Finally, COR suppresses MAMPs-induced stomatal closure, believed to block epiphyte pathogens such as Pst DC3000 from entering the interior of leaves through these natural openings (Melotto et al., 2006). In contrast with leaves, relatively little is known about MAMPs-mediated responses in roots. Recently, the presence of a sensitive MAMPs-triggered immune system was described in Arabidopsis roots (Millet et al., 2010). In the same study, it was shown that MAMPs elicit callose deposition on roots and exudation of the antimicrobial compound camalexin. A microarray analysis of the initial phase of colonization by B. subtilis, showed a suppression of root defense-related gene expression (Niu et al., 2011).
The root-colonizing PGPR, FB17, represents a useful model to study principles of plant root-microbe interactions in terms of mutualism and root colonization. As a mutualist, FB17 confers beneficial traits such as increased abiotic stress tolerance and disease resistance to plants (Rudrappa et al., 2008b, 2010; Zhang et al., 2008, 2010; for review, see Choudhary and Johri, 2009). Herein, we show that foliar applied MAMPs and COR elicited FB17 colonization in Arabidopsis roots through activation of root ALMT1 expression and those FLS2- and COI1-dependent graft-transmissible long-distance signals recruited FB17 belowground. In addition, we show that MAMPs activation of ALMT1 was independent of SA and transcription factor JASMONATE INSENSITIVE1 (JIN1)/MYC2 in JA pathways. Our results reveal that the suppression of MAMPs-activated defense response by B. subtilis (FB17) was independent of biofilm formation and dependent on JIN1/MYC2 in the JA signaling pathway. The demonstration that FB17 selectively down-regulated root-specific defense genes and MAMPs-triggered innate plant responses establishes a strategy adapted by FB17 to effectively colonize host roots and avoid antagonism.
RESULTS
Foliar MAMPs Exposure Elicits Root Colonization by FB17 in Arabidopsis
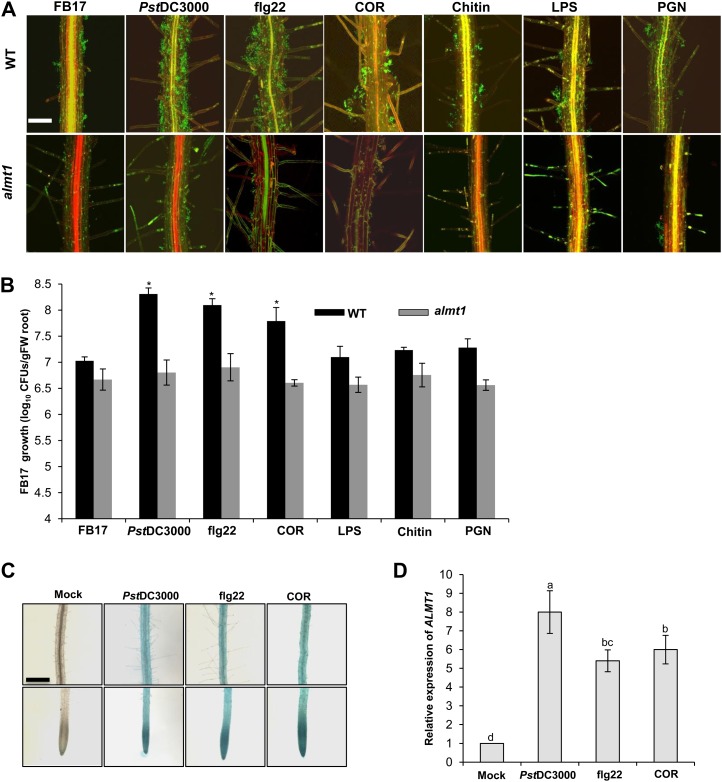
To determine if a foliar spray of MAMPs could influence the recruitment of beneficial rhizosphere bacteria, root-specific colonization was measured in the presence and absence of different MAMPs. Twenty-day-old Arabidopsis wild-type Columbia-0 and ALMT1 knockout (almt1) plants were rhizoinoculated with the beneficial rhizobacteria, FB17, and then subsequently foliar sprayed with different MAMPs and phytotoxin COR. After 3 d of treatments, leaves sprayed with MAMPs/COR or infected with Pst DC3000 stimulated FB17 colonization both qualitatively (confocal microscopy; Fig. 1A) and quantitatively (colony-forming units [cfu]; Fig. 1B). Remarkably, within 3 d of COR (5 µm) or flg22 (1 µm) treatment, there was approximately a 10-fold increase in FB17 colonization compared with the mock treatment (Fig. 1B). To validate the specificity in FB17 root colonization post flg22 or COR treatment, three additional MAMPs, LPS (500 µg mL−1), chitin (500 µg mL−1), and PGN (500 µg mL−1), were also tested for FB17 colonization in Arabidopsis roots. The response to these three MAMPs were much weaker and more variable than flg22-, COR-, or Pst DC3000-elicited responses (Fig. 1, A and B). These data, that aerial MAMPs treatments, specifically flg22 and phytotoxin COR, could induce FB17 root colonization, suggest that plants sense and trigger an intraplant response to recruit beneficial rhizobacteria belowground.
Figure 1.
Foliar MAMPs elicit FB17 colonization on Arabidopsis roots. A, Confocal images showing differential root FB17 colonization. Three-week-old in vitro seedlings of the wild type or almt1 plants were foliar sprayed with flg22 (1 µm), COR (5 µm), chitin (500 µg mL−1), LPS (500 µg mL−1), PGN (500 µg mL−1), Pst DC3000 (OD600 = 0.1) or an equal volume of water as the control and rhizoinoculated with FB17 (OD600 = 0.001) for 24 h. The green fluorescence in the panels shows FB17. Scale bars = 50 µm. B, FB17 growth quantification on the roots by cfu. Three-week-old pellet-grown wild-type or almt1 plants were foliar sprayed with flg22 (1 µm), COR (5 µm), chitin (500 µg mL−1), LPS (500 µg mL−1), PGN (500 µg mL−1), Pst DC3000 (OD600 = 0.1) or an equal volume of water as control and rhizoinoculated with 4 mL/pellet of FB17 (OD600 = 0.5) and incubated for 72 h. Data represent the mean ± se. *P ≤ 0.05; two-tailed Student’s t test (se values are from 24 independent measurements from two experiments). C, flg22 and COR foliar treatment elicits ALMT1 expression in the root central elongation, meristematic, and maturation regions. Three-week-old in vitro-grown transgenic seedlings carrying ALMT1pro:GUS promoter were foliar sprayed with flg22 (1 µm), COR (5 µm) Pst DC3000 (OD600 = 0.1), or an equal volume of water as the experimental control. GUS staining of ALMT1pro:GUS seedlings was performed 24 h post treatment. Scale bars = 50 µm, common to all panels. D, Measurement of ALMT1 expression in the roots of plants foliar sprayed with MAMPs. Total RNA was isolated and sqRT-PCR performed. Data represent mean ± se. Lowercase letters indicate the statistical significance among different treatments according to DMRT at P ≤ 0.05 (se values are three technical replicates of one experiment, repeated twice with similar results). [See online article for color version of this figure.]
Aerial MAMPs Trigger Root ALMT1 Expression
Our laboratory previously reported that Pst DC3000 aerial infection enhanced ALMT1 mediated-MA secretion in the rhizosphere leading to increased FB17 root colonization (Rudrappa et al., 2008b). Concomitantly, the data above also showed that foliar spray with flg22 or COR led to induced FB17 binding, which may be mediated through root ALMT1 expression. To further test this, we employed almt1, which is known to be deficient in root MA secretion (Hoekenga et al., 2006). FB17 showed a significantly lower extent of colonization on the root surface of almt1 under flg22-, COR-, mock- (water control), or positive control Pst DC3000-infected conditions as shown by both microscopic root binding (Fig. 1A) and cfu data (Fig. 1B). To substantiate that ALMT1 expression is activated by flg22 or COR, we employed Arabidopsis transgenic line carrying an ALMT1pro:GUS fusion construct. The in vitro-grown ALMT1pro:GUS and the wild type were foliar sprayed with flg22 (1 µm), COR (5 µm), LPS (500 µg mL−1), chitin (500 µg mL−1), PGN (500 µg mL−1), water, or Pst DC3000 (optical density at 600 nm [OD600] = 0.1; positive control). ALMT1pro:GUS expression was higher in the roots in the flg22 or COR treatments (Fig. 1C). Similarly, semiquantitative reverse transcription (sqRT)-PCR analysis of ALMT1 expression shows increased levels of ALMT1 in the COR- and flg22-treated plants (approximately 6- and 5.5-fold, respectively) compared with the mock treatments (Fig. 1D). However, the other bacterial-derived MAMPs such as chitin, LPS, and PGN did not show any induction of ALMT1 expression (Supplemental Fig. S1, A and B), suggesting that COR or flg22 may be involved in FB17 root-symbiotic colonization response.
In a previous study (Millet et al., 2010), it was shown that bacterial MAMPs flg22, chitin, and PGN were recognized in roots and induced genes involved in the plant immune response. Therefore, to further confirm this finding of a flg22-specific induction of ALMT1 expression, chitin, LPS, PGN, or flg22 were applied to leaves of Arabidopsis transgenic lines carrying an CYP71A12pro:GUS, MYB51pro:GUS, and WRKY11pro:GUS fusion construct and expression of MAMPs responsive defense marker genes CYP71A12, MYB51, and WRKY11 in leaves were quantified by employing GUS staining (Supplemental Fig. S2A) and expression of MAMPs responsive defense marker genes were monitored by sqRT-PCR in wild-type plants (Supplemental Fig. S2, A and B). Out of the three genes, MYB51 and WRKY11 showed MAMPs-responsive induced defense expression in leaves post MAMPs treatment (Supplemental Fig. S2, A and B). The data clearly showed that the concentration of other MAMPs (chitin, LPS, and PGN) used in our study was adequate to induce MAMPs-responsive defense genes locally (Supplemental Fig. S2, A and B). These experiments confirm that all MAMPs and CORs induce defense markers in leaves, but just COR and flg22 are able to induce root ALMT1.
MAMPs Insensitive Mutants, coi1 and fls2, Negate Induction of ALMT1 Expression and FB17 Recruitment
Having shown that a foliar spray of either COR or flg22 triggers FB17 root binding in Arabidopsis, we next evaluated the involvement of FLS2 and COI1 in recruiting FB17 belowground. It has been reported that FLS2 represents a functional flg22 receptor (Gómez-Gómez et al., 2001) and COR blocks SA-signaling and stomatal closure through COI1, an E3 ubiquitin ligase involved in JA signaling and a key component of the defense response against necrotrophic pathogens and insect herbivores (Xie et al., 1998). We compared the levels of ALMT1 expression in the roots of coi1 (for COR insensitive) and fls2 (flagellin receptor mutant) sprayed with COR or flg22. There was a significant reduction (P ≤ 0.05) of ALMT1 in coi1 sprayed with COR and in fls2 sprayed with flg22 compared with the positive control Pst DC3000 (Supplemental Fig. S3A). Moreover, flg22 sprayed on coi1 and COR sprayed on fls2 showed increased root ALMT1 accumulation, although the expression was less than with Pst DC3000-infected plants. To test if a foliar spray of COR or flg22 modulates the FB17 root colonization patterns, root inoculation of FB17 was conducted on wild-type, coi1, and fls2 backgrounds and quantified (Supplemental Fig. S3B). In the fls2 background, the overall root FB17 colonization was lower in the flg22-treated and Pst DC3000-treated plants compared with wild-type controls. Similarly, COR-treated or Pst DC3000-treated coi1 plants had lower root FB17 colonization compared with wild-type plants, indicating that FLS2- and COI1-mediated signaling pathways are required for ALMT1 expression (Supplemental Fig. S3, A and B).
