Abstract
L-selectin plays important roles in lymphocyte homing and leukocyte rolling. Mounting evidence shows that it is involved in many disease entities including diabetes, ischemia/reperfusion injuries, inflammatory diseases, and tumor metastasis. Regulation of L-selectin at protein level has been well characterized. However, the regulation of human L-selectin transcription remains largely unknown. To address transcriptional regulation of L-selectin, we cloned 1088 bp 5′ of the start codon ATG. Luciferase analysis of the serial 5′ deletion mutants located the core promoter region at −288/−1. A major transcription initiation site was mapped at −115 by 5′RACE. Transcription factors Sp1, Ets1, Mzf1, Klf2, and Irf1 bind to and transactivate the L-selectin promoter. Significantly, FOXO1 binds to a FOXO1 motif, CCCTTTGG, at −87/−80, and transactivates the L-selectin promoter in a dose-dependent manner. Over-expression of a constitutive-active FOXO1 increased the endogenous L-selectin expression in Jurkat cells. We conclude that FOXO1 regulates L-selectin expression through targeting its promoter.
Keywords: L-selectin, transcriptional regulation, FOXO1, promoter
Introduction
L-selectin (CD62L, Sell) is a cell surface adhesion molecule that is involved in the cascade of leukocyte rolling,1–3 in lymphocyte homing to lymphoid organs,4–6 and in the formation and maintenance of memory T cells. Deregulation of Sell expression has been correlated with tumor metastasis,7 ischemia/reperfusion related injuries,8 autoimmune diseases, and many other disease entities.9–11
Sell is highly expressed in most leukocytes, including naïve T cells and subsets of memory T cells. Upon T cell activation, cell surface Sell was rapidly shed by membrane metalloproteases,12 which was accompanied by a 3 to 4 folds up-regulation over the resting level by day 2, sustained for 2 days, and then gradually returned to the resting level by day 7.13 Post-translational modifications of Sell, including sulfation, phosphorylation, and glycosylation14 and its shedding from cell surface, have been well characterized.6,15–22 Accumulating evidence shows that Sell is also extensively regulated at transcriptional level. Upon T-cell activation, Sell was rapidly shed from the cell surface, which was accompanied by both increased Sell gene expression and rapid mRNA degradation to maintain the steady state levels of Sell mRNA.13 TNFα up-regulated human Sell mRNA levels in TNFα-sensitive Daudi B cells.23 In adult T-cell leukemia, Leukemic cells express high levels of Sell mRNA, which sustains high levels of cell surface Sell, thus leading to increased endothelial attachment, transmigration, and organ infiltration.24 This makes it clear that regulatory mechanisms governing Sell expression at the transcriptional level are at least as important as those at the translational level.
Similar to mouse Sell gene, human Sell also clusters with E-selectin (Sele) and P-selectin (Selp) on chromosome 125 and consists of ten exons spanning about 21.0 kb. Analysis of the mouse Sell promoter showed that Sp1, Ets1, Mzf1, Irf1, and Klf2 bound to the core promoter region and transactivated the Sell promoter. Alignment of the first 300 bp sequences 5′ of the ATG of human, chimpanzee, rat, and mouse showed that the consensus sequences for these transcription factors were almost identical,26 suggesting the location of human Sell promoter and the similarity of its trans-activation to that of the mouse Sell gene.
In this report, we cloned a 1088 bp genomic fragment 5′ of the ATG of human Sell gene. Luciferase analysis of the serial 5′ deletion mutants located the core promoter region at −288/−1. A major TIS was mapped at −115. Transcription factors, Sp1, Ets1, Klf2, Irf1, and Mzf1 all transactivated human Sell promoter. Significantly, a FOXO1 motif (CCCTTTGG) was mapped at −87/−80, which was confirmed to bind to transcription factor FOXO1 by mutational analysis and EMSA. Furthermore, we demonstrated that FOXO1 transactivated human Sell core promoter in a dose-dependent manner and up-regulated endogenous Sell expression in Jurkat cells. This discovery provides the molecular mechanisms for further addressing the roles of FOXO1—a master regulator of many physiological processes—in regulating the expression of Sell that is important for the homeostasis of our immune system.
