Abstract
The Dobzhansky-Muller model of incompatibilities explains reproductive isolation between species by incorrect epistatic interactions. Although the mechanisms of speciation are of great interest, no incompatibility has been characterized at the gene level in mammals. The Hybrid sterility 1 gene (Hst1) participates in the arrest of meiosis in F1 males of certain strains from two Mus musculus subspecies, e.g., PWD from M. m. musculus and C57BL/6J (henceforth B6) from M. m. domesticus. Hst1 has been identified as a meiotic PR-domain gene (Prdm9) encoding histone 3 methyltransferase in the male offspring of PWD females and B6 males, (PWD×B6)F1. To characterize the incompatibilities underlying hybrid sterility, we phenotyped reproductive and meiotic markers in males with altered copy numbers of Prdm9. A partial rescue of fertility was observed upon removal of the B6 allele of Prdm9 from the azoospermic (PWD×B6)F1 hybrids, whereas removing one of the two Prdm9 copies in PWD or B6 background had no effect on male reproduction. Incompatibility(ies) not involving Prdm9B6 also acts in the (PWD×B6)F1 hybrids, since the correction of hybrid sterility by Prdm9B6 deletion was not complete. Additions and subtractions of Prdm9 copies, as well as allelic replacements, improved meiotic progression and fecundity also in the progeny-producing reciprocal (B6×PWD)F1 males. Moreover, an increased dosage of Prdm9 and reciprocal cross enhanced fertility of other sperm-carrying male hybrids, (PWD×B6-C3H.Prdm9)F1, harboring another Prdm9 allele of M. m. domesticus origin. The levels of Prdm9 mRNA isoforms were similar in the prepubertal testes of all types of F1 hybrids of PWD with B6 and B6-C3H.Prdm9 despite their different prospective fertility, but decreased to 53% after removal of Prdm9B6. Therefore, the Prdm9B6 allele probably takes part in posttranscriptional dominant-negative hybrid interaction(s) absent in the parental strains.
Author Summary
Disturbed gametogenesis in the progeny of two fertile parental forms is called hybrid sterility; it is an important part of reproductive barriers between species. The Dobzhansky-Muller model of incompatibilities explains reproductive isolation between species by incorrect interactions between genes. Hybrid sterility 1 (Hst1) is one of the genes causing meiotic arrest in F1 male hybrids between certain Mus musculus musculus (e.g., the PWD strain) and M. m. domesticus (C57BL/6J etc.) mice. Hst1, the first mammalian candidate for a speciation gene, was identified as a meiotic PR/SET-domain gene, Prdm9, but the mechanism causing sterility has remained unknown. While the F1 male offspring of C57BL/6J males and PWD females produce no sperm, the males from the reciprocal cross using PWD males and C57BL/6J females yield progeny. Here we show that the meiotic progress and fertility of hybrid males from both F1 crosses improved by removal as well as overexpression of the C57BL/6J allele of Prdm9, suggesting that Prdm9 interactions not present in the parental species (incompatibilities) play a role in hybrid sterility. Furthermore, the Prdm9 dosage also controlled fecundity in other F1 hybrids, indicating that this gene is an important regulator of mouse hybrid fertility.
Introduction
Hybrid sterility is a condition in which two fertile parental forms produce progeny with disturbed gametogenesis. In mammals and Drosophila, it affects spermatogenesis more often than oogenesis [1]. Hybrid sterility acts as a reproductive barrier between species [2]. Although its molecular mechanism is of great interest, only five animal genes involved in hybrid sterility have been cloned and characterized, four of them from Drosophila [3]–[9]. The Dobzhansky-Muller model of incompatibilities of genes [10] explains the reproductive isolation between species by their incorrect epistatic interactions. These interactions (or lack of the correct ones) result in hybrid fitness reduction, probably because the combination of the diverged alleles of the interactors did not pass through natural selection.
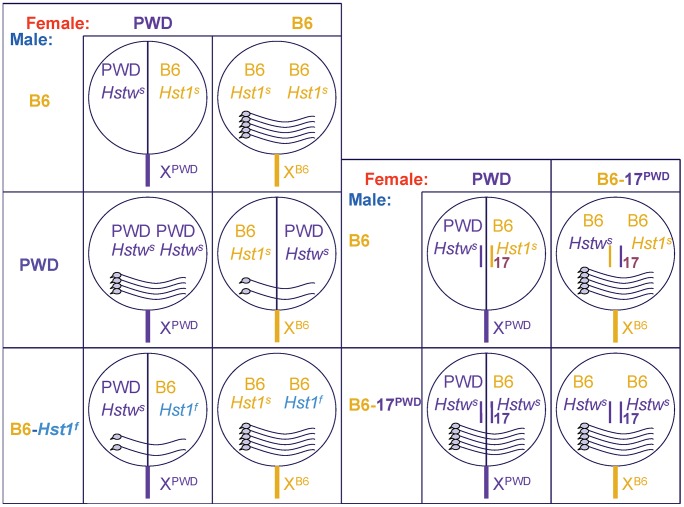
The mouse Hybrid sterility 1 gene (Hst1) is one of the major genes causing meiotic arrest in F1 male hybrids between Mus m. musculus (Mmm) mice harboring the Hstws allele (e.g., the PWD strain) and laboratory strains bearing the Hst1s allele [11]. Strains with the Hst1s allele include Mus m. domesticus (Mmd)-derived C57BL/6J (henceforth B6) and various substrains of the 129 strain. While the male offspring from the crosses of Hst1s strains with PWD females, e.g., (PWD×B6)F1, are azoospermic, the males from the reciprocal crosses using PWD males [(B6×PWD)F1] can sire offspring (Figure 1, [6], [12]). Other laboratory strains (C3H, the congenic B6.C3H-Hst1f, etc.) harbor the Hst1f allele and produce sperm-carrying F1 males in crosses with PWD females [13]. The male offspring of B6 with C3H, as well as F1 females in all PWD, B6, and C3H crosses are fertile.
Figure 1. Fertility of male offspring resulting from various crosses.
The female parents are shown from left to right and male parents from top to bottom. The circles symbolize genomes (chrX sticking out), intersubspecific hybrids have the circles split in halves; the pictures of sperm cells within the circles indicate the degree of fertility. Except for the (PWD×B6)F1 hybrids, all males carry sperm; female offspring from all indicated crosses are fertile.
The Hst1 gene is the first mammalian candidate for a speciation gene. Hst1 was identified by consecutive mapping of the Hst1 alleles [14]–[16], expression profiling [17], allelic sequencing [18], [19], and transgenic rescue [6]. The best candidate was confirmed by comparing the phenotypes of its null allele with the phenotypes of the sterile hybrids [6]. Hst1 is also called Prdm9 (PR-domain containing 9) or Meisetz (Meiotic gene with SET/PR domain and Zinc fingers). The product of this gene trimethylates histone 3 on lysine 4 (H3K4m3; [20]). Spermatocytes of Prdm9−/− animals and sterile hybrids arrest at the pachytene stage of meiotic prophase I, displaying defects in chromosomal pairing and sex body formation, as well as downregulation of meiotic genes. The downregulation correlates with a lower level of H3K4m3 at promoters, at least in the case of the Morc2b gene (4932411A10Rik; [6], [20]). The Morc2b mRNA is elevated in (PWD×B6-C3H.Hst1f)F1 compared to (PWD×B6)F1 prepubertal testis, while the levels of all known Prdm9 transcripts are similar [6].