Both flg22 and COR Trigger a FLS2- and COI1- Dependent Graft-Transmissible Long-Distance Signal to Recruit FB17 Belowground
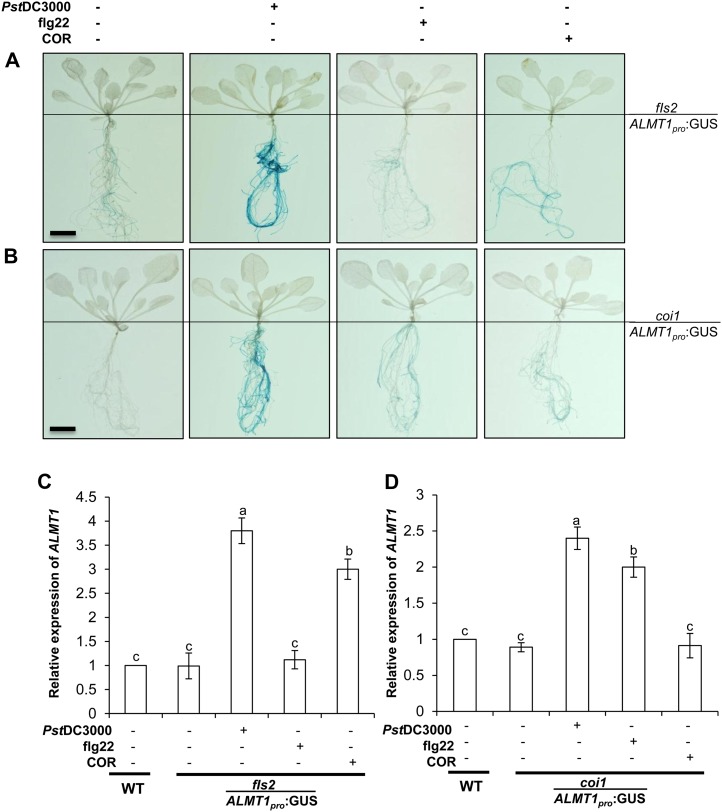
To confirm that both COR- and flg22-induced ALMT1 involves an intraplant shoot-to-root ALMT1 signal, insensitive mutants coi1 and fls2 scions were micrografted onto ALMT1pro:GUS rootstocks. After maturation of the micrografts, the leaves were sprayed with flg22, COR, or Pst DC3000. After 24 h of treatment, whole plants were stained for GUS expression (Fig. 2, A and B), and ALMT1 expression in rootstocks was quantified by sqRT-PCR (Fig. 2, C and D). Untreated grafts were used as controls. ALMT1pro:GUS expression in the roots of fls2-scion micrografts stained deeper blue after being sprayed by COR and Pst DC3000 compared with grafts sprayed with flg22 (Fig. 2A). ALMT1pro:GUS expression in the roots of coi1-scion micrografts were deeper blue after being sprayed with either flg22 or Pst DC3000 compared with grafts sprayed with COR (Fig. 2B). In contrast, wild-type scions grafted over ALMT1pro:GUS rootstocks showed an ALMT1 expression with flg22, COR, or Pst DC3000 treatments (Supplemental Fig. S4). However, in both micrograft sets, Pst DC3000-treated plants displayed the deepest blue staining, suggesting regulation of ALMT1 through FLS2 and COI1. As described above, the ALMT1pro:GUS stocks with fls2 scion grafts sprayed with COR or Pst DC3000 showed increased root ALMT1 expression compared with grafts sprayed with flg22 (Fig. 2C). Similarly, ALMT1pro:GUS stocks with coi1 scion grafts sprayed with flg22 or Pst DC3000 showed increased root ALMT1 expression, compared with grafts sprayed with COR, based on sqRT-PCR analysis (Fig. 2D). These results indicate that in the flagellin receptor mutant, fls2, the induction of root-ALMT1 expression is prevented when sprayed with flg22 but still occurs when sprayed with COR or treated with Pst DC3000.
Figure 2.
MAMPs trigger a FLS2- and COI1-dependent graft transmissible signal to trigger root ALMT1 expression. A and B, Two weeks after graft maturation, micrografts were transferred to liquid medium for 4 d and foliar sprayed with flg22 (1 µm), COR (5 µm), or Pst DC3000 (OD600 = 0.1). Plants were incubated for 24 h and subjected to GUS staining. The images are a representative sample of six plants. Scale bars = 4 mm, common to all panels. Graft notation is fls2/ALMT1pro:GUS, where fls2 is scion and ALMT1pro:GUS is rootstock and coi1/ALMT1pro:GUS, where coi1 is scion and ALMT1pro:GUS is rootstock. C and D, Measurement of root ALMT1 expression in the micrografts fls2/ALMT1pro:GUS and coi1/ALMT1pro:GUS foliar sprayed with flg22 (1 µm), COR (5 µm), or Pst DC3000 (OD600 = 0.1). Total RNA was isolated after 24 h of incubation and sqRT-PCR was performed. Data represent the mean ± se. The lowercase letters represent statistical differences at P ≤ 0.05 according to DMRT (se values are three technical replicates of one experiment). [See online article for color version of this figure.]
Having shown through the chimeric graft experiments that an intraplant mobile signal may be involved to link aerial MAMPs recognition and root ALMT1 components, we next checked the implications of this intraplant long-distance signaling in root FB17 colonization. In accordance, with the increased ALMT1 expression, the fls2/ALMT1pro:GUS grafts sprayed with COR or Pst DC3000 revealed higher FB17 root colonization compared with those sprayed with flg22 (Supplemental Fig. S5A). It is interesting that the same trend was obtained, wherein the coi1/ALMT1pro:GUS grafts sprayed with flg22 or Pst DC3000 showed higher FB17 binding than those sprayed with COR (Supplemental Fig. S5B). These data suggest there is functional regulation of aboveground MAMPs and belowground ALMT1 cross talk with regard to beneficial microbe recruitment belowground.
flg22 and COR Activation of ALMT1 Expression Is SA/JAR1/JIN1/MYC2 Independent
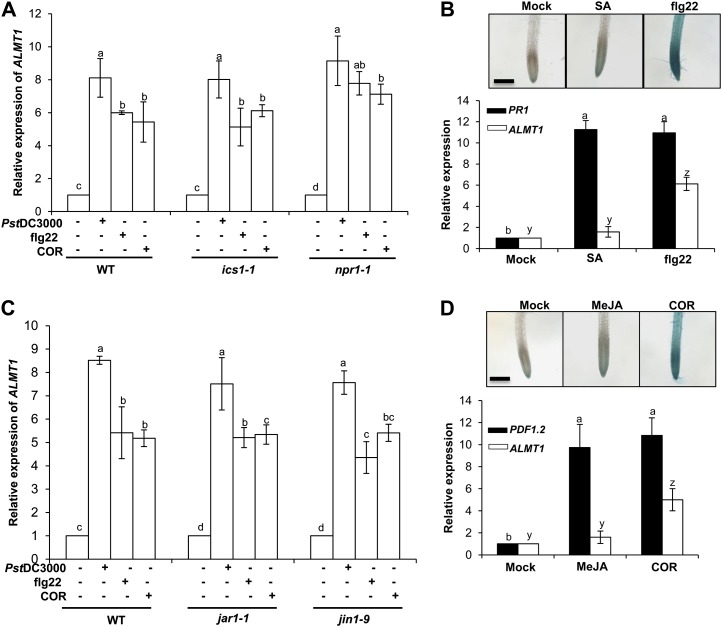
More than a decade of research has shown that SA acts as a major player in systemically transmitting defense signals in many plant systems. Many genes that show induced expression in response to MAMPs are also induced by SA. Tsuda et al. (2008) showed an intimate interaction between MAMPs-triggered responses and SA-mediated signaling mechanisms revealing SA accumulation after flg22 treatment. Herein, we examined the dependence of flg22-triggered ALMT1 expression on SA. To test this, ALMT1 expression in roots was quantified in two SA-impaired lines, ics1-1 and npr1-1, which were foliar treated with flg22 (Fig. 3A). Surprisingly, both ics1-1 and npr1-1 observed normal flg22-elicited root ALMT1 expression compared with root ALMT1 expression in the wild type treated with flg22 (Fig. 3A). For further confirmation of noninvolvement of SA in mediating flg22-triggered ALMT1 expression, the seedlings were treated with exogenous SA, and ALMT1 expression was monitored in ALMT1pro:GUS and quantified by sqRT-PCR. In both experiments, seedlings with exogenous SA did not activate ALMT1 expression (Fig. 3B). In addition, our data clearly showed that SA induced PR1 compared with ALMT1 expression (Fig. 3B), suggesting that the concentration of SA used in our study was adequate for gene expression. Together, these results show that the systemic induction of root ALMT1 expression by flg22 is independent of SA signaling.
Figure 3.
flg22-induced ALMT1 expression is SA/JAR1/JIN1/MYC2 independent. A, Measurement of root ALMT1 expression by sqRT-PCR in the wild type, ics1-1, and npr1-1. Three-week-old in vitro-grown wild type, ics1-1 and npr1-1 were foliar sprayed with flg22 (1 µm), COR (5 µm), Pst DC3000 (OD600 = 0.1), or an equal volume of water as control, and incubated for 24 h. The total root RNA was isolated and was subsequently analyzed for relative expression level of ALMT1. B, Measurement of ALMT1 and PR1 expression by sqRT-PCR in the wild type treated with flg22 (1 µm) or SA (100 µm). RNA isolation was performed 24 h post treatment. Insert: ALMT1 expression in ALMT1pro:GUS plants treated with flg22 (1 µm) or SA (100 µm) was analyzed 24 h post treatment. Scale bar = 50 µm. C, Measurement of root ALMT1 expression by sqRT-PCR in the wild type and mutants jar1-1 and jin1-9. Three-week-old in vitro-grown wild type, jar1-1, and jin1-9 were foliar sprayed with flg22 (1 µm), COR (5 µm), or Pst DC3000 (OD600 = 0.1), an equal volume of water as control and incubated for 24 h. The total root RNA was isolated and was subsequently analyzed for relative expression level of ALMT1. D, Measurement of ALMT1 and PDF1.2 expression by sqRT-PCR in wild-type plants treated with COR (5 µm) and MeJA (100 µm). Insert: ALMT1 expression in ALMT1pro:GUS treated with COR and MeJA. GUS staining and RNA isolation was performed 24 h post treatment. Scale bar = 50 µm. For all the panels, data presented as mean ± se. The lower case letters represent statistical difference at P ≤ 0.05 according to DMRT (se values are three technical replicates of one experiment, repeated twice with similar results). [See online article for color version of this figure.]
It is generally understood that COR and methyl jasmonate (MeJA) are similar in both structure and function (Uppalapati et al., 2005). COR might activate JA signaling downstream of JAR1 (for JASMONIC ACID RESISTANT1; involved in JA-Ile synthesis) and by recruiting transcription factors other than JIN1/MYC2 (Dombrecht et al., 2007). To test the involvement of the JA-signaling pathway in COR-triggered root ALMT1 expression, mutants (jar1-1 and jin1-9) impaired in JA-signaling pathways were tested. The jar1-1 and jin1-9 were foliar treated with COR, and root ALMT1 expression was measured (Fig. 3C). Surprisingly, COR was involved in induction of root ALMT1 expression in both jar1-1 and jin1-9, confirming the noninvolvement of JAR1/JIN1/MYC2 in JA-signaling pathway in COR-triggered root ALMT1 expression. Subsequently, a structural analog of COR, MeJA, was applied exogenously to the wild-type seedlings and root ALMT1 expression was monitored. Interestingly, exogenous treatment of MeJA did not induce ALMT1 expression in roots (Fig. 3D). In contrast, supplementation of MeJA to wild-type plants induced PDF1.2, a marker gene used routinely for characterization of the JA-dependent defense responses (Fig. 3D). These data suggest that although COR mimics MeJA structurally, it may induce a differential intraplant signaling response to trigger ALMT1 expression that may not involve JAR1/JIN1/MYC2 in JA signaling.
Aerial Exposure of MAMPs Induce Root Defense Responses Similar to Live Pathogens
A common root defense mechanism adapted by various plant species is to biosynthesize and exude a plethora of antimicrobial compounds into the rhizosphere (Badri and Vivanco, 2009). Our data above clearly showed that after aerial treatment with MAMPs, an intraplant signal regulates ALMT1 to recruit FB17 belowground. Millet et al. (2010) and Denoux et al. (2008) showed that flg22 treatment up-regulated the expression of three defense related genes, CYP71A12, MYB51, and WRKY11. CYP71A12 encodes a cytochrome P450 that is very similar to CYP71A13, which catalyzes the conversion of indole-3-acetaldoxime to indole-3-acetonitrile during camalexin biosynthesis (Nafisi et al., 2007). MYB51 is a transcription factor essential for the regulation of indole-glucosinolate biosynthesis (Gigolashvili et al., 2007). The transcription factor WRKY11 is a negative regulator of basal resistance in Arabidopsis (Journot-Catalino et al., 2006). Millet et al. (2010) also showed that MAMPs treatment in roots up-regulated CYP71A12, MYB51, and WRKY11 defense genes. To analyze the mutualistic association of FB17 and Arabidopsis roots, we speculated that after MAMPs treatment FB17 may modulate root defense genes for efficient colonization.
To test the hypothesis that FB17 may intervene with MAMPs-triggered plant innate immunity, we evaluated the promoter:GUS transgenic lines of CYP71A12, MYB51, and WRKY11 post treatment with flg22, COR, Pst DC3000, or equal volume of water as a mock. All three GUS reporter genes (CYP71A12, MYB51, and WRKY11) were activated in the roots after foliar flg22 or Pst DC3000 treatment only (Supplemental Fig. S6A). To validate the reporter expression data of root defense response after foliar flg22 or COR treatment, we examined the induction of CYP71A12, MYB51, and WRKY11 gene expression using sqRT-PCR in wild-type roots (Supplemental Fig. S6B). The sqRT-PCR data validated the histochemical GUS expression that both Pst DC3000 and flg22, but not COR, up-regulated root defense responses in plants. These data also suggest that upon aerial infection, plants may elevate the overall innate defense response to mitigate the pathogenesis. The question is how do plants differentiate between pathogens and nonpathogens in terms of modulating defense response and how do beneficial microorganisms intervene with the root defense response to be host associated?