Materials and Methods
Cell lines and reagents
Mouse EL4 cells (mouse lymphoma cell line) and human Jurkat cells, both grown in suspension, were maintained in RPMI 1640 containing 10% heat- inactivated Fetal Bovine Serum (FBS) and 1% penicillin/streptomycin. HeLa cells, an adherent cell line, were cultured in DMEM supplemented with 1% penicillin/streptomycin and 10% FBS. All cell lines were grown in an incubator at 37 °C in a 5% CO2 atmosphere. All antibodies were purchased from Santa Cruz Biotechnology and all chemicals were products of Sigma unless specified otherwise. All restriction and modifying enzymes were purchased from New England Biolab (NEB). γ-32P-ATP was purchased from PerkinElmer (Shanghai, China). Plasmids, pcDNA3-FOXO1 and -FOXO1-3A were all kindly provided by Professor Amnon Altman from the La Jolla Institute for Allergy and Immunology.
5′ rapid amplification of cDNA ends (RACE)
mRNAs were prepared from cultured Jurkat cells using a Genelute Direct mRNA Miniprep Kit (Sigma, St. Louis, Missouri). 5′ RACE was performed with a SMART™ RACE cDNA Amplification Kit as instructed by the vendor (Clontech, Mountain View, California). Briefly, 0.5 μg of mRNA was used as the start material and 5′ RACE products were amplified by standard PCR using the universal primer (UPM) included in the kit, and by a human Sell gene specific primer (GSP) complementary to nucleotides +77/+105 (we define the ‘A’ in the ATG as “+1” position). PCR products were purified and cloned into pCR2.1 (Invitrogen, Carlsbad, California). 5′ ends were identified by sequencing 20 randomly picked colonies (Retrogen, San Diego, California).
Transient transfection
For all transient transfections, HeLa cells were seeded at 5 × 105 per 60 mm dish in complete DMEM the day before and the media were refreshed two hours before transfections with 10% DMEM that was free of antibiotics. The two T cell lines, Jurkat or EL4 cells, were plated at 1 × 106 per well in 10% RPMI1640 free of antibiotics in 12-well plates two hours before transfections. Transfection was performed using Lipofectamine 2000 (Invitrogen). Briefly, for each 100 μL reaction, 2.5 μL of the Lipofectamine 2000 was added into 50 μL OPTI-MEM (Invitrogen), vortexed for seconds, and was then left to stand at room temperature (RT) for 5 minutes. Plasmids mixtures, as indicated in the texts or figure legends, diluted into 50 μl OPTI-MEM was added into the above 50 μl mixture of OPTI-MEM and Lipofectamine 2000, vortexed for seconds, and continued to incubate at RT for 20 minutes. The mixture was then added drop-wise to cells and continued to incubate for 24 to 36 hours.
Cloning 5′ flanking sequence and 5′ serial deletion of human Sell gene
The sequence of the 5′ flanking sequence of human Sell gene was obtained from the NCBI database (ENSG00000188404). The longest fragment −1088/−1 was amplified by DNA Polymerase Chain Reaction (PCR) using genomic DNA from Jurkat cells as template. Sense primer containing an XhoI restriction site and anti-sense primer at BglII site are listed in Table 1. The PCR conditions were 94 °C for 2 minutes, followed by 35 cycles of 94 °C for 30 seconds, 60 °C for 30 seconds, and 72 °C for 1 minute. The PCR products were gel-purified and cloned into pGL3-Basic, and sequence identity was confirmed by DNA sequencing (Retrogen). The resulting plasmid is named human pGL3-Sell1088 (human Sell1088, hSell1088). The serial 5′ deletion mutants starting 5′ at −488, −288, −188, and −108 were also amplified by PCR with the same anti-sense primer and different sense primers as listed in Table 1. All mutants were cloned into pGL3-Basic, which were designated as hSell488, hSell288, hSell188, and hSell108. The sequence of each fragment was confirmed by sequencing. Putative transcription factor binding sites were searched using Genomatix (http://www.genomatix.de) and TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html). All plasmid were prepared using an EndoFree Plasmid Maxi kit from Qiagen.
Table 1.
Primers for constructs and for real time PCR.