The chromatin of mouse meiotic recombination hotspots is marked by H3K4m3 at the start of meiosis [21]. Genetic mapping of a gene acting in trans to influence the activity of recombination hotspots also led to the identification of Prdm9 [22], [23]. PRDM9 binds DNA at recombination hotspots via its zinc-fingers (ZnFs) in vitro, and genetic manipulation of ZnFs changes the localization of the hotspots [22], [24].
The Prdm9 genes from C3H and B6 strains (alleles Hst1f and Hst1s, respectively), display many polymorphisms [6]. The difference that may underlie hybrid sterility is the number of C-terminal ZnFs [6]. The number of ZnFs corresponds to Hst1 alleles in other classical Mmd laboratory strains [19], [25]. The ZnF-encoding region of the PWD allele differs from both C3H and B6 [6], but whether Prdm9PWD is identical to Hstws and whether it contributes to hybrid sterility is not known. Although accelerated evolution of the minisatellite-like ZnF-encoding region of PRDM9 was manifested in human and animals [26], it remains to be shown whether PRDM9 has a more general role in speciation.
The Prdm9B6 allele is necessary but not sufficient for hybrid sterility. The Hstw gene from Mmm is also located on chr17 [12]. In the sterile (PWD×B6)F1 males, chr17 carries the combination Hstws/Prdm9B6, but the same genotype in the B6 background of (B6.PWD-Chr17×B6)F1 yields fertile males (Figure 1; [27]). It was shown recently that Hstws/Prdm9B6 is the only combination resulting in a complete meiotic arrest in (PWD×B6)F1 hybrid males, because both Prdm9B6/Prdm9B6 and Hstws/Hstws homozygotes on the same background were fertile [12]. However, even on the F1 background, the Hstws/Prdm9B6 combination does not always lead to azoospermic males, as the reciprocal (B6×PWD)F1 males carry sperm. Thus, mouse hybrid sterility reflects incompatibilities among multiple hybrid sterility loci, one of them being the Prdm9 gene [12].
Here we manipulated the dosage and allelic combinations of Prdm9 in an attempt to characterize the role of this mouse hybrid sterility gene in the incompatibilities. If Prdm9B6 has a dominant-negative effect(s), its deletion should alleviate the meiotic arrest and rescue the fertility of hybrids. Moreover, a Prdm9-overexpressing transgene might dilute the Prdm9B6 incompatibility(ies), which should rescue fertility regardless of the Prdm9 allelic origin. We show that the fertility of all F1 intersubspecific hybrids tested is proportional to the dosage of Prdm9 regardless of its allele; the only exception is the combination of one Prdm9B6 allele with one Prdm9PWD allele in either type of reciprocal hybrid that results into a more sterile phenotype than the corresponding hybrid harboring only one Prdm9PWD allele. This exception indicates an F1 hybrid-specific dominant-negative interaction(s) of Prdm9B6.
Results
Dosage-dependent, allele-independent rescue of fertility in the (PWD×B6)F1 hybrid
The fertility of azoospermic intersubspecific (PWD×B6)F1 hybrid males is rescued by Prdm9C3H-carrying BACs [6]. Moreover, six copies of the Prdm9C3H allele in a transgene (BAC24) increased reproductive fitness compared to two copies of the same allele (BAC5 transgene) in Prdm9PWD/B6 (PWD×B6)F1 intersubspecific hybrid males ([6] and Table 1), although no effect of increased Prdm9 dosage appeared on the intrasubspecific background (a mix of 129 and B6 genomes, henceforth B6 * 129 [6]). There are three possible explanations for the fertility rescue by the Prdm9C3H BACs: first, by allelic replacement; second, by increased dosage regardless of allelic origin; third, both. To distinguish among these hypotheses, a transgene harboring the Prdm9B6 allele was utilized. The C57BL/6J-Tg(RP23-159N6)75Bdm strain carries two Prdm9-expressing copies of a B6 BAC transgene on B6 background [24]. In this background, the transgene has no effect on fertility (Table S1). After outcrossing the heterozygous transgenic males to PWD, non-transgenic F1 hybrid males were azoospermic (as expected), while their transgenic littermates had an increased testicular weight (TW) and carried sperm (Table 1). The increased copy number of the Prdm9B6 allele also rescued fertility of azoospermic hybrids produced by another Mmm strain, STUS (Table S1). Thus the improvement of fertility by increased Prdm9 dosage is not restricted to the C3H allele.
Table 1. The effect of Prdm9 dosage on hybrid sterility.
| Cross (female first) | Prdm9 | n | TW | SC |
| PWD×B6-BACB6 | PWD/B6 | 9 | 55 | 0.00a |
| PWD×B6-BACB6 | PWD/B6+2B6 | 3 | 167* | 18* a |
| PWD×B6-KO | PWD/B6 | 10 | 59 | 0.00 |
| PWD×B6-KO | PWD/− | 12 | 82* | 0.06* |
| PWD×(B6-Prdm9C3H×KO) | PWD/− | 10 | 94 | 0.10 |
| PWD×(B6-Prdm9C3H×KO) | PWD/C3H | 8 | 140* | 1.06* |
| PWD×B6-Prdm9C3H | PWD/C3H | 8 | 109 | 0.2 |
| PWD×(KO×BAC5) | PWD/− | 13 | 85 | 0.07 |
| PWD×(KO×BAC5) | PWD/−+2C3H | 15 | 168* | 1.6* |
| PWD×(KO×BAC24) | PWD/− | 9 | 77 | 0.03 |
| PWD×(KO×BAC24) | PWD/−+6C3H | 5 | 235* | 3.9* |
B6-BACB6, C57BL/6J-Tg(RP23-159N6)75Bdm mouse heterozygous for a Prdm9B6 transgene; BAC5, transgenic strain with two copies of Prdm9C3H; BAC24, strain carrying six transgenic copies of Prdm9C3H; KO, B6-KO, heterozygote for the Prdm9 knock-out; Prdm9, genotype at the Prdm9 locus (maternal/paternal); −, null; +, transgenic Prdm9 alleles; n, number of males; TW, mean weight of paired testicles in mg; SC, average sperm count (millions) in paired caput epididymides or
in the left epididymis by a different method;
significantly higher (p<0.01) compared to littermates (in the row above).
To further investigate the effect of Prdm9 dosage on hybrid sterility, we used two null alleles of Prdm9, a large deletion of proximal chr17 (Sod2df14J [28]) and a knock-out of Prdm9 that removes the first five coding exons (Prdm9tm1Ymat [20]). Males hemizygous for these alleles on (129 * B6) as well as on B6 background display similar fertility parameters as their littermates ([20], [29], and Table S2). To determine the effect of the null alleles on intersubspecific F1 hybrids, the hemizygous males were outcrossed to PWD. Unlike their azoospermic Prdm9PWD/B6 F1 littermates, most of the Prdm9PWD/− intersubspecific hybrids were semisterile with TW, sperm count (SC), and offspring production (offspring per female per month, OFM) significantly higher than in the (PWD×B6)F1 littermates (Table 1 and Table S3). All Prdm9PWD/− hybrid males resulting from the cross of PWD females with Prdm9B6/− on B6 background carried a low but detectable amount of sperm and most of them produced offspring (0.3±0.2 OFM). As an additional control, we introduced the Prdm9tm1Ymat null allele into the PWD background. Here, one copy of Prdm9PWD was sufficient to maintain the fertility parameters of the Prdm9PWD/PWD littermates (Table S2). Therefore, the (PWD×B6)F1 hybrid genetic background appears to be more sensitive to low Prdm9 dosage than that of either parent. Both addition and removal of Prdm9B6 improves the phenotype of (PWD×B6)F1 males. The fertility rescue of azoospermic hybrid males by the Prdm9 null alleles, albeit partial, suggests that an aberrant interaction(s) of Prdm9B6 occurs in (PWD×B6)F1 hybrids that is not present in the parental strains.