FB17 Suppresses MAMPs-Triggered Root Defense Response
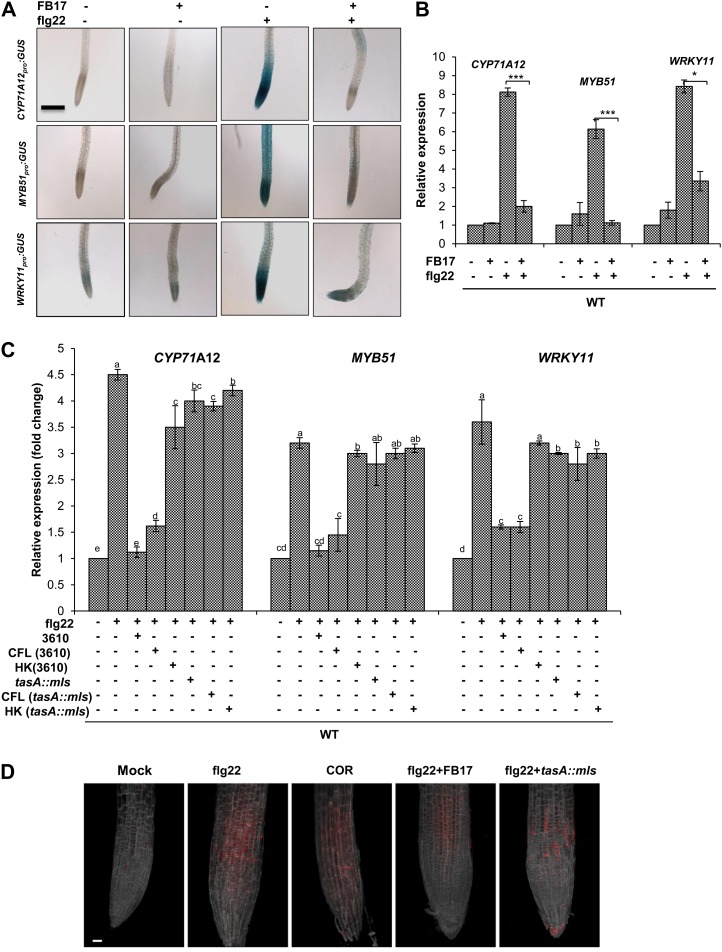
The work above showed that MAMPs trigger a root defense response similar to a foliar pathogen; next, we evaluated how FB17 bypasses the elevated root defense. To test the possible mechanism of FB17 colonization on the root under a primed defense scenario, we examined the promoter:GUS transgenic lines of CYP71A12, MYB51, and WRKY11 subjected to aerial treatment with flg22 and/or root inoculation with FB17. Foliar treatment with flg22 highly induced CYP71A12, MYB51, and WRKY11 compared with the FB17 alone or the mock. It is most interesting that FB17 strongly suppressed the flg22-elicited activation of the CYP71A12, MYB51, and WRKY11 (Fig. 4A). This is validated by measuring the relative expression of CYA71A12, WRKY11, and MYB51 by sqRT-PCR after flg22 treatment and/or FB17 inoculation (Fig. 4B). To test which component of the FB17 is necessary or sufficient for the suppression of innate immunity, the cell free lysate (CFL) and heat-killed (HK) bacteria were tested for their ability to suppress the MAMPs-triggered responses in the roots (Supplemental Fig. S7). The HK fraction failed to suppress CYP71A12pro:GUS or MYB51pro:GUS reporter response in roots elicited by flg22 (Supplemental Fig. S7). In contrast, CFL treatment actively suppressed the flg22-mediated root defense responses (Supplemental Fig. S7). These results suggest that live bacteria or a diffusible bacterial component may suppress the flg22-mediated root defense responses. The results also indicated that beneficial microorganisms may actively block the innate defense responses in the roots to establish a compatible interaction with the host.
Figure 4.
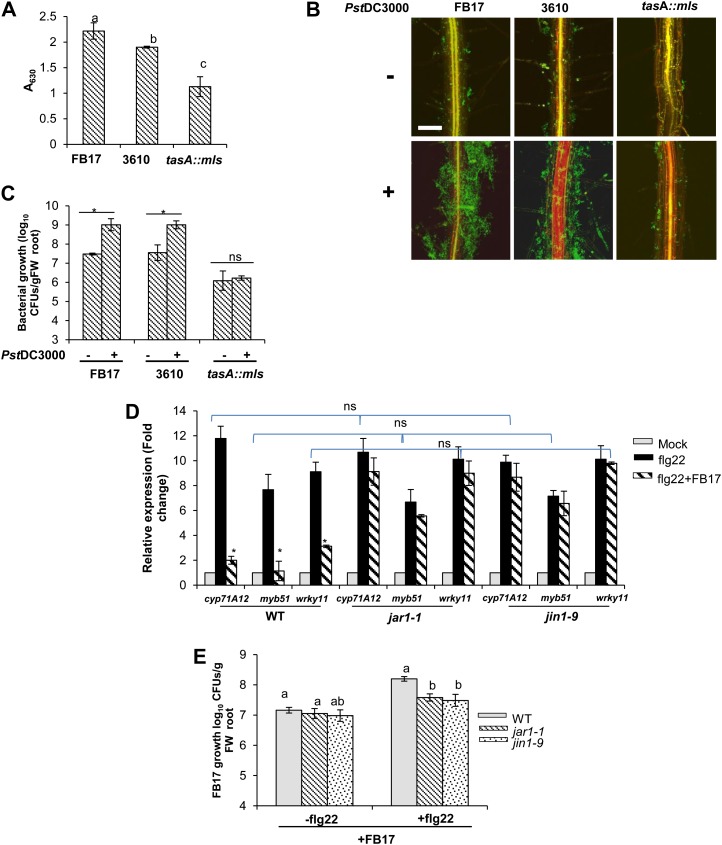
Rhizobacteria FB17 suppresses MAMPs-elicited defense responses in the root. A, Transgenic seedlings carrying a CYP71A12pro:GUS; MYB51pro:GUS or WRKY11pro:GUS reporter construct in wild-type background were grown in vitro, simultaneously treated with flg22 (1 µm) or with equal volume of water as control and FB17 (OD600 = 0.001), and incubated for 24 h before GUS staining. Scale bars = 4 mm, common to all panels. B, Three-week-old in vitro-grown wild-type plants were simultaneously treated with flg22 (1 µm), or with an equal volume of water as control, and FB17 (OD600 = 0.001), and incubated for 24 h. Total RNA from roots was isolated, and sqRT-PCR was performed. Data represent the mean ± se. *P ≤ 0.05, ** *P ≤ 0.001; two-tailed Student’s t test, n = 3. C, Three-week-old in vitro-grown wild-type plants, simultaneously treated with flg22 (1 µm) + 3610 or tasA::mls (OD600 = 0.001), flg22 (1 µm) + CFL of 3610 or tasA::mls and flg22 (1 µm) + HK (3610 or tasA::mls) and incubated for 24 h before RNA isolation. Real-time PCR was performed and expression of CYP71A12, MYB51, and WRKY11 was quantified. Data are presented as mean ± se. The lowercase letters represent statistical difference at P ≤ 0.05 according to DMRT (se values are three technical replicates of one experiment, repeated twice with similar results). D, flg22-elicited deposition of callose in Arabidopsis roots. Wild-type seedlings were cotreated with flg22 (1 µm), or with COR (5 µm), and root treated with FB17 or tasA::mls and incubated for 24 h. The red punctate spots on roots depict callose deposition. Scale bar = 50 µm for all the panels. [See online article for color version of this figure.]
B. subtilis Suppression of MAMPs-Triggered Root Defense Response Is Independent of Biofilm Formation
The work above showed that a B. subtilis-derived component actively suppressed the flg22-mediated defense response in Arabidopsis roots. It is known that the biofilm formation and colonization ability of Bacillus spp. and other endospore-forming Gram-positive bacteria is dependent on extracellular matrix and TasA (Branda et al., 2006). We hypothesized that TasA protein may play a role in the suppression of root defense genes in Arabidopsis. To test this, the wild type and the promoter:GUS transgenic lines of CYP71A12, MYB51, and WRKY11 were subjected to foliar treatment with flg22 and/or root inoculated with parental B. subtilis strain 3610 (a parental strain for tasA mutant) or tasA::mls mutant. Quantitative real-time (qRT)-PCR was also performed using root tissues to estimate the expression levels of CYP71A12, MYB51, and WRKY11. Like FB17, reporter lines treated with wild type 3610 showed active suppression of flg22-mediated CYP71A12, MYB51, and WRKY11 (Fig. 4C). Interestingly, a CFL from 3610 also suppressed the flg22-mediated CYP71A12 and MYB51 defense responses. In contrast, a HK component of 3610 was impaired in suppressing the flg22-mediated CYP71A12 and MYB51 root defense responses (Fig. 4C). In marked contrast, the tasA::mls was impaired in suppressing the flg22-mediated CYP71A12 and MYB51 defense responses in roots. As expected and previously reported (Millet et al., 2010), COR suppressed callose deposition in Arabidopsis roots (Fig. 4D). To study further MAMPs signaling activation and its response on callose deposition, we tested plants root treated with FB17/tasA::mls and flg22. FB17 suppressed the flg22-elicited callose deposition in Arabidopsis roots (Fig. 4D). To further test whether the 3610 and tasA::mls strains differ in their ability to form biofilms with FB17, we used the methods of Hamon and Lazazzera (2001) to measure adherence of the bacterium to the wells of a microtiter plate. As shown in Figure 5A, the tasA::mls formed significantly less biofilm compared with its parental strains, 3610 and FB17. To determine if tasA had a colonization ability on biotic surfaces, we measured colonization on roots. Our results showed that tasA::mls is impaired in root colonization compared with FB17 and 3610 (Fig. 5, B and C).
Figure 5.
tasA::mls failed to form root colonization and FB17-mediated suppression of flg22-elicited root defense response is JAR1/JIN1/MYC2 dependent. A, Surface attachment and biofilm formation by FB17, 3610 wild-type, and tasA::mls strains. Optical density at 630 nm of solubilized CV from solid surface assays over time for tested B. subtilis strains; data are presented as mean ± se. The lowercase letters represent statistical difference at P ≤ 0.05 according to DMRT (se values are three technical replicates of one experiment, repeated twice with similar results). B, Confocal images showing differential root colonization by FB17, 3610 wild-type, and tasA::mls mutant strains. Three-week-old in vitro seedlings of the wild type were foliar sprayed with Pst DC3000 (OD600 = 0.1) or an equal volume of water as the control and rhizoinoculated with FB17 or the 3610 wild type and tasA::mls (OD600 = 0.001) for 24 h. The green fluorescence in the panels shows biofilm. Scale bars = 50 µm. C, Evaluation of rhizobacterial growth on the Arabidopsis roots by cfu. Three-week-old pellet-grown wild-type plants were foliar sprayed with Pst DC3000 (OD600 = 0.1) or an equal volume of water as control and rhizoinoculated with 4 mL/pellet of FB17 or the 3610 wild type and tasA::mls (OD600 = 0.5) and incubated for 72 h. Data represent the mean ± se. *P ≤ 0.05, ** *P ≤ 0.001; two-tailed Student’s t test, n = 3. D, Measurement of root CYP71A12, MYB51, and WRKY11 expression by sqRT-PCR in the wild type and mutants jar1-1 and jin1-9. Three-week-old in vitro-grown wild type, jar1-1, and jin1-9 plants were treated with 1 µM of flg22, or flg22 + FB7, or an equal volume of water as the control, and incubated for 24 h. The total root RNA was isolated and was subsequently analyzed for relative expression level. Data are presented as mean ± se. ns represents nonsignificant within the compared means; statistical difference at P ≤ 0.05 according to DMRT. E, FB17 growth quantification on the roots by cfu. Plants (the wild type, jin1-9, and jar1-1) were foliar sprayed with flg22 (1 µM) or an equal volume of water as the control and rhizoinoculated with 4 mL/pellet of FB17 (OD600 = 0.5) and incubated for 72 h. Data represent the mean ± se. The lowercase letters represent statistical difference at P ≤ 0.05 according to DMRT (se values are three technical replicates of one experiment, repeated twice with similar results). [See online article for color version of this figure.]