| Primers for constructs | |
| For.1088 | 5′-ATAGCTCGAGTAACCTCTTTGAGACTCT-3′ |
| For.488 | 5′-ATAGCTCGAGGAAGGAGGAAGAGGA-3′ |
| For.288 | 5′-ATAGCTCGAGCTGATCAGCAGTTCATT-3′ |
| For.188 | 5′-ATAGCTCGAGAAAAGGGGAGGAGGAGGA-3′ |
| For.108 | 5′-ATAGCTCGAGTCTACCTGCAGCACAGCA-3′ |
| Rev. | 5′-CTACAGATCTGGCTTTGCTTGGTCCT-3′ |
| FOXO1m | 5′-GGGTCTCAGGTCCTTGCCTTCGTTGAGTGTGCTGTGCTGCAG-3′ |
| Primers for real time PCR | |
| For.Sell | 5′-GGCAGCCCTCTGTTACACA-3′ |
| Rev.Sell | 5′-ACATCACAGTTGCAGGTGTA-3′ |
| For.GAPDH | 5′-CATGAGAAGTATGACAACAGCCT-3′ |
| Rev.GAPDH | 5′-AGTCCTTCCACGATACCAAAGT-3′ |
| Probes (shown only sense strand) | |
| APOC3 | 5′-CCTTTACTCCAAACACCCCCCA-3′ |
| FOXO1 | 5′-GCACACTCCCTTTGGGCAAGGA-3′ |
| FOXO1m | 5′-GCACACTCAACGAAGGCAAGGA-3′ |
Luciferase activity analysis
Thirty hours after transient transfections, cells were harvested and washed once with PBS. Cell pellets were lysed with Passive Lysis Buffer (Promega, Madison, Wisconsin), re-suspended by vortexing for a few seconds, and incubated at RT for 30 minutes. The lysates were spun down and the supernatants were saved for a Dual luciferase assay. Plasmid pRL-CMV-expressing Renilla luciferase was always co-transfected, at one fiftieth of the luciferase constructs, as an internal control for transfection efficiency. Luciferase activity was analyzed on AutoLumate Plus LB 953 (Berthold, Oak Ridge, Tennessee) using Dual-Luciferase Reporter Assay System. The luciferase activity was normalized to that of Renilla activity. Data presented were from at least three independent experiments in triplicate.
Site-directed mutagenesis
Mutagenesis of the putative FOXO1 binding sites was performed using the GeneEditor In Vitro Site-directed Mutagenesis System (Promega) with 5′ phosphorylated anti-sense primer, FOXO1m, listed in Table 1. Mutation, CCCTTTGG ➔ CAACGAAG, was designed not to introduce any alternative putative transcription factor binding sites in the context of hSell108, and the resulting plasmid was designated as hSell108Fmut. The desired point mutations were confirmed by DNA sequencing.
Nuclear extract preparation and EMSA
HeLa cells were transfected as described above and nuclear extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents from Pierrce (Rockford, Illinois). Protein concentration was determined using a Bio-Rad Protein Assay Kit (Hercules, California) following manufacturer’s instructions; one μg of the extract was used for EMSA. Probes for APOC3 and human Sell wild-type and FOXO1 site mutant (FOXO1 and FOXO1m respectively) are listed in Table 1. APOC3 probe was generated by annealing sense and antisense APOC3 oligos, labeled with T4 Polynucleotide Kinase in a 50 μL volume in the presence of γ32P-ATP, and purified through Sephadex G50 column. For the EMSA assay, one μg of nuclear extract was incubated with γ32P-APOC3 on ice for 30 minutes in binding buffer containing 40 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 50 mM KCl, 10% glycerol, 0.1% bovine serum albumin, and 1 μg of poly (dI: dC). For a competition assay, 10× or 100× cold probes annealed from sense and anti-sense oligos were added before adding γ32P-APOC3. DNA-protein complexes were resolved on a 6% native polyacrylamide gel, which was dried and exposed to X-ray films overnight at −80 °C.
Western blot
Thirty hours after transient transfection, cells were lysed in lysis buffer containing 1% (w/v) SDS, 20 mM Tris-HCl, pH 8.0, 50 mM NaCl, 5 mM EDTA, and 1× protease inhibitor cocktail (Sigma, St. Louis, Missouri). The cell lysates were sonicated three times for 3 seconds with a 30 second interval on a Branson 450 Sonifier 02 with the setting at 2 and constant power, the samples were boiled for 5 minutes, and the protein concentration was determined using a BioRad Protein Assay as instructed by the manufacturer. Ten μg of each sample was resolved on 4–14% Tris-Bis gel (Invitrogen) and then transferred to PVDF membrane. The membrane was first blocked with TBST (138 mM NaCl, 2.6 mM KCl, 24.7 mM Tris, and 0.05% Tween20) containing 10% non-fat milk powder for 1 hour at RT, followed by incubation with rabbit anti-human FOXO1 at 1:1500 dilution in TBST containing 1% non-fat milk powder for 1 hour at RT, washed 3 times for 10 minutes with TBST containing 1% non-fat milk powder. The membrane was then incubated with goat anti-rabbit HRP-conjugated secondary antibody at 1:10,000 dilutions in TBST containing 1% milk for 1 hour at RT, washed 3 times for 10 minutes with TBST at RT, and protein bands were detected with an Enhanced Chemiluminescence Kit (Pierce). To show equal loading of each sample, the same membrane was stripped with stripping buffer (100 mM β-mercaptoethanol, 2% SDS, 62.5 mM Tris·Cl, pH 6.7) at 60 °C for 30 minutes, and reprobed with mouse anti β-actin (Abcam, Cambridge, Massachusetts) at 1:20,000 dilution.