Incompatibility of Prdm9B6 in (PWD×B6)F1 males
The Prdm9C3H allele rescues the fertility phenotype of the (PWD×B6)F1 hybrid in a dosage-dependent manner. To compare the effect of a single C3H allele and a null allele on hybrid males resulting from a cross segregating these alleles, we utilized the congenic strain B6-Prdm9C3H. After outcrossing animals hemizygous for Prdm9 to this congenic and then crossing the preselected Prdm9C3H/− males to PWD females, the resulting Prdm9PWD/− hybrids had a significantly lower TW and SC than their Prdm9PWD/C3H littermates (Table 1). The Prdm9PWD/− hybrids produced markedly less progeny (0.3±0.2 OFM) in comparison with the Prdm9PWD/C3H hybrids (3.6±0.5 OFM, Table S4). Thus the intersubspecific F1 hybrid males carrying Prdm9PWD/− were not just more fertile than Prdm9PWD/B6, but they were also less fertile than Prdm9PWD/C3H.
To analyze the sensitivity of Prdm9PWD/− F1 background, the dosage of Prdm9C3H was increased by utilizing the BAC5 (two copies of Prdm9C3H) or the BAC24 (six copies of Prdm9C3H) transgenes. Again, the fertility parameters of the F1 transgenic hybrids improved with Prdm9C3H dosage (Table 1; pSC = 0.006, pTW = 0.008, but no significant difference in relative testes weight: 6.6 versus 7.3, p = 0.33). The Prdm9 dosage effect is therefore observed in both Prdm9PWD/B6 and Prdm9PWD/− F1 hybrids. In conclusion, the B6 allele displays different properties than the C3H allele when present in one copy in the Prdm9PWD/B6 F1 hybrid male, because it decreases the fertility compared to Prdm9PWD/− F1 hybrid. This could be the result of a dominant-negative interaction of Prdm9B6 with specific loci in (PWD×B6)F1 and/or with the Prdm9PWD allele.
The semisterility of Prdm9PWD/− maintains characteristics of hybrid sterility
Hybrid sterility is most often sex-specific [1] and dependent on the origin of parents. The (PWD×B6)F1 hybrid displays a complete male-specific arrest of gametogenesis that is at least partially alleviated in reciprocal (B6×PWD)F1 and in 94% of males from the backcross ((PWD×B6)×B6) [12]. To determine whether the semisterility of the Prdm9PWD/− intersubspecific males displays the same features, additional phenotyping was performed. All four tested Prdm9PWD/− females produced offspring, in a number (4.8±0.8 OFM) similar to their Prdm9PWD/B6 littermates (4.5±0.2) and (PWD×B6-Prdm9C3H)F1 females (5.5±0.6). The semisterility of Prdm9PWD/− F1 hybrids is thus male-specific. When the Prdm9PWD/− F1 females were backcrossed to PWD, the resulting BC1 Prdm9PWD/− hemizygous males displayed a four-fold increase of average SC compared to Prdm9PWD/− F1 hybrid males (p = 0.013), indicating that the compromised fertility is most pronounced in the F1 generation. The TW and SC of the reciprocal (B6-Prdm9−/wt×PWD)F1 Prdm9−/PWD males (Table 2) was markedly higher than that of the (PWD×B6-Prdm9−/wt)F1 Prdm9PWD/− hybrids (Table 1), displaying an asymmetry similar to the F1 cross of B6 and PWD. Thus, the fertility of Prdm9PWD/− hybrid likely suffers from an incompatibility(ies), but to a lesser degree than in the presence of Prdm9B6.
Table 2. Effects of Prdm9 alleles and dosage on reciprocal hybrids.
| Cross (female first) | Prdm9 | n | BW | TW | SC |
| B6-KO×PWD | B6/PWD | 15 | 25 | 105 | 0.5 |
| B6-KO×PWD | −/PWD | 11 | 27 | 174* | 4.1* |
| (B6×B6-Prdm9C3H)×PWD | B6/PWD | 7 | 24 | 97 | 0.3 |
| (B6×B6-Prdm9C3H)×PWD | C3H/PWD | 5 | 25 | 171* | 2.2* |
| BAC5/BAC5×PWD | B6+2C3H/PWD | 7 | 25 | 181* | 3.1* |
| B6.PWD-Chr17×PWD | PWD/PWDa | 7 | 24 | 220* | 4.2* |
| B6×B6 | B6/B6 | 3 | 28 | 195* | 3.2* |
| PWD×PWD | PWD/PWD | 4 | 20 | 119b | 1.9* |
B6-KO, heterozygote for the Prdm9 knock-out; B6-BACB6, C57BL/6J-Tg(RP23-159N6)75Bdm male heterozygous for a Prdm9B6 transgene; Prdm9, genotype at the Prdm9 locus (maternal/paternal); −, null; +, transgenic Prdm9 alleles; n, number of males; BW, body weight (g); TW, mean weight of paired testicles in mg; SC, average sperm count (millions) in paired caput epididymides;
genotype at the entire chr17 (the rescue can also be due to a different gene, Hstw);
significantly higher than in Prdm9B6/PWD reciprocal hybrids (rows 1 and/or 3);
significantly higher relative testis weight (TW per body weight) compared to animals in rows 1 and 3 (p<0.002).
Prdm9 dosage and alleles affect spermatogenesis of reciprocal (B6×PWD)F1 hybrids
Previously, the fertility of the azoospermic (PWD×B6)F1 males resulting from the cross of PWD females with B6 males was rescued by Prdm9C3H overexpression [6], but Prdm9 dosage has not been studied in the reciprocal, sperm-carrying (B6×PWD)F1 hybrids, although these males do not reach the reproductive fitness of fully fertile males (Table 2 and Table 3). To analyze the Prdm9 dosage effect in the reciprocal hybrids, we crossed PWD males with females carrying a variable number of four different Prdm9 alleles on B6 background. The fertility parameters of the Prdm9−/PWD hybrid males were superior to those of their (B6×PWD)F1 Prdm9B6/PWD littermates (Table 2). The parameters of the Prdm9B6/PWD transgenics carrying BAC5 were also better than those of the Prdm9B6/PWD control (Table 2). In contrast, BAC21 overlapping most of BAC5 but carrying truncated Prdm9 [6] did not improve the fertility of Prdm9B6/PWD hybrids (Table S5). To discern whether a single copy of Prdm9C3H can improve the fertility of reciprocal hybrids, males from the cross ((B6×B6-Prdm9C3H)×PWD) were inspected. The increased fecundity of a subset of these males could be ascribed to the presence of Prdm9C3H (Table 2, pTW<0.001, pSC = 0.003). The reciprocal Prdm9C3H/PWD F1 males displayed superior fertility parameters than the Prdm9PWD/C3H hybrids (Table S4; pTW = 0.004, pSC = 0.03, pOFM = 0.04).