To distinguish whether biofilm formation was critical for suppressing the MAMPs-activated defense responses in host plants, we tested mutants in B. subtilis defective in the production of the extracellular polysaccharide (EPS) matrix. Two mutants of FB17 defective in the epsG (DFB79) and epsO (DFB82) genes predicted to be required for EPS synthesis, failed to form biofilms compared with the parental strain FB17 (Supplemental Fig. S8A). To test for suppression of MAMPs-triggered defense responses, the biofilm deficient mutants DFB79, DFB82, and tasA::mls were presented to flg22-treated CYP71A12pro:GUS, MYB51pro:GUS, and WRKY11pro:GUS plant cell lines. Unlike the tasA mutant, the EPS-defective epsG (DFB79) and epsO (DFB82) mutants actively suppressed the flg22-triggered CYP71A12, MYB51, and WRKY11 defense responses comparably with FB17 or PY79-treated plants (Supplemental Fig. S8B). These results suggest that biofilm formation in B. subtilis is not critical for suppression of MAMPs-triggered immunity (MTI).
B. subtilis-Mediated Suppression of flg22-Triggered Root Defense Response Is JAR1/JIN1/MYC2 Dependent
It is widely accepted that the flg22-triggered defense response in roots is suppressed by pathogen-secreted phytotoxin COR or beneficial microbes primarily dependent on the JA pathway (Millet et al., 2010; Jacobs et al., 2011). To evaluate if B. subtilis-mediated suppression of flg22 is JA dependent, we used 3-week-old seedlings of the wild type, jar1-1, and jin1-9. Seedlings were sprayed with flg22 or mock treated and root inoculated with FB17. Subsequently, expression of root defense genes (CYP71A12, MYB51, and WRKY11) and root FB17 colonization were quantified (Fig. 5, D and E). Interestingly, in JA-signaling mutants (jar1-1 and jin1-9), flg22-mediated CYP71A12, MYB51, and WRKY11 expression remained unchanged compared with wild-type plants (Fig. 5D). To substantiate whether reduced FB17 colonization in jar1-1 and jin1-9 roots was associated with elevated immunity, we checked total cfu in JA mutants. We found reduced FB17 colonization in flg22-treated jar1-1 and jin1-9 compared with wild-type roots (Fig. 5E), suggesting that inability of FB17 to suppress MTI may result in reduced FB17 root titers. These data suggest that flg22-activated root defense in Arabidopsis is independent of JIN1/MYC2 in JA-signaling, but FB17-mediated suppression of flg22-activated root defense is JAR1/JIN1/MYC2 dependent. These findings are consistent with the general convention that the plant growth-promoting rhizobacterial colonization in host plant roots may require the suppression of MAMPs signaling to protect the beneficial bacteria against MAMPs-elicited defense in the early phase of root colonization (Zamioudis and Pieterse, 2012).
Regulation of Root Defense Response and ALMT1 Expression by MAMPs
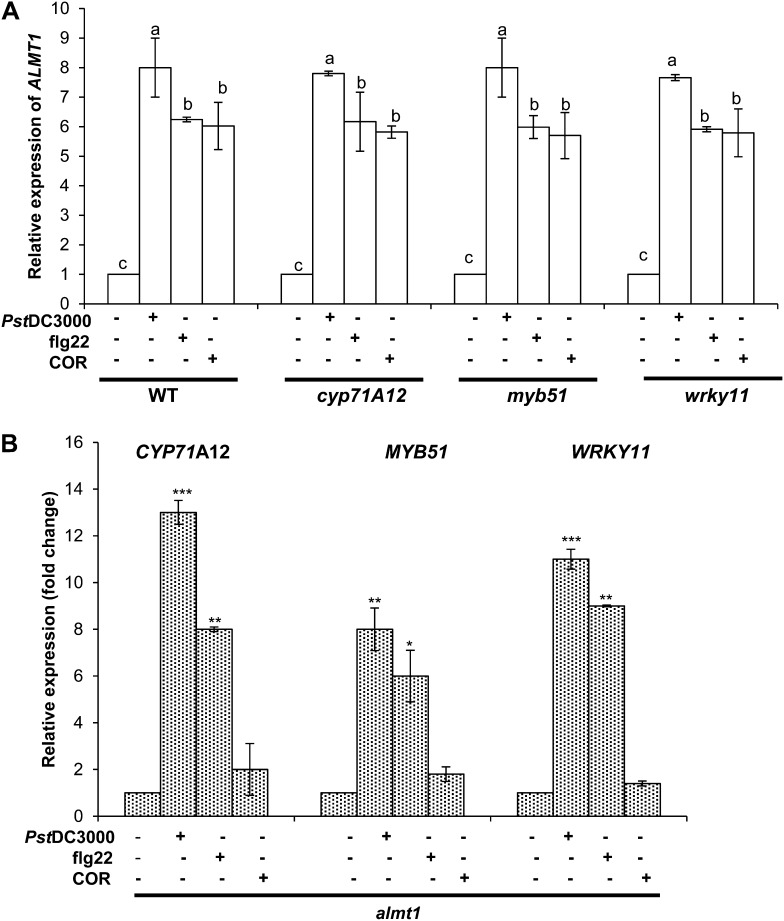
Because foliar MAMPs treatment elicited root defense genes and also induced ALMT1 expression, we tested the epistatic relationship between ALMT1 and root defense genes. For this, we tested the ALMT1 expression analyses on cyp71A12, myb51, and wrky11 insertion mutants in the absence and presence of foliar MAMPs. In brief, 20-d-old seedlings of insertion mutants cyp71A12, myb51, and wrky11, were treated with Pst DC3000, COR, or flg22. After 24 h of treatment, ALMT1 root expression was evaluated using sqRT-PCR. As per our previous report, wild-type plants treated with Pst DC3000 showed enhanced root ALMT1 expression (Rudrappa et al., 2008b; Fig. 6A). The wild-type plants treated with COR or flg22 also showed increased root ALMT1 expression (Fig. 6A). Interestingly, the mutant lines cyp71A12, myb51, and wrky11 also revealed similar trends in ALMT1 expression when treated with Pst DC3000, MAMPs, or COR as compared with wild-type plants (Fig. 6A). Inversely, we also evaluated the MAMPs-triggered root defense response in almt1 plants subjected to aerial Pst DC3000 and MAMPs treatments. The data showed similar expression levels of root defense response genes as shown for wild-type plants (Fig. 6A). These data confirm that MAMPs activation of root defense response genes functions in parallel with and independent of ALMT1 responses. We next evaluated the functional response of ALMT1 expression by visualizing the FB17 colonization in cyp71A12, myb51, and wrky11 mutants. The leaves of the wild type, cyp71A12, myb51, and wrky11 plants were inoculated with Pst DC3000 or MAMPs and subsequently root drenched with FB17. After 72 h, both wild-type and mutant plants treated with Pst DC3000 or MAMPs showed enhanced FB17 colonization (Supplemental Fig. S9).
Figure 6.
Independent regulation of root defense response and ALMT1 expression by MAMPs. A, Measurement of ALMT1 expression in cyp71A12, myb51, wrky11, and wild-type plants. Three-week-old Arabidopsis mutants cyp71A12, myb51, wrky11, and the wild type were treated with flg22 (1 µM), COR (5 µM), Pst DC3000 (OD600 = 0.1), or an equal volume of water as the control, and total RNA was collected from roots after 24 h of incubation. sqRT-PCR was performed, and relative expression of ALMT1 was quantified. Data represent the mean ± se. The lowercase letters represent a statistical difference at P ≤ 0.05 according to DMRT (se values are three technical replicates of one experiment, repeated twice with similar results). B, Measurement of CYP71A12, MYB51, and WRKY11 expression in almt1 plants. Three-week-old Arabidopsis mutants almt1 were treated with flg22 (1 µM), COR (5 µM), Pst DC3000 (OD600 = 0.1), or an equal volume of water as the control, and total RNA was collected from roots after 24 h of incubation. sqRT-PCR was performed, and relative expression of CYP71A12, MYB51, and WRKY11 was quantified. Data represent the mean ± se. *P ≤ 0.05, ** *P ≤ 0.001; two-tailed Student’s t test (se values are three technical replicates of one experiment, repeated twice with similar results).
Our previous results and those of others (Hoekenga et al., 2006; Rudrappa et al., 2008b) have shown the specificity of root ALMT1 activation to Pst DC3000 and aluminum (Al3+) treatments. To further verify that ALMT1 expression is independent of MAMPs-triggered root-defense responses, we used the promoter:GUS transgenic lines of CYP71A12, MYB51, and WRKY11 against MA and AlCl3. It has been shown that MAMPs and AlCl3 trigger ALMT1 expression. We wondered that if MAMPs could trigger ALMT1 expression leading to increased MA titers helping FB17 association, how would an increase in MA affect root defense responses? Interestingly, a direct treatment of MA (100 nm) and AlCl3 (4 µm) to promoter:GUS transgenic plants of CYP71A12, MYB51, and WRKY11 showed unaltered expression of all the three root-specific defense genes (Supplemental Fig. S10), suggesting the noninterference of the MAMPs-triggered metabolite (MA) with the root defense genes. These data suggest that xenobiotic agents, which are able to induce ALMT1 expression in roots, do not disturb immunity in roots. Moreover, ALMT1 does not interfere with the root immune responses.
Defense-Suppressive Function of COR Is Abolished upon Root Colonization by FB17
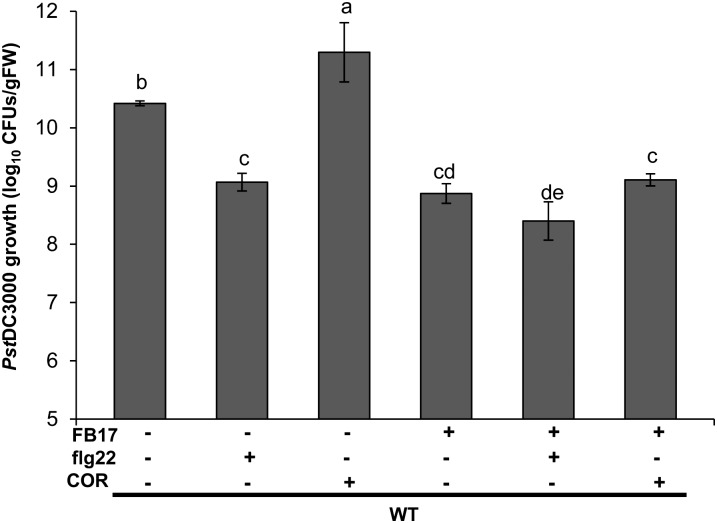
Having shown that FB17 abolishes the flg22-mediated root defense, we next wanted to see how cotreatment with MAMPs and FB17 modulates Pst DC3000 pathogenesis in Arabidopsis. To initiate this experiment, 3-week-old wild-type plants were foliar pretreated with COR and flg22, and after 24 h Pst DC3000 was coinoculated in the absence or presence of root-inoculated FB17. After 72 h of treatment, the growth of Pst DC3000 was monitored by cfu (Rudrappa et al., 2008b; Millet et al., 2010). Previous experiments showed that application of flg22-treated plants triggered resistance against Pst DC3000 (Zipfel et al., 2004). In contrast, treatment of plants with COR resulted in down-regulation of flg22-triggered immune response in Arabidopsis roots (Millet et al., 2010). In accordance with the published data, our results showed flg22-induced resistance against Pst DC3000 and that resistance was more pronounced during cotreatment with flg22 and FB17 inoculation (Fig. 7). In parallel, plants treated with COR showed enhanced susceptibility to Pst DC30000 (Fig. 7). In addition, plants cotreated with COR and FB17 showed resistance to Pst DC3000 (Fig. 7). In light of the fact that both COR and FB17 suppress the flg22-induced root defense responses (Supplemental Fig. S12) with FB17, abolishing COR-mediated susceptibility, FB17 may act through an unknown pathway to up-regulate disease resistance against Pst DC3000.
Figure 7.
Defense-suppressive function of COR is abolished upon root colonization by FB17. Three-week-old Arabidopsis wild-type plants were foliar sprayed with flg22 (1 µM), COR (5 µM), or an equal volume of water as the control. After 24 h of incubation, plants were infected with Pst DC3000 by leaf dip. After 72 h of infection, Pst DC3000 growth was measured as cfu. Data represent the mean ± se. The lowercase letters represent a statistical difference at P ≤ 0.05 according to DMRT (se values are three technical replicates of one experiment, repeated twice with similar results).