Reverse transcription and real time PCR
Total RNA was isolated from Jurkat or transfected Jurkat cells using the RNeasy kit (QIAGEN, Valencia, California). One μg of total RNA was reverse transcribed using a iScript cDNA Synthesis Kit (BioRad) at the conditions recommended by the vendor in a 20 μL volume. Of the 20 μL of the cDNA, one μL was used to quantify the gene expression by real time PCR (BioRad, iQ5 cycler) in a 25 μL of reaction containing 200 μM each of sense and antisense primers and iQ SYBR Green Supermix (BioRad). The primers for human Sell and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) used as the reference gene are listed in Table 1. Amplification efficiency was >95% for both pairs of primers and the relative gene expression was calculated by the ΔΔCt method as described in the BioRad’s Real-Time PCR Applications Guide.
Results
Bioinformatic analysis of the 5′ flanking region of human Sell gene
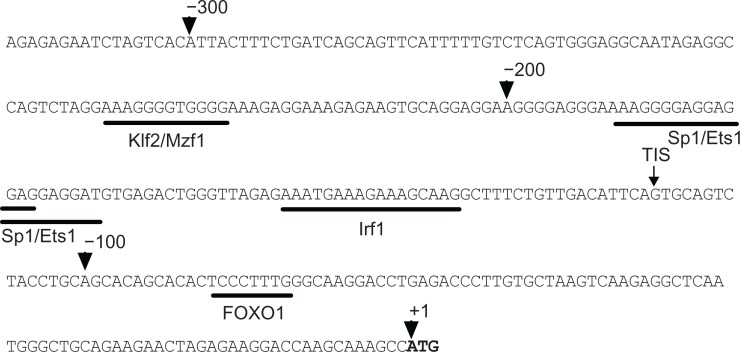
Similar to mouse Sell gene, human Sell is also clustered with Sele upstream and Selp downstream on Chromosome 1, which consists of ten exons interrupted by 9 introns that spans about 21.0 kb. We have shown that the 5′ flanking sequences immediately upstream of the translational start codon ATG (in bold) are well conserved among mouse, rat, chimpanzee, and human.26 As shown in Figure 1, ‘A’ in the start codon ATG was numbered +1 and nucleotides upstream of the ATG were designated as negative with a hundredth indicated by arrowheads. The sequence is GC-rich and contains no CpG islands and no TATA-boxes or CAAT-boxes, typical features for housekeeping genes. Using TFSEARCH, Genomatix suite and Alibaba2.1, we identified, in addition to the four characterized motifs for Mzf1/Klf2, Ets1, Sp1, and Irf1 in mouse Sell promoter,26 a potential FOXO1 binding sequence, CCCTTTGG, at −87/−80. This sequence is conserved between chimpanzee and human (Fig. 6A) and is designated as FOXO1 motif.
Figure 1.
Nucleotide sequence of the 5′ flanking sequence of human Sell gene.
Notes: Annotated sequence is the first ∼317 bp of 5′-flanking region of human Sell. The ‘A’ in the start codon ATG (in bold) was defined as +1 and nucleotides upstream were numbered negative. Arrowheads indicate the position of the nucleotides relative to +1. Small arrow indicates the major TIS mapped by 5′ RACE. Putative transcription factor binding sites are underlined and labeled underneath.
Abbreviations: Mzf1, Myeloid Zinc Finger Protein 1; Klf2, Kruppel-Like Factor 2; Ets1, E26 transformation specific sequence; Sp1, Specificity protein 1; Irf1, Interferon Response Factor 1; FOXO1, Forkhead box protein O1.