Table 3. Overview of male reproductive phenotypes.
| Prdm9 | −/− | PWD/B6 | PWD/− | B6/PWD | PWD/C3H | PWD/PWD | B6/B6 |
| Background | B6 | F1 | F1 | F1 | F1 | PWD | B6 |
| Sex body | 21% | 31% | 67% | 71% | 88% | 96% | 99% |
| Diplotene | 0.3% | 5% | 16% | 17% | 16% | 18% | 21% |
| Spermatids | <1% | <2% | 30% | 45% | 45% | 80% | 74% |
| SC | 0.000 | 0.000 | 0.06 | 0.4 | 0.4 | 1.9 | 3.2 |
| TW | 54 | 61 | 85 | 105 | 110 | 119a | 195 |
| OFM | 0.00 | 0.00 | 0.3 | 3.6 | 3.4 | 6.3 | 6 |
Prdm9, genotype at Prdm9 (maternal/paternal); Sex body, % pachytene spermatocytes that form a sex body; Diplotene, % diplotene of all primary spermatocytes; Spermatids, % round spermatids counted from the total of round spermatids and primary spermatocytes; SC, sperm count in paired caputs (millions); TW, testicular weight (mg); OFM, offspring per female per month;
significantly higher relative TW (TW per body weight) than for males in columns 1 through 5. See Table S4 for more details and statistics.
To determine whether the fertility rescue of the reciprocal hybrids is limited to the Prdm9C3H allele and the PWD Mmm strain, we again used the C57BL/6J-Tg(RP23-159N6)75Bdm (carrying Prdm9B6) and STUS strains. The Mmm-derived strain STUS produces sterile (azoospermic or oligospermic) male offspring with B6 females [30], [31]. In agreement with this finding, the non-transgenic F1 male progeny of STUS males with B6 females heterozygous for the Prdm9B6 BAC were sterile; however, the transgenic littermates were fertile (Table S1). Thus, a single copy of the Prdm9B6 allele caused a decrease in fertility of F1 hybrids compared to the males having this allele removed, while the increased dosage of the same allele improved fertility in all F1 intersubspecific hybrid backgrounds tested.
The substitution of chr17B6 with chr17PWD using the B6.PWD-Chr17 strain rescued fertility of (PWD×B6)F1 azoospermic hybrids, because the (PWD×B6.PWD-Chr17)F1 males are fertile [12]. To assess the role of chr17 heterozygosity and Prdm9PWD dosage in the semifertile reciprocal Prdm9B6/PWD hybrids, (B6.PWD-Chr17×PWD)F1 males were phenotyped. The replacement of chr17B6 with chr17PWD in (B6×PWD)F1 hybrids led to full fertility (Table 2, pTW<0.001, pSC<0.001).
Thus, the reciprocal hybrids are sensitive to the same combinations of Prdm9 alleles and dosage as the (PWD×B6)F1 hybrids, but with a shift towards higher fertility (Table 4 and Table S4). Because one copy of Prdm9B6 reduced fecundity when added to Prdm9−/PWD F1 hybrids, the semifertility of the (B6×PWD)F1 hybrids appears to be affected by a dominant-negative interaction(s) of Prdm9B6.
Table 4. Males differing by the Prdm9 allele, its dosage, or background divided into classes according to fertility.
| Class | Sterile | Semisterile | Semifertile | Fertile | Fertile |
| TW (mg) | 45 to 70 | 70 to 90 | 90 to 140 | above 140 | above 180 |
| SC (×106) | 0.00 | 0.01 to 0.2 | 0.2 to 1.1 | above 1.1 | above 3.5 |
| OFM | 0.00 | 0.1 to 1 | 3 to 4 | above 4 | above 5 |
| Background: | Prdm9 a: | Prdm9 a: | Prdm9 a: | Prdm9 a: | Prdm9 a: |
| PWD×B6 | PWD/B6 | PWD/− | PWD/B6+2B6 | PWD/PWD | |
| PWD×B6 | PWD/C3H | PWD/−+2C3H | PWD/−+6C3H | ||
| PWD×B6 | PWD/B6+2C3H | PWD/B6+6C3H | |||
| B6×PWD | B6/PWD | −/PWD | PWD/PWD | ||
| B6×PWD | C3H/PWD | B6+2C3H/PWD | |||
| STUS×B6 | STUS/B6 | STUS/B6+2B6 | |||
| B6×STUS | B6/STUS | B6+2B6/STUS | |||
| B6×B6 | −/− | B6/− | |||
| B6×B6 | B6/B6 | ||||
| B6×B6 | PWD/B6 | ||||
| B6×B6 | B6+2C3H/B6 |
TW, mean testicular weight; SC, mean sperm count in paired caput epididymides, OFM, offspring per female per month;
genotype at Prdm9 (maternal/paternal); −, null; +, added transgenic copies of Prdm9; Background: maternal×paternal background. Note that the two fertile classes display overlapping parameters.
The overall fertility phenotypes correlate with the strength of meiotic arrest
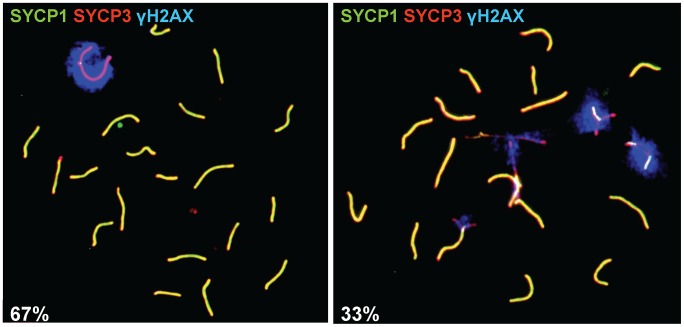
The Prdm9PWD/B6, Prdm9PWD/−, and Prdm9PWD/C3H F1 hybrids show a progressive increase in overall fertility, yet even the Prdm9PWD/C3H F1 hybrid does not reach the parameters of other fertile males (Table 1). The fecundity defects in these hybrids could either represent different degrees of the same arrest or multiple breakdowns affecting different stages of spermatogenesis. To compare the progress of spermatogenesis in these hybrids, indirect immunofluorescence microscopy was performed on surface-spread nuclei of adult testicular cells (chromosome spreads, Table 3 and Table S4). In agreement with the SC data but in contrast to sterile hybrids carrying Prdm9PWD/B6, the chromosome spreads revealed the presence of spermatids in (PWD×B6-Prdm9wt/−)F1 Prdm9PWD/− males. The relative number of round spermatids in these Prdm9PWD/− testes did not reach the number observed in Prdm9PWD/C3H hybrids (Table 3 and Table S4, p<0.001). Due to an arrest at pachynema, the relative number of the four stages of primary spermatocytes in Prdm9PWD/B6 was different from Prdm9PWD/− and Prdm9PWD/C3H F1 intersubspecific hybrids (Table 3 and Table S4, Figure S1). A sex body was formed in 67% of pachytene spermatocytes of Prdm9PWD/− hybrids (Figure 2), a higher proportion in comparison to Prdm9PWD/B6 (p<0.001) but lower compared to Prdm9PWD/C3H hybrids (p = 0.001). The Prdm9PWD/C3H hybrid carried a lower ratio of pachytene spermatocytes displaying a sex body than the B6 (p = 0.004) and PWD (p = 0.03) fertile controls. The staining of spermatocyte chromosome spreads with MLH1 and SYCP1 revealed that the proportion of nuclei with fully synapsed pachytene chromosomes carrying over 20 recombination nodules is higher in the Prdm9PWD/− than in the Prdm9PWD/B6 hybrids (p<0.001). The Prdm9PWD/− hybrids were similar in this respect to Prdm9PWD/C3H, but both carried less pachytene spermatocytes with completed recombination than B6 and PWD (Table S4). The meiotic phenotypes thus correlate with the TW-SC-OFM data; the Prdm9PWD/B6, Prdm9PWD/−, and Prdm9PWD/C3H F1 hybrids display a gradual increase in meiotic progress, yet even the Prdm9PWD/C3H F1 hybrid does not reach the parameters of B6 or PWD.