DISCUSSION
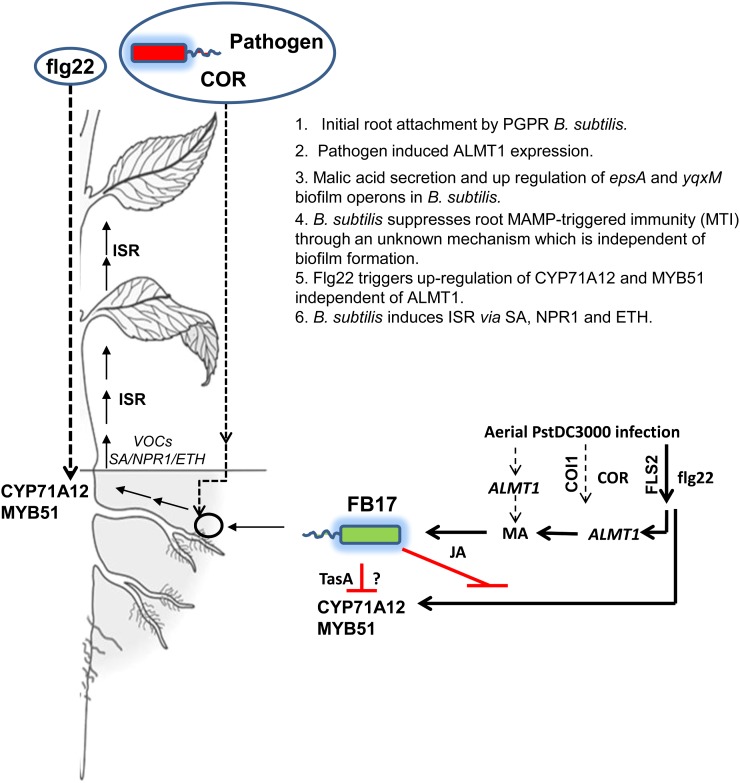
Recently, several reports have established the role of beneficial PGPRs in improving plant health by increasing tolerance to pathogens, insect pests, abiotic stress, including drought and salinity (Zhang et al., 2008, 2010; for review, see Choudhary and Johri, 2009). Among these PGPR strains, FB17 rhizoinoculated onto the roots of Arabidopsis plants reduced disease severity, thus inhibiting the proliferation of foliar pathogen Pst DC3000 through induction of JA/ET-mediated ISR and SA-mediated SAR (Rudrappa et al., 2010). The magnitude of colonization on the root by beneficial microbes is limited by several factors that include root surface biochemistry and composition of root exudates (Rudrappa et al., 2008a; Doornbos et al., 2011; Chen et al., 2012). It has been speculated that components of root exudates may play a critical role in establishing a beneficial microbiome in the rhizosphere (Lugtenberg et al., 1999; Bais et al., 2006). The composition of root exudates depends on plant species and cultivar, development stage, and stress factors (Uren 2000). There is little knowledge in the field as to how plant roots respond to biotic stress, i.e. foliar pathogen attack. It is known that plants, upon foliar attack, modulate their primary and secondary metabolic pathways to augment pathogen defense. This is shocking given that plant roots respond to various belowground biotic and abiotic stresses by synthesizing complex toxins and also play an active part in environmental sensing. In this report, we demonstrated that MAMPs elicit potential root responses to trigger beneficial rhizobacterial recruitment. We show that foliar MAMPs treatments induce a graft-transmissible intraplant shoot-to-root signal to elicit a malate transporter leading to increase in FB17 colonization. We also found that FB17 suppresses MAMPs-triggered innate immune responses in roots (Fig. 8). The ability of plant roots to activate a malate-driven beneficial microbe recruitment by recognizing that specific MAMPs may play a critical role in efficient colonization by rhizobacteria and in turn may limit access of pathogenic microbes to the roots.
Figure 8.
A schematic depicting the long-distance intraplant signaling involving foliar MAMPs treatment and root ALMT1 to recruit rhizobacteria B. subtilis FB17. The schematic shows that MAMPs-triggered signal recruits FB17 through ALMT1 belowground along with elicitation of root defense with foliar MAMPs treatments. In contrast, FB17 suppresses MAMPs-elicited root defense response. The dashed lines show the pathogen and MAMP-induced responses. The schematic shows that FB17 in the initial phase of root colonization suppresses root defense genes possibly for biofilm formation. [See online article for color version of this figure.]
Foliar MAMPs Trigger Root FB17 Colonization
In plants, increased accumulation of MA is triggered by elevated biosynthesis of one or more MA precursors such as fumarate, oxaloacetate, or pyruvate (Casati et al., 1999). Alternatively, up-regulation of ALMT1 elicits root MA secretions (Kobayashi et al., 2007). It has been argued that most of the Gram-positive and Gram-negative microbes prefer simple sugars as a carbon source compared with complex carboxylic acids. A recent work shows that B. subtilis prefers MA as a carbon source and inflicts metabolite repression to take up other carbon sources for an efficient MA uptake (for review, see Fernie and Martinoia, 2009; Kleijn et al., 2010). This recent data validates the specificity and preference of B. subtilis for MA. Our previous studies showed that aerial plant pathogenesis involved the induction of MA in root secretions that effectively recruited beneficial rhizobacteria (Rudrappa et al., 2008a, 2010). We also showed that the pathogenic phyllosphere pathogen, Pst DC3000, induced root ALMT1 expression (Rudrappa et al., 2008b). This study reveals that pathogen-derived MAMPs, especially flg22 and phytotoxin COR, are sufficient to mimic the live pathogen response of induced root ALMT1 expression in Arabidopsis (Fig. 1). Unlike COR and flg22, other tested MAMPs (chitin, PGN, and LPS), failed to elicit ALMT1 expression, thus raising a hypothesis that plants have evolved root responses to specific MAMPs depending on the nature of the invading pathogen. It is known that several species of Bacillus biosynthesize PGNs (Daniel and Errington, 2003). The specificity of MAMPs to elicit ALMT1 expression, thereby enhancing FB17 colonization, suggests that plants recognize pathogenic and nonpathogenic MAMPs differently. Surprisingly, FB17 failed to elicit ALMT1 expression in roots (Supplemental Fig. S11). The fact that aerial treatment with flg22 or COR induced root ALMT1 expression suggests the involvement of various downstream signaling components of FLS2 and COI1 pathways to target ALMT1.
Foliar MAMPs Induce Root ALMT1 Signal
Genetic approaches can be useful for dissection of signaling pathways that might shed light on the recruitment of beneficial microbes in the rhizosphere. It is known that there exists a root-to-shoot signaling pathway mediated through BYPASS1 that regulates leaf venation patterning (van Norman et al., 2004). The roots of BYPASS1 overproduce a mobile component that functions non-cell autonomously to arrest shoot growth. Several plant hormones are known to travel long distances, from the root to the shoot, and to affect shoot processes. An excellent example is the control of shoot branching, which is regulated by the root-derived signaling molecule recently identified as strigolactone (Gomez-Roldan et al., 2008). Similarly, cytokinins have been implicated as long-distance signals that communicate the nitrogen status from the root to the shoot (Takei et al., 2001; see also Kudo et al., 2010). The signal that communicates drought conditions from the root to the shoot is widely believed to be mediated through abscisic acid (Bahrun et al., 2002). Although there are some examples of root-to-shoot signaling regulating developmental phenotype and hormone translocation, there is no information that dictates the existence of a shoot-to-root intraplant signal relaying a plant’s response with beneficial microbes. Our results show the connection between foliar spray of MAMPs and root ALMT1 components to regulate a positive feedback response in plants. The fact that FB17 root treatment failed to increase root ALMT1 expression suggests a pathogen-specific induction of malate efflux for rhizobacterial recruitment (Supplemental Fig. S11). This also strengthens our model (Fig. 8) and provides a molecular validation that foliar treatment launched an intraplant signal connecting shoot to root to attract beneficial microbes belowground. Using fls2 or coi1 scions and ALMT1pro:GUS rootstocks showed the activation of ALMT1, suggesting the existence of a mobile signal (Fig. 2; Supplemental Fig. S5, A and B).
SA/JAR1/JIN1/MYC2-Independent Regulation of flg22 or COR-Induced ALMT1 Expression
Different bacterial MAMPs, including flg22, induce overlapping genes (Zipfel et al., 2004), and further studies showed that flg22-induced gene expression and regulation is SA-independent and -dependent in early and late phases after flg22 administration (Vlot et al., 2009). This intriguing observation suggests that the perception of MAMPs by plants leads to the induction of early responsive genes through an SA-independent signaling mechanism resulting in SA accumulation (Tsuda et al., 2008). When SA has accumulated to a sufficiently high level, expression of some of the early responsive genes is kept high by an SA-dependent signaling mechanism (Tsuda et al., 2008). This suggests that plants exploit SA-mediated signaling to maintain MAMPs-triggered defense expression (Tsuda et al., 2008). However, our observations showed that MAMPs-triggered root ALMT1 expression was independent of the SA signaling pathway (Fig. 3). Our studies clearly indicate that MAMPs triggered ALMT1 expression involves the FLS2 pathway. It is surprising as flg22 has been shown to involve SA, JA, and ET pathways in parallel for induction of resistance against foliar pathogens (Zipfel et al., 2004). There is a possibility that downstream components of FLS2 may hold a key in MAMPs-triggered regulation of ALMT1. In parallel, it is known that Pst DC3000 utilizes COR, a structural mimic of the signaling molecule JA-Ile, to suppress MAMPs-activated defense response in Arabidopsis roots (Millet et al., 2010). Our data show that regulation of root ALMT1 by COR is dependent on ubiquitin ligase COI1, a key regulator of JA signaling. These data and the fact that COR modulates the genes involved in the pathways to JA (Uppalapati et al., 2005) prompted us to evaluate the importance of JA in COR-COI1-mediated regulation of ALMT1. In contrast, MeJA failed to induce root ALMT1 expression and COR regulation of ALMT1 was found independent of JA signaling. This is surprising because COR and JA are considered structural analogs and are thought to induce similar plant responses and have impact on multiple phytohormone pathways (Uppalapati et al., 2005). Although both COR and MeJA are involved in various physiological responses, we do not understand to what extent COR mimics MeJA. There are striking structural differences in COR and MeJA, and this may evade the regulation of ALMT1 by JA.
B. subtilis Suppresses MTI in Roots to Confer Beneficial Symbiotic Interactions
In beneficial symbiotic interactions, especially legume-rhizobia and mycorrhizal fungi, rhizobium and mycorrhizae have evolved mechanisms to efficiently suppress the host immune systems to establish successful infections (for review, see Zamioudis and Pieterse, 2012). Similarly, the beneficial symbiotic microbes such as PGPR, that often grow endophytically inside roots or on root surfaces, may also minimize stimulation of the host’s immune system. It was recently reported that the beneficial bacterium P. fluorescens WCS417r suppresses the MAMPs-triggered response in Arabidopsis roots that is responsible for deposition of callose in the root elongation zone elicited by the flagellar peptide flg22 (Millet et al., 2010). In this study, applying FB17 to Arabidopsis roots resulted in the early suppression of defense genes that may facilitate FB17 colonization on Arabidopsis roots. This is surprising because general convention dictates that beneficial rhizobacteria are known to elicit root defense responses (Verhagen et al., 2004). However, it is possible that the suppression of MAMPs signaling is necessary for successful root colonization by PGPRs. We speculate that the early phases of root colonization by PGPRs require the suppression of MAMPs signaling to protect the bacteria against MAMPs-elicited antimicrobial exudates. Interestingly, FB17 showed resistance against MAMPs-elicited Arabidopsis root exudates (data not shown), explaining the efficiency of FB17 to colonize Arabidopsis roots. There is a tempting possibility that FB17 may down-regulate the root defense response in the initial phases of root-microbe interaction for the establishment of the biofilm community. It is also known that FB17 specifically induces the expression of PR1 and PDF1.2 in Arabidopsis leaves but not in the roots (Rudrappa et al., 2010). Similarly, B. subtilis strain AR156, when applied to Arabidopsis roots, resulted in the expression of four defense-related genes (PR1, PR2, PR5, and PDF1.2) in the leaves but not in the roots (Niu et al., 2011).
In addition, the observation that FB17 suppresses MTI in the roots is at odds with the prevailing view that MAMPs are the molecular determinants responsible for ISR (Millet et al., 2010). To our knowledge, FB17 does not produce COR or compounds with related structures. Therefore, it is likely that FB17 suppresses the MAMPs-triggered response in roots via a different mechanism. The fact that the HK FB17 treatment negated the suppression of MAMPs-triggered root response supports the view that a small diffusible proteinaceous molecule may mediate this suppression. Similarly, the inability of the TasA mutant (tasA::mls) to suppress flg22-mediated innate responses indicates the importance of amyloid fibers in down-regulating plant defense response. Functional amyloids that participate in normal biological processes in various organisms include harpins in Xanthomonas campestris and Pst DC3000 (Oh et al., 2007), and fimbriae described in Mycobacterium tuberculosis (Alteri et al., 2007). Of all the amyloid fibers, type III-dependent harpins in plant pathogenic bacteria are known to interact with plants to cause a hypersensitive response type of programmed cell death (Oh et al., 2007). Interestingly, a transcriptomic analysis of Arabidopsis plants treated with harpins revealed genes related to cell wall biogenesis, cellular communication, and signaling (Livaja et al., 2008). In addition, harpin-treated Arabidopsis plants also showed down-regulation of important plant defense genes such as WRKY transcription factors and oxidative burst-associated genes like NADPH oxidases (Livaja et al., 2008). B. subtilis has recently been shown to produce amyloid fibers that were important to regulate the structural integrity in B. subtilis biofilms (Romero et al., 2010). To our knowledge, other than harpins, no other amyloid fibers have been reported to interact with plants. It is surprising, given the abundance of amyloid fibers in biofilms of both Gram-positive and Gram-negative bacteria, how little we know about the role of amyloid fibers in plant-microbe interactions. Interestingly, TasA protein was described as having antibiotic activity (Stöver and Driks, 1999).