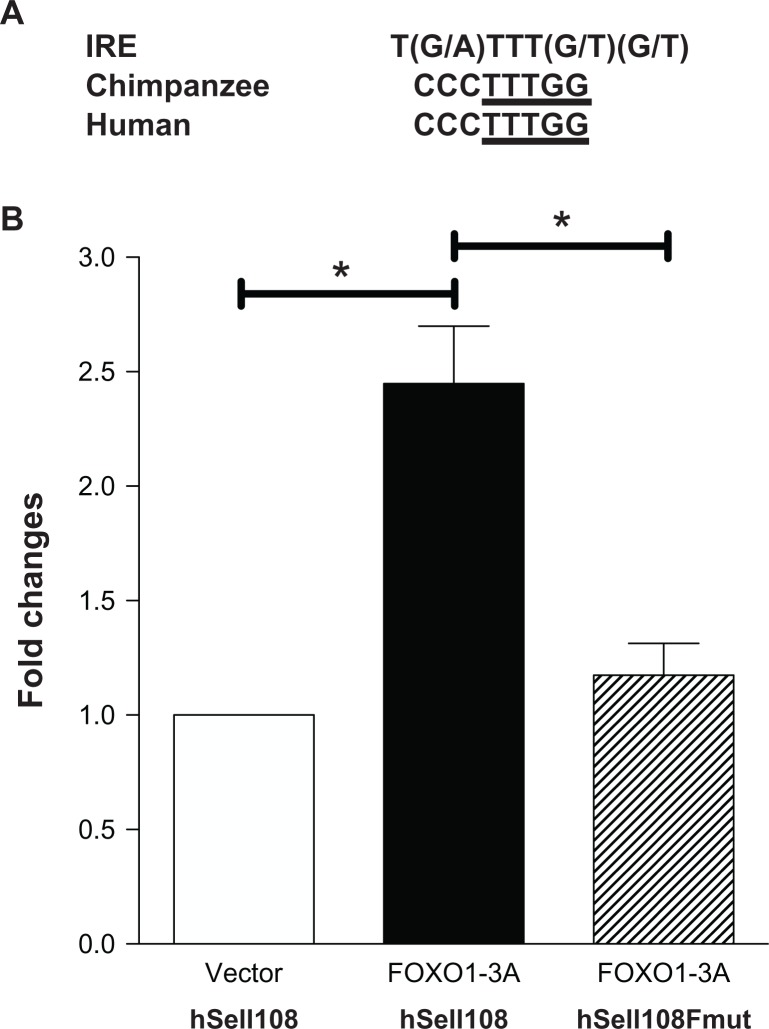
Figure 6.
FOXO1 transactivates human Sell promoter through binding to the motif. (A) Alignment of the IRE with the FOXO1 motifs in the Sell promoter of both human and chimpanzee. (B) Jurkat cells were transiently co-transfected with the combination of plasmids, pcDNA3 and hSell108, FOXO1-3A and hSell108, or FOXO1-3A and hSellFmut, as labeled on the bottom.
Notes: Luciferase activity was analyzed 30 hours after transfection. Luciferase activity expressed as fold increase relative to that of pcDNA3 transfected cells. Data shown are mean ± SD of three independent transfections in one experiment. Each experiment was repeated at least three times in triplicates.
Determination of transcription initiation sites
To determine the transcription initiation sites (TISs) of human Sell gene, 5′-RACE was performed on mRNA from human Jurkat cells with a GSP complementary to the nucleotides +77/+105 and the UPM primer. The PCR products were gel purified and cloned into pCR2.1. TISs were identified by sequencing 20 randomly picked colonies. The 5′ ends ranged from nucleotides −150 to −91. However one TIS at position −115 was identified in 8 colonies, whereas other TISs appeared in less than 3 colonies. We define the nucleotide ‘G’ at −115 as the major TIS of human Sell gene (Fig. 1, arrow).