Figure 2. Sex body formation in the Prdm9PWD/− F1 hybrid male.
Surface-spread nuclei of adult testicular cells treated with a hypotonic solution were indirectly labeled using antibodies marking the synaptonemal complex (anti-SYCP1 and anti-SYCP3) to discern the stage of primary spermatocytes and the phosphorylated form of the histone variant H2AX (anti-γH2AX) to visualize the sex body and then observed under a fluorescent microscope. Left, pachytene carrying a sex body (67 cases found per total of 100 nuclei from four biological replicates); right, pachytene without a sex body (33/100 nuclei).
To determine whether the partial arrest of spermatogenesis of the semifertile reciprocal (B6×PWD)F1 hybrids involves meiosis I, spermatocyte chromosome spreads were analyzed. The relative number of pachytene cells carrying a sex body in the Prdm9B6/PWD hybrid was lower than in B6 and PWD (Table 3 and Table S4) and it was elevated by removing the Prdm9B6 allele from the Prdm9B6/PWD hybrid (p = 0.03, Table 3 and Table S4). The number was higher in the Prdm9C3H/PWD intersubspecific male in comparison to the Prdm9B6/PWD F1 hybrid (p = 0.03, Table S4), and increased in the BAC5-carrying reciprocal hybrid compared to the same hybrid harboring BAC21 (p = 0.003, Table S5). Analysis of MLH1 recombination nodules indicated that Prdm9B6/PWD hybrid testes carried less pachytene spermatocytes with completed recombination than B6 or PWD (Table S4). The relative number of the four stages of primary spermatocytes in Prdm9B6/PWD differed from that in fertile males (Table S4, Figure S1). The number of offspring (OFM) correlated with meiotic phenotypes in all investigated hybrids (Table S4), thus postmeiotic incompatibilities may play a minor role in our model of F1 mouse hybrid sterility.
Hybrid incompatibility(ies) of Prdm9B6 is not due to a change in Prdm9 transcript levels
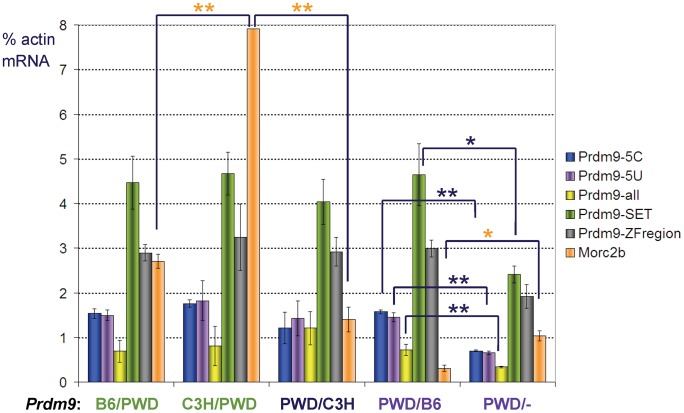
The fertility of the azoospermic Prdm9PWD/B6 F1 hybrids can be rescued by Prdm9 overexpression [6]; however, the amounts of the Prdm9 mRNAs are similar in prepubertal Prdm9PWD/B6 and Prdm9PWD/C3H hybrids that differ in prospective fertility but not in Prdm9 dosage [6]. To understand the mechanism of the partial fertility rescue inflicted by the Prdm9 null alleles in PWD hybrids, the transcript levels of Prdm9 were investigated in prepubertal hybrid testes. The expression of Prdm9 was analyzed using five qRT-PCR amplicons along the gene to account for all alternative transcripts [6]. The mRNA levels of Prdm9 were similar in four investigated types of 14-day-old F1 hybrid testes carrying Prdm9PWD/B6, Prdm9PWD/C3H, Prdm9B6/PWD, and Prdm9C3H/PWD, but were significantly decreased to 52.9±2.3% in the prospectively sperm-carrying Prdm9PWD/− F1 hybrids compared to the prospectively azoospermic Prdm9PWD/B6 littermate controls (Figure 3). In other words, the transcription from the PWD, C3H, and B6 alleles seems to be similar and dosage-dependent. Therefore, the dominant-negative interaction(s) of the Prdm9B6 allele contributing to sterility in the (PWD×B6)F1 hybrid is most likely not a consequence of a change in the Prdm9 transcript level.
Figure 3. Expression of Prdm9 and Morc2b in prepubertal hybrid testis.
Real-time qRT-PCR was performed using RNAs of 14-day-old F1 intersubspecific hybrids of the indicated Prdm9 genotypes (maternal/paternal); −, null. The animals carrying genotypes labeled by the same color were littermates. The columns indicate mRNA expression (mean ± standard deviation) relative to β-actin mRNA for five amplicons in Prdm9 (arranged from the 5′ to 3′ of the gene) and one in Morc2b (orange); the asterisks mark significantly different values (*, p<0.05; **, p<0.01); the expression of Prdm9 was similar in all except the Prdm9PWD/− F1 hybrids.
Discussion
Our “digital genetics“ approach (additions and subtractions of Prdm9 copies) brings a new insight into the interactions of Prdm9 and other hybrid sterility genes participating in the genetic Dobzhansky-Muller incompatibilities (DMIs) and controlling the reproductive fitness of intersubspecific mouse hybrids.
The phenotype of intersubspecific hybrid males is affected by Prdm9 allelic combination and dosage (summarized in Table 4). One copy of Prdm9B6 on multiple F1 intersubspecific hybrid backgrounds is one of the causes of reduced fertility, but a rescue can be achieved with a transgene carrying multiple copies of Prdm9B6 or Prdm9C3H. The replacement of Prdm9B6 with Prdm9C3H in (PWD×B6)F1 males significantly improves fecundity, but it nevertheless leads to semifertility that can be improved by an increased Prdm9 dosage. The fertility rescue of hybrids by Prdm9 transgenes is also dependent on the Prdm9 copy number.
The F1 background is sensitive to Prdm9 dosage, indicating DMIs between PWD and B6 genomes. These DMIs could involve genetic interactions between Prdm9 and other loci, but they might also be explained by interactions between Prdm9-independent loci appearing as the consequence of sensitization by the Prdm9 dosage. Although the overexpression of both Prdm9B6 and Prdm9C3H alleles improved the fertility of F1 hybrids, the variation of effects among the Prdm9 alleles when in one or two copies suggests either variation in the strength of the same interaction(s) or specific DMIs for each Prdm9 allele. Prdm9B6 was the only allele that resulted in a worse phenotype when one copy was added to either type of F1 reciprocal hybrids carrying one Prdm9PWD allele, indicating a dominant-negative effect of Prdm9B6 specific for (PWD×B6)F1 and (B6×PWD)F1. The beneficial effect of the increased copy number of Prdm9 transgenes irrespective of the transgenic allele suggests that the “toxic“ effect of the Prdm9B6 incompatibility can be diluted by the overabundance of Prdm9B6.
The fertility of F1 hybrid males harboring chrXB6 was always better than that of the comparable reciprocal chrXPWD-carrying males, suggesting a chrXPWD DMI(s) occur(s) in intersubspecific hybrids. Although theoretical options also include the interactions of chrY, mitochondrial genome or genomic imprinting, the interaction of chrXPWD and chr17PWD/B6 was revealed by mapping hybrid sterility loci in ((PWD×B6)×B6) backcross, as well as in F1 using chrX subconsomics [12]. The decreased fertility of the reciprocal hybrid males Prdm9B6/PWD compared to Prdm9−/PWD and Prdm9C3H/PWD could be explained by the incompatibility of Prdm9B6-Hstws with chrXB6 or autosomal loci.