Why would Bacillus spp. produce amyloid fibers in its association with plants? The widespread existence of amyloid-producing rhizobacteria suggests that the production of antibiotic-rich amyloid fibers could be a strategy to outcompete other microbes in the rhizosphere. It could also be speculated that amyloid fiber producing Bacillus spp. could colonize better on plant roots. It is interesting that our data showed that TasA plays a critical role in regulating colonization on plant roots. The importance of root colonization on B. subtilis-mediated MTI suppression was immediately ruled out as B. subtilis mutants defective in another structural component of biofilm formation, specifically biosynthesis of the EPS matrix, actively suppressed MTI in roots. Thus, MTI suppression is coregulated with biofilms, but biofilm formation per se is not required for MTI. Because association of Bacillus spp. induces defense response in plants against pathogens, it could be speculated that B. subtilis-derived extracellular components may not elicit defense responses in the root, but may trigger a long-distance signal from the roots to leaves, mediating ISR in the leaves by simultaneously activating the SA- and JA/ET-signaling pathways in an NPR-1-dependent manner (Rudrappa et al., 2010; Niu et al., 2011). Conversely, B. subtilis-derived extracellular components may not play any role in ISR-mediated signaling against foliar pathogens and only involve host suppression for efficient colonization. On a similar line, it would be interesting to check the importance of known Bacillus spp. biofilm mutants on induction of ISR response in host plants.
FB17 Down-Regulates flg22-Mediated Innate Defense Responses in Arabidopsis Roots
Our data showed that flg22 up-regulated root ALMT1 expression leading to increased colonization by FB17. In parallel, the data presented here also reveal that FB17 abolishes the flg22-mediated root-defense response, which may explain FB17 efficiency to bind to roots. Our data falls in line with the published data showing that plants pretreated with flg22 were more resistant to Pst DC3000 (Zipfel et al., 2004). It was also shown that flg22-mediated resistance requires the activation of SA, JA, and ET pathways in parallel and that knocking out a single pathway alone does not abolish the induction of resistance (Zipfel et al., 2004). The increased protection against Pst DC3000 under mixed treatment of FB17 and flg22 may be explained by the added ability of both MAMPs and FB17 to modulate multiple defense pathways (Zipfel et al., 2004; Rudrappa et al., 2008b, 2010).
In parallel, COR, a polyketide and a phytotoxin produced by various species of Pst DC3000, down-regulates innate defense responses in plants through ubiquitin ligase COI1, a key regulator of JA (Melotto et al., 2006; Tsuda et al., 2008; Millet et al., 2010). Various lines of studies have shown that COR intervenes with the SA/JA antagonistic pathway to suppress a defense response in plants leading to susceptibility against Pst DC3000 (Brooks et al., 2005; Melotto et al., 2006). In contrast, a recent study showed that COR-induced suppression of plant defense is independent of SA-JA antagonism (Brooks et al., 2005; Katsir et al., 2008; Millet et al., 2010). Our data and that of Millet et al. (2010) showed that COR suppresses the root defense response in Arabidopsis. It was shown that COR abolishes the flg22-mediated root defense response in Arabidopsis that is dependent on COI1 and JIN1/MYC2 (Millet et al., 2010). At the molecular level, JA-SA antagonism is known from studies in coi1 and jin1-9, where both mutants displayed increased SA signaling after bacterial challenge (Kloek et al., 2001). Similarly, JA signaling is recruited by beneficial mycorrhizal fungi Piriformospora indica to suppress the early root MTI responses (Jacobs et al., 2011). Because both jin1-9 and jar1-1 mutants exhibited an enhanced MAMPs-induced immune response in response to FB17 colonization (Fig. 5), JA signaling contributes at least partially to the suppression of root MTI. This was further validated by decreased FB17 colonization under MAMPs treatment in JA mutants, suggesting that JAR1/JIN1/MYC2-mediated immune activation may restrict FB17 root colonization.
At this juncture, it is hard to explain how and why FB17 suppresses root innate immune response. On the contrary, from the published work, it is known that FB17 mediates resistance against Pst DC3000 through JA/ET-mediated ISR and SA-mediated SAR by triggering PDF1.2 and PR1 expression (Rudrappa et al., 2010). It would be interesting to determine how FB17, in spite of abolishing MAMPs-triggered root responses, protects plants against aerial pathogens. We speculate that the FB17 may adapt a temporal suppression of MAMPs-triggered root response to facilitate root colonization.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) wild-type seeds were procured from Lehle Seeds (Round Rock, TX). Arabidopsis insertion lines coi1 (SALK_45434C), fls2 (SALK_093905C), jar1-1 (SALK_030821C), jin1-9 (SALK_061267C), npr1-1 (CS3726), almt1 (SALK_005672C), myb51-1 (SM_3_16332), cyp71A12 (GABI_127H03-1), and wrky11 (SALK_141511C) were obtained from the Arabidopsis Biological Resource Center. Arabidopsis transgenic lines CYP71A12pro:GUS, MYB51pro:GUS and WRKY11pro:GUS were obtained from Dr. Frederick M. Ausubel (Harvard Medical School, Boston, MA). About 50 to 100 seeds were sown in 90-mm petri plates containing two layers of filter paper (Whatman 70-mm diameter (Ø), Cat No. 1001–070), wetted with 4 mL of sterile Nano water, and allowed to germinate for 3 to 4 d until the root and shoot emerged by incubating at 23°C ± 2°C under a 16-h-light/8-h-dark photoperiod. The plates were illuminated with cool-white fluorescent light with an intensity of 110 µmol m−2 s−1. Five uniform seedlings were transferred onto sterile wire mesh in Magenta boxes containing sterile liquid medium 0.5× Murashige and Skoog (MS; Murashige and Skoog, 1962) basal media, 2.5 mm 2-(N-morpholino) ethane sulphonic acid, pH 5.8, for 2 weeks. Each box with five seedlings was considered as one biological replicate.
Treatment of Seedlings with MAMPs
MAMPs were used at the following concentrations unless otherwise specified: For foliar spray experiments, 5 µm of COR, 1 µm of flg22, 500 µg mL−1 of chitin, 500 µg mL−1 of PGN, and 500 µg mL−1 of LPS were used. Plants grown on wire mesh were carefully removed and sprayed with COR or other MAMPs and transferred to a new sterile Magenta box containing basal media as described above. After 24 h, experiments were terminated and roots were collected and used for RNA isolation or GUS staining. A 10 mg mL−1 chitin solution was prepared by autoclaving 250 mg of chitin (Sigma-Aldrich) suspended in 25 mL of water for 30 min. The solution was then centrifuged and the supernatant collected.
Bacterial Strains and Infections
Pst DC3000 Infection Assays
Pseudomonas syringae pv tomato (Pst) DC3000, obtained from Dr. Jorge M. Vivanco (Colorado State University, Fort Collins, CO) was maintained on Luria-Bertani (LB) plates with 50 mg L−1 rifampicin. A single colony from a freshly streaked plate with antibiotic selection was used to grow overnight cultures from which approximately OD600 = 0.02 to 0.5 inoculum was prepared and used in all the experiments. For routine plant-based studies, cells were grown in LB medium at 37°C, 220 rpm. Cells collected by centrifugation were washed twice with sterile 10 mm MgCl2 and were resuspended in 10 mm MgCl2 to a final density of OD600 = 0.1 for foliar dip of Arabidopsis. Fully expanded leaves of Arabidopsis, plants grown on sterile peat pellets, were dipped in 25 mL of OD600 = 0.1 culture of Pst DC3000 with 0.0125% (v/v) Silwet l-77 for 5 min.
Bacillus subtilis Root Colonization
Bacillus subtilis strain FB17 (obtained from Dr. Ray Fall, University of Colorado, Boulder, CO) was maintained on LB plates, wild-type strain 3610, tasA::mls (erythromycin and lincomycin resistant; obtained from Dr. Roberto Kolter, Harvard Medical School, Boston, MA) and PY79, DFB79 (epsG::TnYLB-1, kanamycin resistant), DFB82 (epsO::TnYLB-1, kanamycin resistant; obtained from Dr. Daniel B. Kearns, Indiana University, Bloomington, IN), were maintained on LB plates with specific antibiotic selections. All strains were streaked from a freezer stock onto low-salt LB plates (10 g L−1 tryptone, 5 g L−1 yeast extract, 5 g L−1 NaCl) with appropriate antibiotics. An LB liquid culture of 10 mL was started with bacteria from the plates that had been stored at –4°C for less than two weeks. After 12 h at 30°C, a subculture using 1:100 dilution was incubated further at 30°C, when the OD600 reached 0.8 to 1.0, bacterial cells were spun down and resuspended in sterile 10 mm MgCl2 to obtain appropriate density of OD600 = 0.5. Treatments included Arabidopsis roots inoculated with FB17, Pst DC3000 + FB17, foliar-sprayed MAMPs and COR with root-inoculated FB17. The plants were incubated in the growth chamber for an additional 3 d when the experiment was terminated. Bacterial populations were monitored by serial dilution assays.
Treatment of Seedlings with Bacterial Strains for GUS Assays
Pst DC3000, FB17, 3610, tasA::mls, PY79, DFB79, and DFB82 were cultured on LB medium with appropriate antibiotics; for infection of seedlings grown in Magenta boxes, bacteria were grown overnight in LB supplemented with an appropriate antibiotic at 30°C, centrifuged, washed with water, and resuspended in 0.5× MS medium to a final OD600 of 0.002. Three-week-old seedlings were treated by adding 1 mL of bacterial suspension and incubated for 24 h. For experiments using CFL and HK bacteria, FB17, 3610, tasA::mls were grown overnight and diluted with water to OD600 of 2. For CFL preparation, bacteria were filtered using 0.22-µm syringe filters and, for HK, the bacteria were maintained at 65°C for 6 h. Three-week-old seedlings were treated by adding 1:1 of HK or CFL with 0.5× MS liquid medium with 2.5 mm 2-(N-morpholino) ethane sulphonic acid into each Magenta box and flg22. After 24 h of incubation, the experiments were terminated and seedlings were checked for GUS activity.
Root Exudation Collection and Microtiter Assay
Individual wild-type and transgenic plants were grown sterilely on wire mesh in liquid basal medium. Fully expanded leaves of 3-week-old plants were sprayed with different MAMPs or with Pst DC3000 (prepared as above), aseptically. The spent liquid medium from the infected and uninfected plants containing root exudates were collected after 2 d and lyophilized. The lyophilized samples were diluted with sterile water to get a 2× concentration. FB17 and Pst DC3000 were suspended in the exudate to OD600 = 0.001 and distributed to 96-well plates and incubated 30°C for 24 h. Bacterial populations were monitored by serial dilution assays.
In Vitro Assay for Solid Surface Biofilm Formation
In vitro biofilm formation of FB17, 3610, tasA::mls, PY79, DFB79, and DFB82 was monitored based on the methods of O’Toole and Kolter (1998), with slight modification. The biofilm growth medium based on Hamon and Lazazzera (2001) was LB medium plus 0.015 mm (NH4)2SO4, 10 mm K2HPO4, pH 7, 3.4 mm Na2H2C6H5O7, 1 mm MgSO4, and 0.1% (w/v) Glc. The inoculum was obtained by growing the cells in biofilm growth medium and shaking to midexponential growth and then diluting the cells to OD600 = 0.02 in fresh biofilm growth medium. Samples of 700 µL of the diluted cells were aliquoted to each of 12 sterile round-bottom culture tubes with caps (12 × 75 mm; VWR Scientific), and the cultures grown at 37°C without agitation for 48 h. Cells that had adhered to the tubes were treated with 0.1% crystal violet (CV) for 10 to 15 min at 25°C without agitation, the tubes were drained of liquid via pipet, gently rinsed several times with water, and allowed to dry at 25°C overnight and photographed. The CV that had stained the cells was solubilized in 1 mL of 80% (v/v) ethanol and 20% (v/v) acetone. Biofilm formation was quantified by measuring the optical density at 630 nm for each well using an Opsys MR-Dynex plate reader. The assay was performed on three separate occasions.