Transcriptional analysis of the 5′ regulatory sequence of human Sell gene
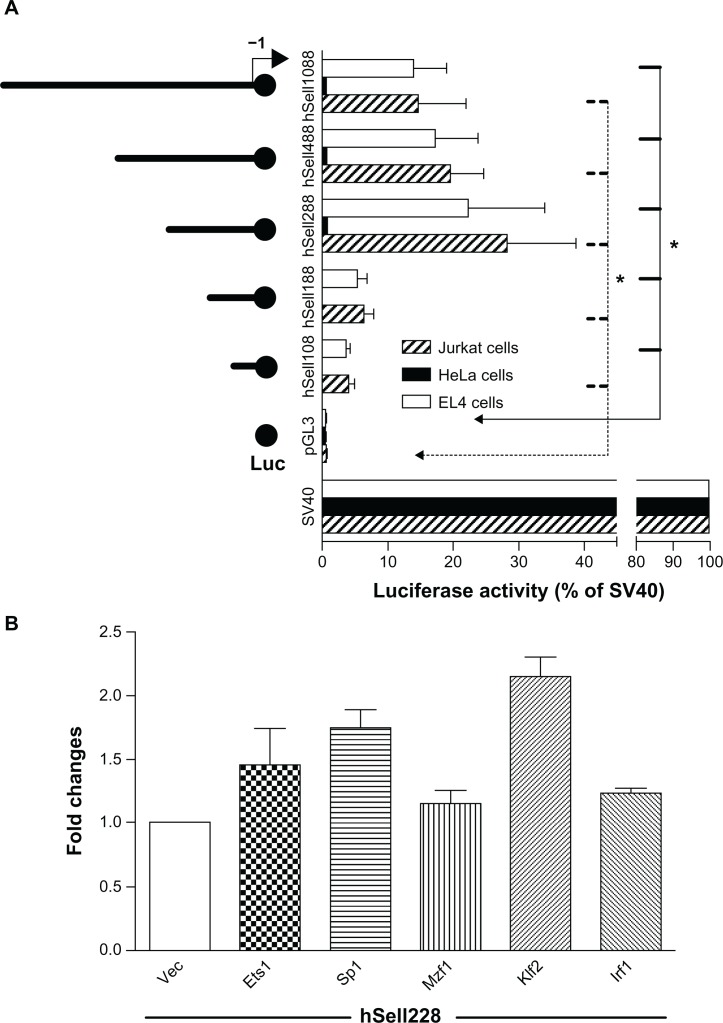
To test whether the 5′-flanking region of human Sell gene had promoter activity, luciferase activity was analyzed 30 hours after co-transfection of pRL-CMV-expressing Renilla luciferase and one of the serial 5′ truncated plasmids (Fig. 2A, left panel) into two lymphocyte cell lines, human Jurkat cells (stripped bars) or mouse EL4 cells (open bars) that express high level of Sell, and HeLa cells (solid bars) that do not express Sell. As shown in the right panel of Figure 2A, human Sell288 drove the highest luciferase activity, reaching more than 22% of that of pGL3-Promoter (a SV40 promoter-driven luciferase) in both lymphocyte cell lines; further 5′ deletion mutants hSell188 and hSell108 showed basal level of Luciferase activity. However, all constructs showed negligible luciferase activity in Sell-negative HeLa cells. To assure that the two lymphocyte cell lines can be transfected efficiently, pGL3-Promoter was always included in the experiments and its luciferase activity was set as 100. To test whether transcription factors Sp1, Ets1, Klf2, Mzf1, and Irf1 trans-activate human Sell promoter, plasmids expressing these transcription factors26 were co-transfected with core promoter construct Sell288 into Jurkat cells and luciferase activity was analyzed 30 hours after the transfection. As shown in Figure 2B, over-expression of Ets1, Sp1, Mzf1, Klf2, and Irf1 increased the core promoter activity compared to that of vector co-transfection by 46%, 75%, 15%, 115%, and 24% respectively. Taken together, transcription factors, Ets1, Sp1, Mzf1, Klf2, and Irf1 transactivated human Sell promoter.
Figure 2.
Mapping the core promoter region of human Sell gene. (A) 5′ serial deletion mutants shown on the left side were transiently transfected into Jurkat (stripped bars), EL4 (open bars), or HeLa cells (solid bars) and Luciferase activity shown on the right side was analyzed 30 hours after transfection. (B) Jurkat cells were co-transfected with core promoter construct, hSell288, with plasmids expressing Sp1, Mzf1, Klf2, Irf1, Ets1.
Notes: Luciferase activity was analyzed 30 hours after the co-transfection. Luciferase activity was expressed as percentage of that of pGL3-Promoter in 2A and as fold changes relative to that of pGL3 vector transfected Jurkat cells in 2B. Data shown are mean ± SD of three independent transfections in one experiment. Each experiment was repeated at least three times.