Another DMI(s) not involving Prdm9B6 probably also acts in F1 hybrids, since the null Prdm9 alleles do not restore complete fertility. As the fertility of the reciprocal Prdm9−/PWD hybrids was superior to that of the Prdm9PWD/− F1 males, this DMI (or one of these DMIs) independent of Prdm9B6 could involve chrXPWD. Both the elimination of Prdm9B6 and Prdm9 overexpression rescued fecundity in the reciprocal (B6×PWD)F1 hybrids, suggesting that Prdm9B6 also participate(s) in a DMI(s) not involving chrXPWD. Supposing the same DMIs also work in the (PWD×B6)F1 male, a hypothesis supported by its complete sterility, there seems to be at least three sets of incompatibilities affecting the meiotic arrest in this male: Prdm9B6 with chrXPWD, Prdm9B6 with an unknown autosomal locus or loci, and chrXPWD with an unknown autosomal locus or loci. Alternatively, the sets of incompatibilities could be: interautosomal B6 versus PWD sensitive to the dosage of any Prdm9 allele; chrXPWD with B6 autosomes; Prdm9B6 with Prdm9PWD and/or with PWD autosomal loci. Although backcrosses using the Prdm9 null alleles could reveal the number and map positions of the unknown autosomal loci, we already have a good candidate for one of these loci, Hstws on chr17PWD.
The Hst1s-Hstws (Prdm9B6-chr17PWD) incompatibility in F1 hybrids is alleviated by deletion of Prdm9B6 and substitution of chr17B6 with chr17PWD leading to increased fertility. An epistatic interaction of chr17PWD/B6 with chrXPWD is necessary, albeit not sufficient for sterility of ((PWD×B6)×B6)BC1 males [12]. However, at the moment we cannot distinguish between the effects of intergenic and interallelic interactions, also because the impact of Prdm9PWD on hybrid sterility has not been directly investigated. While there is strong evidence that Prdm9 is identical with Hst1 in Mmd [6], we are unable to exclude that the Hstw locus in Mmm is linked to but different from Prdm9. On the other hand, Prdm9 carries the fastest evolving ZnF domain in metazoans [26] and Prdm9PWD/− hybrids display a reduced number of pachytene spermatocytes harboring sex bodies, as well as other features of partial meiotic arrest. Therefore, the incompleteness of the rescue of hybrid fertility by Prdm9B6 deletions in the (PWD×B6)F1 hybrid can be interpreted as the consequence of Prdm9PWD haploinsufficiency in the context of the F1 hybrid background, because Prdm9PWD/− F1 males were more affected than Prdm9PWD/− backcross males.
The hybrid sterility phenotype shows the features of spermatogenesis seen in the Prdm9−/− male and it can be alleviated by Prdm9 transgenes [6]. Prdm9B6 thus appears to participate in a loss-of-function DMI [32]. However, hybrid sterility can also be partially corrected by Prdm9 null alleles, suggesting a gain-of-function DMI. Thus Prdm9B6 might take part in multiple DMIs, both gain- and loss-of-function. Alternatively, the increased dosage of Prdm9 could leave its part not participating in a gain-of-function DMI(s) for the normal function.
The viability of certain Drosophila hybrids is affected by the gene dosage of Hmr (Hybrid male rescue) [33] and by the DNA-regulatory divergence of the Lhr (Lethal hybrid rescue) gene [34]. Although the fertility of mouse hybrids can be rescued by the increased dosage of Prdm9, we excluded that the key difference between the Prdm9C3H and Prdm9B6 alleles lies in increased transcription, because the expression of Prdm9 mRNAs in Prdm9PWD/B6, Prdm9PWD/C3H, Prdm9B6/PWD, and Prdm9C3H/PWD prepubertal hybrid testes of the same age were similar despite the different prospective fertility (Figure 3). Increased translational efficiency remains a possibility for the key allelic difference, as Prdm9C3H and Prdm9B6 differ in the 5′-untranslated region [6]. However, the polymorphism in the ZnF region of the protein products could also provide an explanation for the functional allelic difference.
As hybrid sterility can be overcome by the increased dosage of any Prdm9 allele, one must control the number of copies in experiments designed to discern the functional sequence differences in the Hst1 alleles. Grey et al [24] successfully used transgenesis to learn that the distribution of meiotic recombination hotspots is affected by the ZnF domain allele of Prdm9; no difference in the distribution was seen in the control Prdm9B6 BAC transgenics. It might seem that the correction of hybrid sterility caused by the same Prdm9B6 BAC could be caused by a different mechanism than redistribution of recombination hotspots. However, the increased dosage of Prdm9 in F1 hybrids may overcome the DMIs and change the localization of hotspots. Nevertheless, it is unknown how a changed distribution of hotspots could lead to sterility, especially when considering that Prdm9 function is dispensable for fertility in the dog [35], [36]. Thus (an)other function(s) of Prdm9 may be involved in hybrid sterility, e.g., transactivation of meiotic genes.
While azoospermia was rare or absent, fertility reduced below the range found in pure species was found in one third of males in the Bavarian part of the natural house mouse hybrid zone [37]. The lack of azoospermic males can be explained by the absence of F1-like animals in the zone [37], [38]. F1 male sterility may thus be more important for establishing than for maintaining the hybrid zone. Although most of the fertility differences detected in our study were robust enough to affect the number of offspring, they are likely to have even greater impact in nature considering the sperm competition during multiple mating [39].
Prdm9B6 plays a role in the complete meiotic arrest of (PWD×B6)F1 hybrids [6]. The importance of this finding for mouse speciation could be somewhat limited considering that only males carrying a certain allele resulting from one direction of a cross between two subspecies are affected. In this report, we demonstrated that Prdm9 also participates in the sterility of the reciprocal (B6×STUS)F1 and in the partial meiotic failure of the (B6×PWD)F1 males. The meiosis of (PWD×B6-Prdm9C3H)F1 hybrids harboring another Mmd Prdm9 allele is adversely affected by a DMI that can be alleviated by an increased Prdm9 dosage or using the reciprocal cross, (B6-Prdm9C3H×PWD)F1. The reciprocal crosses of PWD and of the wild-Mmd-derived strain WSB/Ei also display differences in hybrid sterility [40]. Although many quantitative trait loci were detected in (WSB×PWD)F2 intercross males, heterozygosity in a region overlapping the genomic position of Prdm9 decreases SC and relative TW; regions associated with fertility were also found on chrXPWD [40]. The Prdm9 allele of WSB is similar to C3H, being the same in the ZnF domain [23], yet WSB differs from C3H in other parts of Prdm9 [19], [41]. Therefore, the semisterility of (PWD×WSB)F1 males seems to involve Prdm9. Admittedly, the degree of importance of Prdm9 for mouse speciation also depends on the frequency of alleles causing reduced fertility near the hybrid zone that is currently unknown; however, the relevance of Prdm9 for hybrid sterility now appears to be greater than shown previously.
Materials and Methods
Ethics statement
The mice were kept at the Specific Pathogen-Free Facility of the Institute of Molecular Genetics, Prague, and in a conventional breeding facility of the Institute of Vertebrate Biology in Studenec. Principles of laboratory animal care obeyed the Czech Republic Act for Experimental Work with Animals (Decree No. 207/2004 Sb, and the Acts Nos. 246/92 Sb, and 77/2004 Sb) fully compatible with the corresponding EU regulations and standards, namely Council Directive 806/609/EEC and Appendix A of the Council of Europe Convention ETS123.