RNA Isolation, RT-PCR
Total RNA was isolated from roots 24 h post treatment. RNA was extracted using PureLink RNA isolation buffer according to the manufacturer’s instruction manual (Invitrogen). Possible contaminant genomic DNA in RNA extract was removed using Turbo DNAfree kit (Ambion). The gene-specific primers for the genes ALMT1 (forward: 5′-GGCCGACCGTGCTATACGAG-3′, reverse: 5′-GAGTTGAATTACTTACTGAAG-3′), CYP71A12 (forward: 5′-GATTATCACCTCGGTTCCT-3′, reverse: 5′-CCACTAATACTTCCCAGATTA-3′), MYB51 (forward: 5′-ACAAATGGTCTGCTATAGCT-3′, reverse: 5′-CTTGTGTGTAACTGGATCAA-3′), PR1 (forward: 5′-TTC TTC CCT CGA AAG CTC AA-3′, reverse: 5′-CGT TCA CAT AAT TCC CAC GA-3′), PDF10.2 (forward: 5′-GGTGGAAGCACAGAAGTTGT-3′, reverse: 5′-AATACACACGATTTAGCACC-3′), WRKY11 (forward: 5′-ACGGACAAAAACCGATCAAG-3′; reverse: 5′-AAGCCGAGGCAAACACTAAA-3′), and UBQ1 (forward: TCGTAAGTACAATCAGGATAAGATG, reverse: CACTGAAACAAGAAAAACAAACCCT), were synthesized (Invitrogen). First-strand complementary DNAs were synthesized from 500 ng of total RNA in 20 μL final volume using Moloney Murine Leukemia Virus reverse transcriptase and oligo(18-mer) primer (Fermentas GmbH). PCR amplifications were performed using PCR mixture (15 μL) that contained 1 μL of RT reaction product as template, 1× PCR buffer, 200 μm dNTPs mix (Fermentas GmbH), 1 IU of Taq DNA polymerase (Promega), and 0.1 μm of each primer depending on the gene. PCR was performed at initial denaturation at 94°C for 4 min, 22 or 26 cycles (30 s at 94°C, 30 s at 60°C, and 30 s at 94°C), and final elongation (8 min at 72°C) using a thermal cycler (Bio-Rad). The PCR products were separated on 1.4% agarose gel, stained with ethidium bromide (0.001%), and documented in a gel documentation system; the bands were quantified using E.A.S.Y. WIN 32. Each band was normalized against the intensity obtained with the same complementary DNA using the UBQ1 primers. For qRT-PCR, 1 µg of total RNA was reverse transcribed using ProtoScript first strand complementary DNA synthesis kit from New England Biolabs. qRT-PCR was performed using a Mastercycler ep realplex machine (Eppendorf) and 5 PRIME-RealMaster Mix SYBR ROX (5-PRIME). The program used for qRT-PCR was as follows: 3 min at 95°C, 45 cycles of 15 s at 95°C and 30 s at 53°C, followed by a melt curve from 70°C to 95°C. Expression values were normalized to that of the UBQ1.
Micrografting
Seeds were sterilized for 5 min in 3% (v/v) bleach 5.25% (w/v) sodium hypochlorite followed by 1 min in 70% ethanol (v/v), rinsed four times with sterile Nano water, and resuspended in 1 mL of sterile Nano water. Twenty to 30 seeds were sown in 90-mm petri plates containing six layers of filter paper (Whatman 70-mm Ø, Cat No. 1001–070) overlaid with a nitrocellulose membrane (Whatmann OPTITRAN BA-S 85, 0.45 µm, 82-mm Ø) and wetted with 4 mL of sterile Nano water. Plates were oriented vertically under 24 h of 140 µmol m−2 s−1 at 22°C (Arabidopsis chamber AR36L2; Percival Scientific). Three to 5 d post sowing, seedlings were butt grafted (Turnbull et al., 2002). Grafts healed in plates oriented vertically under 10 h of 50 µmol m−2 s−1 at 27°C (Arabidopsis chamber AR22L, Percival Scientific), and for 2 to 4 d after, grafted seedlings were placed in 90-mm petri plates containing 0.5× MS and 1% (w/v) agar and returned to the 24-h 140-µmol m−2 s−1 22°C incubator. Adventitious root growth from scions was monitored and removed periodically. The rootstocks used were ALMT1pro:GUS and the wild type, and the scions used were ALMT1pro:GUS, the wild type, fls2, and coi1. GUS and ALMT1 expression in grafts and FB17 growth in the roots were measured as described herein.
ALMT1pro:GUS Expression Assays in Grafts
Micrografted plants were grown sterilely on wire mesh in 6-well microtiter plates and leaves were sprayed with different MAMPs. Twenty-four hours post treatment plants were stained for GUS (Sigma-Aldrich) according to the manufacturer’s instructions. The plants were vacuum infiltrated for 5 min and then incubated at 37°C for 1 h. Tissues were cleared in 95% ethanol and photographed using a Nikon D90 camera.
Callose Staining
Roots were harvested 1 d post inoculation with COR or flg22 or flg22 + FB17 or flg22 + tasA::mls or mock (water control), and samples were fixed in 3:1 ethanol:acetic acid solution for several hours. The fixative was changed several times to ensure both thorough fixing and clearing of the tissues, which was essential for good callose detection in the roots. Tissues were rinsed with water several times and rehydrated in 70% ethanol for 2 h, 50% ethanol for an additional 2 h, and water overnight. Decolorization was done by adding 10% NaOH and being placed at 37°C for 1 to 2 h. Tissue was rinsed several times with water prior to incubation for 30 min in darkness in 0.01% (w/v) aniline blue dissolved in 150 mm K2HPO4, pH 9.5. Callose deposits were detected using Zeiss meta spectral detector on the LSM5 DUO microscope (excitation, 365 nm; emission, 420 nm).
Microscopy
To view adherent FB17 cells on the root surface by laser scanning confocal microscopy, the roots were stained with SYTO13 (Invitrogen, Molecular Probes). Experiments were performed 24 h post inoculation and post treatment with FB17 to 3-week-old plants grown in Magenta boxes. Images were captured with a 25× C-Apochromat objective (numerical aperture of 1.2) on a Zeiss LSM 510 NLO attached to an Axiovert 200M with Zeiss AIM software (Rel. 3.2). Images were acquired with the 488-nm line excitation of an Argon laser using a 505-nm-long pass emission filter. GUS-stained plants were viewed with a Zeiss M2Bio Fluorescence Stereomicroscope with the 1.6× lens and digitally acquired with a Zeiss Axio Cam color camera using AxioVision software.
Data Analysis
All data presented are the mean values of at least six replicates unless mentioned otherwise, and the data have been presented as means ± se. The data were analyzed by one-way ANOVA using Microsoft Excel 2010 (Microsoft Corporation), and post hoc mean separations were performed by Duncan’s Multiple Range Test (DMRT) at P ≤ 0.05 using software SPSS version 12.0.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analyses of GUS staining in ALMT1pro:GUS and ALMT1 expression.
Supplemental Figure S2. GUS staining and RT-PCR analyses in leaves after different with different MAMPs and COR treatments.
Supplemental Figure S3. Measurement of ALMT1 expression and FB17 colonization in wild-type and mutant fls2 and coi1.
Supplemental Figure S4. GUS expression in micrograft wild type/ALMT1pro:GUS.
Supplemental Figure S5. FB17 colonization on roots of micrografts fls2/ALMT1pro:GUS and coi1/ALMT1pro:GUS.
Supplemental Figure S6. GUS staining and RT-PCR analyses of CYP71A12, MYB51, and WRKY11.
Supplemental Figure S7. GUS staining in the roots of CYP71A12pro:GUS, MYB51pro:GUS, and WRKY11pro:GUS promoter:GUS reporters.
Supplemental Figure S8. Surface biofilm formation and suppression of flg22-activated innate immunity by different B. subtilis biofilm-deficient mutants.
Supplemental Figure S9. FB17 colonization in roots of cyp71A12, myb51, and wrky11.
Supplemental Figure S10. GUS staining in the roots of CYP71A12pro:GUS, MYB51pro:GUS, and WRKY11pro:GUS lines post treatment with AlCl3 or MA.
Supplemental Figure S11. GUS analyses in roots of ALMT1pro:GUS after root FB17 treatment.
Supplemental Figure S12. Suppression of flg22-activated root immune responses by foliar application of COR.
Supplementary Material
Acknowledgments
We thank Drs. Frederick M. Ausubel and Roberto Kolter for Arabidopsis transgenic lines CYP71A12pro:GUS, MYB51pro:GUS, and WRKY11pro:GUS reporter lines, and the tasA::mls mutant. We also thank both anonymous reviewers for insightful comments.
Glossary
- MA
malic acid
- MAMP
microbe-associated molecular pattern
- COR
coronatine
- PGPR
plant growth-promoting rhizobacteria
- SAR
systemic acquired resistance
- ISR
induced systemic resistance
- SA
salicylic acid
- JA
jasmonic acid
- ET
ethylene
- LPS
lipopolysaccharides
- PGN
peptidoglycan
- cfu
colony-forming units
- sqRT
semiquantitative reverse transcription
- MeJA
methyl jasmonate
- CFL
cell free lysate
- HK
heat killed
- qRT
quantitative real-time
- EPS
extracellular polysaccharide
- MTI
MAMPs-triggered immunity
- MS
Murashige and Skoog
- LB
Luria-Bertani
- CV
crystal violet
- OD600
optical density at 600 nm
- DMRT
Duncan’s Multiple Range Test
References
- Alteri CJ, Xicohténcatl-Cortes J, Hess S, Caballero-Olín G, Girón JA, Friedman RL. (2007) Mycobacterium tuberculosis produces pili during human infection. Proc Natl Acad Sci USA 104: 5145–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri DV, Loyola-Vargas VM, Broeckling CD, De-la-Peña C, Jasinski M, Santelia D, Martinoia E, Sumner LW, Banta LM, Stermitz F, et al. (2008) Altered profile of secondary metabolites in the root exudates of Arabidopsis ATP-binding cassette transporter mutants. Plant Physiol 146: 762–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri DV, Vivanco JM. (2009) Regulation and function of root exudates. Plant Cell Environ 32: 666–681 [DOI] [PubMed] [Google Scholar]
- Bahrun A, Jensen CR, Asch F, Mogensen VO. (2002) Drought-induced changes in xylem pH, ionic composition, and ABA concentration act as early signals in field-grown maize (Zea mays L.). J Exp Bot 53: 251–263 [DOI] [PubMed] [Google Scholar]
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57: 233–266 [DOI] [PubMed] [Google Scholar]
- Benhamou N, Kloepper JW, Quadt-Hallman A, Tuzun S. (1996) Induction of defense-related ultrastructural modifications in pea root tissues inoculated with endophytic bacteria. Plant Physiol 112: 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block A, Li G, Fu ZQ, Alfano JR. (2008) Phytopathogen type III effector weaponry and their plant targets. Curr Opin Plant Biol 11: 396–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Chu F, Kearns DB, Losick R, Kolter R. (2006) A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol 59: 1229–1238 [DOI] [PubMed] [Google Scholar]
- Brooks DM, Bender CL, Kunkel BN. (2005) The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defenses in Arabidopsis thaliana. Mol Plant Pathol 6: 629–639 [DOI] [PubMed] [Google Scholar]
- Casati P, Drincovich MF, Edwards GE, Andreo CS. (1999) Malate metabolism by NADP-malic enzyme in plant defense. Photosynth Res 61: 99–105 [Google Scholar]
- Chen C, Belanger RR, Benhamou N, Paulitz TC. (2000) Defense enzymes induced in cucumber roots by treatment with plant growth-promoting rhizobacteria (PGPR) and Pythium aphanidermatum. Physiol Mol Plant Pathol 56: 13–23 [Google Scholar]
- Chen Y, Cao S, Chai Y, Clardy J, Kolter R, Guo J-H, Losick R. (2012) A Bacillus subtilis sensor kinase involved in triggering biofilm formation on the roots of tomato plants. Mol Microbiol 85: 418–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary DK, Johri BN. (2009) Interactions of Bacillus spp. and plants—with special reference to induced systemic resistance (ISR). Microbiol Res 164: 493–513 [DOI] [PubMed] [Google Scholar]
- Conrath U, Beckers GJ, Flors V, García-Agustín P, Jakab G, Mauch F, Newman M-A, Pieterse CM, Poinssot B, Pozo MJ, et al. (2006) Priming: getting ready for battle. Mol Plant Microbe Interact 19: 1062–1071 [DOI] [PubMed] [Google Scholar]
- Conrath U, Pieterse CM, Mauch-Mani B. (2002) Priming in plant-pathogen interactions. Trends Plant Sci 7: 210–216 [DOI] [PubMed] [Google Scholar]
- Daniel RA, Errington J. (2003) Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113: 767–776 [DOI] [PubMed] [Google Scholar]
- De Meyer G, Capieau K, Audenaert K, Buchala A, Métraux J-P, Höfte M. (1999) Nanogram amounts of salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 activate the systemic acquired resistance pathway in bean. Mol Plant Microbe Interact 12: 450–458 [DOI] [PubMed] [Google Scholar]
- Denoux C, Galletti R, Mammarella N, Gopalan S, Werck D, De Lorenzo G, Ferrari S, Ausubel FM, Dewdney J. (2008) Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol Plant 1: 423–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doornbos RF, Geraats BPJ, Kuramae EE, van Loon LC, Bakker PAHM. (2011) Effects of jasmonic acid, ethylene, and salicylic acid signaling on the rhizosphere bacterial community of Arabidopsis thaliana. Mol Plant Microbe Interact 24: 395–407 [DOI] [PubMed] [Google Scholar]