FOXO1 up-regulates endogenous Sell expression
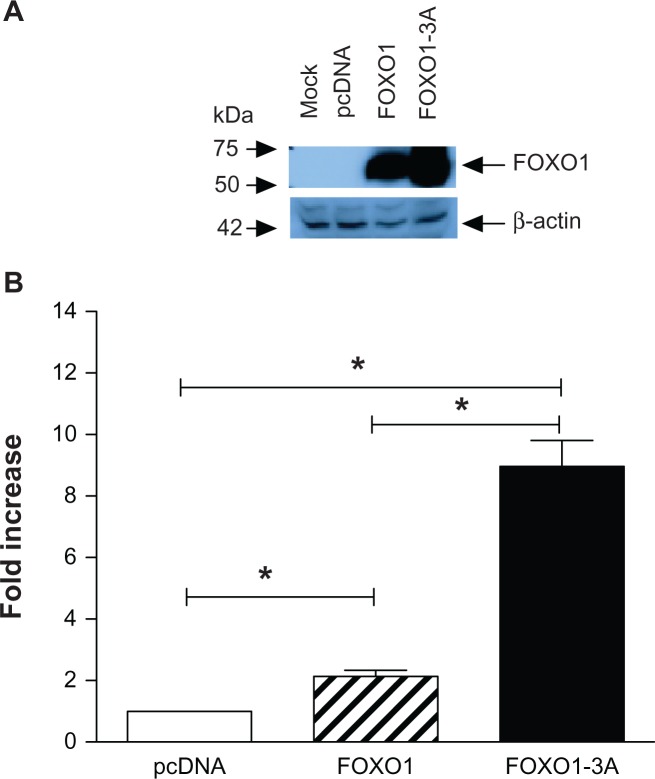
FOXO1 maintained Sell expression during Th1 polarization27 and constitutive active FOXO1 up- regulated Sell expression in Jurkat cells.28 To re-evaluate that FOXO1 increases Sell expression in our model, Jurkat cells were transiently transfected with plasmids expressing either native FOXO1 or a constitutive active mutant, FOXO1-3A, where three PI3K/Akt phosphorylation sites (Thronine24Alaine/Serine256Alaine/Serine319Alaine) were mutated, thus leading to its constitutive nuclear localization, for 30 hours. Forced expression of FOXO1 and FOXO1-3A were confirmed by Western blot (Fig. 3A). Over-expression of FOXO1 and FOXO1-3A increased endogenous Sell expression more than 2-folds (Fig. 3B, stripped bar) and 8-folds (solid bar) respectively, compared to vector (open bar) transfected cells. The lower trans-activation activity of FOXO1 compared to that of FOXO1-3A was consistent with the fact that deficiency of phosphatase and tensin homolog (PTEN), a phosphatase, caused the constitutive cytosolic localization of native FOXO1 in Jurkat cells.
Figure 3.
Constitutive active FOXO1 up-regulates endogenous human Sell expression. Jurkat cells were transiently transfected with pcDNA3 (white bar), pcDNA3-FOXO1 (striped bar), or pcDNA3-FOXO1-3A (solid bar) for 30 hours. Of the two sets of transfected cells, one set was lysed with SDS buffer and equal amount of lysate from each treatment was analyzed for expression of FOXO1 by Western blot, where β-actin was used as loading control (A); the other set was used to analyze the expression of human Sell by real time PCR, which was normalized to that of GAPDH (B).
Notes: Data were presented as mean ± SD of at least three independent experiments in triplicate on each transfection. Data were graphed as fold increase relative to that of pcDNA3 transfected Jurkat cells, which was set as 1.
FOXO1 up-regulates human Sell expression through trans-activating its promoter
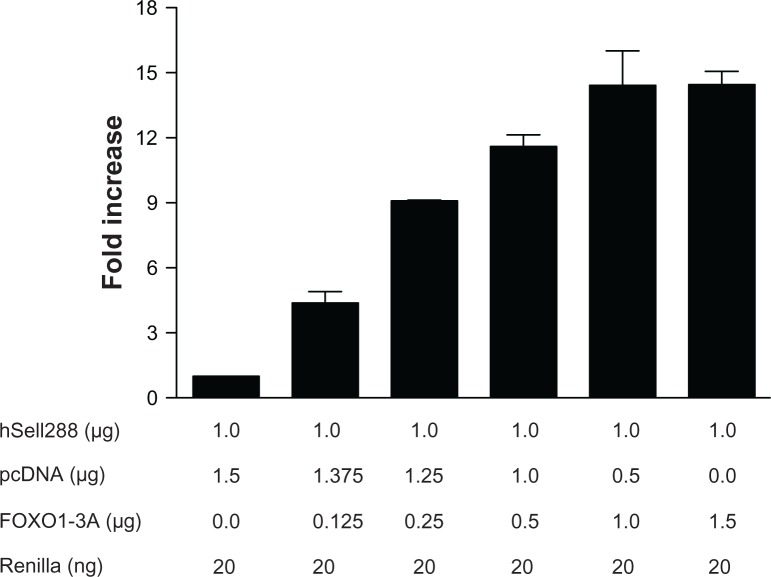
To explore the mechanisms of FOXO1-induced up-regulation of human Sell, the core promoter construct, Sell288, was co-transfected with increasing amounts of FOXO1-3A into Jurkat cells for 30 hours. To exclude the effect of plasmid itself, the amount of total plasmids in each transfection were kept the same by adjusting the amount of plasmid pcDNA3. As shown in Figure 4, FOXO1-3A increased core promoter activity in a dose-dependent manner. These results suggest that FOXO1 induces Sell up-regulation through trans-activation of its promoter.