Mice
The STUS and PWD/Ph strains are derived from wild mice of Mmm subspecies [30], [42]. Mice carrying a deletion of chr17, Sod2df14J, were generated through embryonic stem cells [28] harboring Prdm9B6 on (129×B6)F1 background and were transfected with BAC5. The deletion causes lethality when homozygous, it is several Mbp in length, and it includes the Hst1 region [6]. The BAC5, BAC21, and BAC24 C3H/HeJ transgenes have no effect on fertility in non-intersubspecific hybrid males, but BAC5 and BAC24 rescue fertility of sterile hybrids [6]. The results of quantitative PCR [6] indicate that BAC24 line contains six and BAC5 two copies of Prdm9C3H; BAC21 carries two copies of truncated Prdm9 (only the last, ZnF-encoding exon). BAC5 and BAC24 transgenes rescue fertility in Prdm9−/− (data not shown). All three transgenic lines were transferred to B6 background through 10 generations of backcrossing. The B6-Prdm9C3H (B6*B10.C3H-Hst1f) congenic carries the C3H polymorphisms at Prdm9 and the differential segment is 3.3 Mbp (position in the mm9 genome assembly 12.5 to 15.9 Mbp) to 6.4 Mbp (10.2 to 16.5 Mbp) in length. The knock-out line Prdm9tm1Ymat was generated in 129P2/OlaHsd ES cells by replacement of the first five coding exons with LacZ [20] and maintained on mixed 129P2/OlaHsd * C57BL/6 background. The C57BL/6J-Tg(RP23-159N6)75Bdm strain (transgene Accession ID: MGI:5311012) was generated by injection of a circular BAC DNA into zygotic male pronuclei; it carries four to five copies of RP23-159N6 BAC harboring Prdm9B6 (Hst1s) on B6 background, and the Prdm9 steady-state mRNA level in primary spermatocytes is about 1.3- to 2.5-times increased compared to non-transgenic animals [24], suggesting that two Prdm9 copies are expressed from the transgene.
Genotyping, phenotyping, and statistics
See Text S1 for the PCR primers and conditions used for genotyping. Body weight (BW) and testicular weight (TW, from paired testicles) were determined in adult males (9 to 12- week-old). Sperm count (SC) was obtained from paired caput epididymides at room temperature [6], except for the experiments using the Prdm9B6 transgene, where the entire left epididymis was extracted at 37°C [43]. Multiple biological replicates of each genotype were also analyzed for cellular phenotypes and RNA expression. Slides with surface-spread nuclei (chromosome spreads) were obtained from adult testicular cells using isotonic [44] or hypotonic [45] treatment; see Text S1 for the antibodies. Semiquantitative real-time RT-PCR was performed using total testicular RNAs of 14-day-old F1 intersubspecific hybrids exactly as described previously [6]. The significance of BW, TW, and qRT-PCR values was analyzed using Welsch's t-test, SC and OFM with Wilcoxon rank sum test, and cellular phenotypes with χ2 test. Unless stated otherwise, the comparison significant for TW was also significant for relative TW (TW/BW).
Supporting Information
The proportions of four stages of primary spermatocytes determined by SYCP3-SYCP1-γH2AX staining of spread nuclei of adult testicular cells. −/−, Prdm9−/− on B6 background; PWD/B6, (PWD×B6)F1; PWD/−, hemizygous (PWD×B6-Prdm9B6/−)F1; B6/PWD, (B6×PWD)F1; PWD/C3H, (PWD×B6-Prdm9C3H)F1; Fert, fertile males (pooled data from B6, B6-Prdm9B6/−, and (BAC5×PWD)F1). Diplo, diplotene; Pach, pachytene; Zygo, zygotene; Lept, leptotene spermatocytes. The number above each column designates the total number of counted cells (average of 3.6 males per column); the asterisks indicate significant differences (χ2 test): *, p<0.05; **, p<0.01; ***, p<0.001.
(TIF)
The effect of Prdm9 dosage on hybrids of the STUS strain.
(DOC)
The fertility of males hemizygous for the Prdm9m1Ymat knock-out.
(DOC)
The effect of the Sod2df14J deletion and Prdm9tm1Ymat knock-out (KO) on hybrid sterility.
(DOC)
Details of reproductive phenotypes of various males.
(DOC)
Details of reproductive phenotypes.
(DOC)
Supporting Materials and Methods. Antibodies, primers, PCR conditions.
(DOC)
Acknowledgments
We thank K. Stein, E. Škaloudová, and J. Perlová for technical assistance; M. Lišáková for mouse keeping; and P. Jansa, D. Homolka, and T. Bhattacharyya for helpful discussions. We are also grateful to P. Petkov and three anonymous reviewers for useful comments and to Š. Takáčová for editing English.
Funding Statement
The work was supported by grants 1M6837805002 and AV0Z50520514 from the Ministry of Education, Youth, and Sports of the Czech Republic and by grants P305/10/1931, 206/08/0640, and P305/11/P630 from the Czech Science Foundation. FB and BdM were supported by grants from the Centre National de la Recherche Scientifique (CNRS), the Association pour la Recherche sur le Cancer (ARC 3939), and the Agence Nationale de la Recherche (ANR-09-BLAN-0269-01). PF is a Ph.D. student supported in part by the Faculty of Science, Charles University, Prague. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Haldane JBS (1922) Sex ratio and unisexual sterility in animal hybrids. J Genetics 12: 101–109. [Google Scholar]
- 2. Orr HA, Masly JP, Presgraves DC (2004) Speciation genes. Curr Opin Genet Dev 14: 675–679. [DOI] [PubMed] [Google Scholar]
- 3. Ting CT, Tsaur SC, Wu ML, Wu CI (1998) A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282: 1501–1504. [DOI] [PubMed] [Google Scholar]
- 4. Sun S, Ting CT, Wu CI (2004) The normal function of a speciation gene, Odysseus, and its hybrid sterility effect. Science 305: 81–83. [DOI] [PubMed] [Google Scholar]
- 5. Masly JP, Jones CD, Noor MA, Locke J, Orr HA (2006) Gene transposition as a cause of hybrid sterility in Drosophila . Science 313: 1448–1450. [DOI] [PubMed] [Google Scholar]
- 6. Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J (2009) A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323: 373–375. [DOI] [PubMed] [Google Scholar]
- 7. Phadnis N, Orr HA (2009) A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323: 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bayes JJ, Malik HS (2009) Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science 326: 1538–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sawamura K, Maehara K, Mashino S, Kagesawa T, Kajiwara M, et al. (2010) Introgression of Drosophila simulans nuclear pore protein 160 in Drosophila melanogaster alone does not cause inviability but does cause female sterility. Genetics 186: 669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dobzhansky T (1951) Experiments on sexual isolation in Drosophila: X. Reproductive isolation between Drosophila pseudoobscura and Drosophila persimilis under natural and under laboratory conditions. Proc Natl Acad Sci U S A 37: 792–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Forejt J, Ivanyi P (1974) Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.). Genet Res 24: 189–206. [DOI] [PubMed] [Google Scholar]
- 12. Dzur-Gejdosova M, Simecek P, Gregorova S, Bhattacharyya T, Forejt J (2012) Dissecting the genetic architecture of F1 hybrid sterility in house mice. Evolution in press. doi: 10.1111/j.1558-5646.2012.01684.x. [DOI] [PubMed] [Google Scholar]
- 13. Forejt J (1996) Hybrid sterility in the mouse. Trends Genet 12: 412–417. [DOI] [PubMed] [Google Scholar]
- 14. Forejt J, Vincek V, Klein J, Lehrach H, Loudova-Mickova M (1991) Genetic mapping of the t-complex region on mouse chromosome 17 including the Hybrid sterility-1 gene. Mamm Genome 1: 84–91. [DOI] [PubMed] [Google Scholar]