- Erb M, Lenk C, Degenhardt J, Turlings TCJ. (2009) The underestimated role of roots in defense against leaf attackers. Trends Plant Sci 14: 653–659 [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Martinoia E. (2009) Malate. Jack of all trades or master of a few? Phytochemistry 70: 828–832 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Berger B, Mock HP, Müller C, Weisshaar B, Flügge UI. (2007) The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J 50: 886–901 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Bauer Z, Boller T. (2001) Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13: 1155–1163 [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Gust AA, Biswas R, Lenz HD, Rauhut T, Ranf S, Kemmerling B, Götz F, Glawischnig E, Lee J, Felix G, et al. (2007) Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J Biol Chem 282: 32338–32348 [DOI] [PubMed] [Google Scholar]
- Hamon MA, Lazazzera BA. (2001) The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol 42: 1199–1209 [DOI] [PubMed] [Google Scholar]
- Harrison MJ. (2005) Signaling in the arbuscular mycorrhizal symbiosis. Ann Rev Microbiol 59: 19–42 [DOI] [PubMed] [Google Scholar]
- Hoekenga OA, Maron LG, Piñeros MA, Cançado GMA, Shaff J, Kobayashi Y, Ryan PR, Dong B, Delhaize E, Sasaki T, et al. (2006) AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc Natl Acad Sci USA 103: 9738–9743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iriti M, Faoro F. (2003) Benzothiadiazole (BTH) induces cell-death independent resistance in Phaseolus vulgaris against Uromyces appendiculatus. J Phytopathol 151: 171–180 [Google Scholar]
- Jacobs S, Zechmann B, Molitor A, Trujillo M, Petutschnig E, Lipka V, Kogel KH, Schäfer P. (2011) Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol 156: 726–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journot-Catalino N, Somssich IE, Roby D, Kroj T. (2006) The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18: 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT. (2001) Defensive function of herbivore-induced plant volatile emissions in nature. Science 291: 2141–2144 [DOI] [PubMed] [Google Scholar]
- Kleijn RJ, Buescher JM, Le Chat L, Jules M, Aymerich S, Sauer U. (2010) Metabolic fluxes during strong carbon catabolite repression by malate in Bacillus subtilis. J Biol Chem 285: 1587–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloek AP, Verbsky ML, Sharma SB, Schoelz JE, Vogel J, Klessig DF, Kunkel BN. (2001) Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J 26: 509–522 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Hoekenga OA, Itoh H, Nakashima M, Saito S, Shaff JE, Maron LG, Piñeros MA, Kochian LV, Koyama H. (2007) Characterization of AtALMT1 expression in aluminum-inducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiol 145: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo T, Kiba T, Sakakibara H. (2010) Metabolism and long-distance translocation of cytokinins. J Integr Plant Biol 52: 53–60 [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5: 325–331 [DOI] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lin H, Zhang W, Zou Y, Zhang J, Tang X, Zhou JM. (2005) Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc Natl Acad Sci USA 102: 12990–12995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633 [DOI] [PubMed] [Google Scholar]
- Livaja M, Zeidler D, von Rad U, Durner J. (2008) Transcriptional responses of Arabidopsis thaliana to the bacteria-derived PAMPs harpin and lipopolysaccharide. Immunobiology 213: 161–171 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, de la Vega OM, Guevara-García A, Herrera-Estrella L. (2000) Enhanced phosphorus uptake in transgenic tobacco plants that overproduce citrate. Nat Biotechnol 18: 450–453 [DOI] [PubMed] [Google Scholar]
- Lugtenberg BJJ, Dekkers L, Bloemberg GV. (2001) Molecular determinants of rhizosphere colonization by Pseudomonas. Annu Rev Phytopathol 39: 461–490 [DOI] [PubMed] [Google Scholar]
- Lugtenberg BJJ, Kravchenko LV, Simons M. (1999) Tomato seed and root exudate sugars: composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ Microbiol 1: 439–446 [DOI] [PubMed] [Google Scholar]
- Marschner P, Neumann G, Kania A, Weiskopf L, Lieberei R. (2002) Spatial and temporal dynamics of the microbial community structure in the rhizosphere of cluster roots of white lupin (Lupinus albus L.). Plant Soil 246: 167–174 [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He S-Y. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Meziane H, van Der Sluis I, van Loon LC, Höfte M, Bakker PAHM. (2005) Determinants of Pseudomonas putida WCS358 involved in inducing systemic resistance in plants. Mol Plant Pathol 6: 177–185 [DOI] [PubMed] [Google Scholar]
- Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, Ausubel FM. (2010) Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22: 973–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nafisi M, Goregaoker S, Botanga CJ, Glawischnig E, Olsen CE, Halkier BA, Glazebrook J. (2007) Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19: 2039–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu DD, Liu HX, Jiang CH, Wang YP, Wang QY, Jin HL, Guo JH. (2011) The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling pathways. Mol Plant Microbe Interact 24: 533–542 [DOI] [PubMed] [Google Scholar]
- O’Toole GA, Kolter R. (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28: 449–461 [DOI] [PubMed] [Google Scholar]
- Oh JJ, Kim JG, Jeon E, Yoo CH, Moon JS, Rhee S, Hwang I. (2007) Amyloidogenesis of type III-dependent harpins from plant pathogenic bacteria. J Biol Chem 282: 13601–13609 [DOI] [PubMed] [Google Scholar]
- Ongena M, Daayf F, Jacques P, Thonart P, Benhamou N, Paulitz TC, Bèlanger RR. (2000) Systemic induction of phytoalexins in cucumber in response to treatments with fluorescent Pseudomonas. Plant Pathol 49: 523–530 [Google Scholar]
- Pieterse CMJ, van Wees SCM, van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC. (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10: 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda A, Zheng SJ, van Loon JJA, Pieterse CMJ, Dicke M. (2010) Helping plants to deal with insects: the role of beneficial soil-borne microbes. Trends Plant Sci 15: 507–514 [DOI] [PubMed] [Google Scholar]
- Pozo MJ, Azcón-Aguilar C. (2007) Unraveling mycorrhiza-induced resistance. Curr Opin Plant Biol 10: 393–398 [DOI] [PubMed] [Google Scholar]
- Ramamoorthy V, Viswanathan R, Raguchander T, Prakasam V, Samiyappan R. (2001) Induction of systemic resistance by plant growth promoting rhizobacteria in crop plants against pests and diseases. Crop Prot 20: 1–11 [Google Scholar]
- Romero D, Aguilar C, Losick R, Kolter R. (2010) Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci USA 107: 2230–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrappa T, Biedrzycki ML, Bais HP. (2008a) Causes and consequences of plant-associated biofilms. FEMS Microbiol Ecol 64: 153–166 [DOI] [PubMed] [Google Scholar]
- Rudrappa T, Biedrzycki ML, Kunjeti SG, Donofrio NM, Czymmek KJ, Paré PW, Bais HP. (2010) The rhizobacterial elicitor acetoin induces systemic resistance in Arabidopsis thaliana. Commun Integr Biol 3: 130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrappa T, Czymmek KJ, Paré PW, Bais HP. (2008b) Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol 148: 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink HP. (2000) Root nodulation and infection factors produced by rhizobial bacteria. Annu Rev Microbiol 54: 257–288 [DOI] [PubMed] [Google Scholar]
- Stöver AG, Driks A. (1999) Secretion, localization, and antibacterial activity of TasA, a Bacillus subtilis spore-associated protein. J Bacteriol 181: 1664–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Taniguchi M, Sugiyama T. (2001) Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: implication of cytokinin species that induces gene expression of maize response regulator. Plant Cell Physiol 42: 85–93 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F. (2008) Interplay between MAMP-triggered and SA-mediated defense responses. Plant J 53: 763–775 [DOI] [PubMed] [Google Scholar]
- Turnbull CGN, Booker JP, Leyser HMO. (2002) Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J 32: 255–262 [DOI] [PubMed] [Google Scholar]
- Uppalapati SR, Ayoubi P, Weng H, Palmer DA, Mitchell RE, Jones W, Bender CL. (2005) The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J 42: 201–217 [DOI] [PubMed] [Google Scholar]
- Uren NC. (2000) Types, amounts, and possible functions of compounds released into the rhizosphere by soil-grown plants. In Pinton R, Varanini Z, Nannipieri P, eds, The Rhizosphere: Biochemistry and Organic Substances at the Soil-Plant Interface. Marcel Dekker, New York, pp 19–40 [Google Scholar]
- van der Ent S, van Hulten M, Pozo MJ, Czechowski T, Udvardi MK, Pieterse CM, Ton J. (2009) Priming of plant innate immunity by rhizobacteria and beta-aminobutyric acid: differences and similarities in regulation. New Phytol 183: 419–431 [DOI] [PubMed] [Google Scholar]
- van Loon LC, Bakker PA, Pieterse CMJ. (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36: 453–483 [DOI] [PubMed] [Google Scholar]
- van Loon LC, van Strien EA. (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55: 85–97 [Google Scholar]
- van Norman JM, Frederick RL, Sieburth LE. (2004) BYPASS1 negatively regulates a root-derived signal that controls plant architecture. Curr Biol 14: 1739–1746 [DOI] [PubMed] [Google Scholar]
- van Oosten VR, Bodenhausen N, Reymond P, van Pelt JA, van Loon LC, Dicke M, Pieterse CMJ. (2008) Differential effectiveness of microbially induced resistance against herbivorous insects in Arabidopsis. Mol Plant Microbe Interact 21: 919–930 [DOI] [PubMed] [Google Scholar]
- Verhagen BW, Glazebrook J, Zhu T, Chang HS, van Loon LC, Pieterse CMJ. (2004) The transcriptome of rhizobacteria-induced systemic resistance in arabidopsis. Mol Plant Microbe Interact 17: 895–908 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. (2009) Salicylic acid, a multifaceted hormone to combat disease. Ann Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Wan J, Zhang X-C, Neece D, Ramonell KM, Clough S, Kim S-Y, Stacey MG, Stacey G. (2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yan Z, Reddy MS, Ryu C-M, McInroy JA, Wilson M, Kloepper JW. (2002) Induced systemic protection against tomato late blight elicited by plant growth-promoting rhizobacteria. Phytopathology 92: 1329–1333 [DOI] [PubMed] [Google Scholar]
- Yang JW, Yi H-S, Kim H, Lee B, Lee S, Ghim S-Y, Ryu C-M. (2011) Whitefly infestation of pepper plants elicits defense responses against bacterial pathogens in leaves and roots and changes the below-ground microflora. J Ecol 99: 46–56 [Google Scholar]
- Zamioudis C, Pieterse CMJ. (2012) Modulation of host immunity by beneficial microbes. Mol Plant Microbe Interact 25: 139–150 [DOI] [PubMed] [Google Scholar]
- Zhang H, Murzello C, Sun Y, Kim M-S, Xie X, Jeter RM, Zak JC, Dowd SE, Paré PW. (2010) Choline and osmotic-stress tolerance induced in Arabidopsis by the soil microbe Bacillus subtilis (GB03). Mol Plant Microbe Interact 23: 1097–1104 [DOI] [PubMed] [Google Scholar]
- Zhang H, Xie X, Kim M-S, Kornyeyev DA, Holaday S, Paré PW. (2008) Soil bacteria augment Arabidopsis photosynthesis by decreasing glucose sensing and abscisic acid levels in planta. Plant J 56: 264–273 [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhou J-M. (2010) Plant immunity triggered by microbial molecular signatures. Mol Plant 3: 783–793 [DOI] [PubMed] [Google Scholar]
- Zeidler D, Zähringer U, Gerber I, Dubery I, Hartung T, Bors W, Hutzler P, Durner J. (2004) Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc Natl Acad Sci USA 101: 15811–15816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley E, Jones JDG, Felix G, Boller T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.