- 15. Trachtulec Z, Vincek V, Hamvas RM, Forejt J, Lehrach H, et al. (1994) Physical map of mouse chromosome 17 in the region relevant for positional cloning of the Hybrid sterility 1 gene. Genomics 23: 132–137. [DOI] [PubMed] [Google Scholar]
- 16. Gregorova S, Mnukova-Fajdelova M, Trachtulec Z, Capkova J, Loudova M, et al. (1996) Sub-milliMorgan map of the proximal part of mouse Chromosome 17 including the hybrid sterility 1 gene. Mamm Genome 7: 107–113. [DOI] [PubMed] [Google Scholar]
- 17. Trachtulec Z, Mihola O, Vlcek C, Himmelbauer H, Paces V, et al. (2005) Positional cloning of the Hybrid sterility 1 gene: fine genetic mapping and evaluation of two candidate genes. Biol J Linn Soc 84: 637–641. [Google Scholar]
- 18. Mihola O, Forejt J, Trachtulec Z (2007) Conserved alternative and antisense transcripts at the programmed cell death 2 locus. BMC Genomics 8: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trachtulec Z, Vlcek C, Mihola O, Gregorova S, Fotopulosova V, et al. (2008) Fine haplotype structure of a chromosome 17 region in the laboratory and wild mouse. Genetics 178: 1777–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hayashi K, Yoshida K, Matsui Y (2005) A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438: 374–378. [DOI] [PubMed] [Google Scholar]
- 21. Buard J, Barthes P, Grey C, de Massy B (2009) Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J 28: 2616–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, et al. (2010) PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parvanov ED, Petkov PM, Paigen K (2010) Prdm9 controls activation of mammalian recombination hotspots. Science 327: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grey C, Barthes P, Chauveau-Le Friec G, Langa F, Baudat F, et al. (2011) Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol 9: e1001176 doi:10.1371/journal.pbio.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forejt J, Pialek J, Trachtulec Z (2012) Hybrid male sterility genes in the mouse subspecific crosses. In: Macholan M, Baird SJE, Muclinger P and Pialek J, editors. Evolution of the House Mouse. Cambridge: Cambridge University Press, pp. 482–503.
- 26. Oliver PL, Goodstadt L, Bayes JJ, Birtle Z, Roach KC, et al. (2009) Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet 5: e1000753 doi:10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gregorova S, Divina P, Storchova R, Trachtulec Z, Fotopulosova V, et al. (2008) Mouse consomic strains: exploiting genetic divergence between Mus m. musculus and Mus m. domesticus subspecies. Genome Res 18: 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bergstrom DE, Bergstrom RA, Munroe RJ, Lee BK, Browning VL, et al. (2003) Overlapping deletions spanning the proximal two-thirds of the mouse t complex. Mamm Genome 14: 817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Browning VL, Bergstrom RA, Daigle S, Schimenti JC (2002) A haplolethal locus uncovered by deletions in the mouse T complex. Genetics 160: 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pialek J, Vyskocilova M, Bimova B, Havelkova D, Pialkova J, et al. (2008) Development of unique house mouse resources suitable for evolutionary studies of speciation. J Hered 99: 34–44. [DOI] [PubMed] [Google Scholar]
- 31. Vyskocilova M, Prazanova G, Pialek J (2009) Polymorphism in hybrid male sterility in wild-derived Mus musculus musculus strains on proximal chromosome 17. Mamm Genome 20: 83–91. [DOI] [PubMed] [Google Scholar]
- 32. Maheshwari S, Barbash DA (2011) The genetics of hybrid incompatibilities. Annu Rev Genet 45: 331–355. [DOI] [PubMed] [Google Scholar]
- 33. Barbash DA, Roote J, Ashburner M (2000) The Drosophila melanogaster Hybrid male rescue gene causes inviability in male and female species hybrids. Genetics 154: 1747–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maheshwari S, Barbash DA (2012) Cis-by-Trans regulatory divergence causes the asymmetric lethal effects of an ancestral hybrid incompatibility gene. PLoS Genet 8: e1002597 doi:10.1371/journal.pgen.1002597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Munoz-Fuentes V, Di Rienzo A, Vila C (2011) Prdm9, a major determinant of meiotic recombination hotspots, is not functional in dogs and their wild relatives, wolves and coyotes. PLoS ONE 6: e25498 doi:10.1371/journal.pone.0025498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Axelsson E, Webster MT, Ratnakumar A, Ponting CP, Lindblad-Toh K (2012) Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res 22: 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Turner LM, Schwahn DJ, Harr B (2011) Reduced male fertility is common but highly variable in form and severity in a natural house mouse hybrid zone. Evolution 66: 443–458. [DOI] [PubMed] [Google Scholar]
- 38. Macholan M, Munclinger P, Sugerkova M, Dufkova P, Bimova B, et al. (2007) Genetic analysis of autosomal and X-linked markers across a mouse hybrid zone. Evolution 61: 746–771. [DOI] [PubMed] [Google Scholar]
- 39. Dean MD, Ardlie KG, Nachman MW (2006) The frequency of multiple paternity suggests that sperm competition is common in house mice (Mus domesticus). Mol Ecol 15: 4141–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. White MA, Steffy B, Wiltshire T, Payseur BA Genetic dissection of a key reproductive barrier between nascent species of house mice. Genetics 189: 289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Keane TM, Goodstadt L, Danecek P, White MA, Wong K, et al. (2011) Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gregorova S, Forejt J (2000) PWD/Ph and PWK/Ph inbred mouse strains of Mus m. musculus subspecies–a valuable resource of phenotypic variations and genomic polymorphisms. Folia Biol (Praha) 46: 31–41. [DOI] [PubMed] [Google Scholar]
- 43. Vyskocilova M, Trachtulec Z, Forejt J, Pialek J (2005) Does geography matter in hybrid sterility in house mice? Biol J Linn Soc 84: 663–674. [Google Scholar]
- 44. Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, et al. (2005) Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet 37: 41–47. [DOI] [PubMed] [Google Scholar]
- 45. Anderson LK, Reeves A, Webb LM, Ashley T (1999) Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151: 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The proportions of four stages of primary spermatocytes determined by SYCP3-SYCP1-γH2AX staining of spread nuclei of adult testicular cells. −/−, Prdm9−/− on B6 background; PWD/B6, (PWD×B6)F1; PWD/−, hemizygous (PWD×B6-Prdm9B6/−)F1; B6/PWD, (B6×PWD)F1; PWD/C3H, (PWD×B6-Prdm9C3H)F1; Fert, fertile males (pooled data from B6, B6-Prdm9B6/−, and (BAC5×PWD)F1). Diplo, diplotene; Pach, pachytene; Zygo, zygotene; Lept, leptotene spermatocytes. The number above each column designates the total number of counted cells (average of 3.6 males per column); the asterisks indicate significant differences (χ2 test): *, p<0.05; **, p<0.01; ***, p<0.001.
(TIF)
The effect of Prdm9 dosage on hybrids of the STUS strain.
(DOC)
The fertility of males hemizygous for the Prdm9m1Ymat knock-out.
(DOC)
The effect of the Sod2df14J deletion and Prdm9tm1Ymat knock-out (KO) on hybrid sterility.
(DOC)
Details of reproductive phenotypes of various males.
(DOC)
Details of reproductive phenotypes.
(DOC)
Supporting Materials and Methods. Antibodies, primers, PCR conditions.
(DOC)