Figure 4.
Constitutive active FOXO1 up-regulates human Sell core promoter activity in a dose-dependent manner.
Notes: Jurkat cells were transiently co-transfected with the combination of plasmids as labeled in the figure. Luciferase activity normalized to that of Renilla activity was analyzed 30 hours after transfection. Data were presented as mean ± SD of at least three independent experiments in triplicate on each transfection. Data were graphed as fold increase relative to that of pcDNA3 transfected Jurkat cells, which was set as 1.
Mapping FOXO1 motif
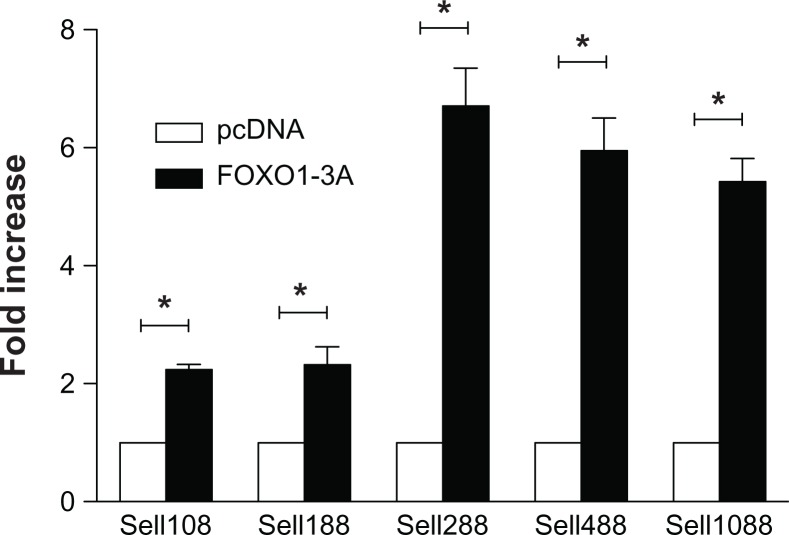
To locate the FOXO1 binding motif, serial 5′ deletion mutants were co-transfected with either pcDNA3 (Vector, Vec.) or FOXO1-3A for 30 hours. Luciferase analysis showed that FOXO1-3A increased the luciferase activity of both Sell108 and Sell188 more than 2-folds, whereas it increased the luciferase activity of Sell288, Sell488, and Sell1088 by a factor of 6.7, 5.9, and 5.4 respectively (Fig. 5, solid bars), compared to that of pcDNA3 co-transfection (open bars). These results indicate that at least fragment −108/−1 harbors a FOXO1 motif and that either fragment −228/−188 contains additional FOXO1 motifs that transactivate promoter constructs Sell288, Sell488, and Sell1088, or FOXO1 may transactivate these three promoter constructs indirectly through binding to other bound transcription factor(s) in the region from −288 to −188.
Figure 5.
Locating the FOXO1 motif.
Notes: Jurkat cells were transiently co-transfected with either pcDNA3 (open bars) or FOXO1-3A (solid bars) and one of the 5′ serial deletion mutants as labeled. Luciferase activity normalized to that of Renilla activity was analyzed 30 hours after the co-transfection. Data were presented as mean ± SD of at least three independent experiments in triplicate on each transfection. Data were graphed as fold increase relative to that of pcDNA3 co-transfected Jurkat cells, which was set as 1.
To map the FOXO1 motif on fragment −108/−1, alignment of chimpanzee and human promoter regions immediately upstream of ATG were performed and compared to the well-characterized FOXO1 binding sequences, including Insulin Response Element (IRE). Sequence CCCTTTGG at −87/−80 is conserved between the two species and bears a high degree of similarity to the IRE (Fig. 6A). Point mutations were introduced into the potential FOXO1 motif (CCCTTTGG ➔ CAACGAAG) in the context of hSell108 and the resulting mutant was confirmed by sequencing and designated as hSell108Fmut. As shown in Figure 6B, co-transfection of FOXO1-3A with hSell108 into Jurkat cells increased its luciferase activity to 2.5 times (Fig. 6B, solid bar) that of vector co-transfected cells (Fig. 6B, open bar), which was almost abolished by co-transfection of hSell108Fmut (Fig. 6B, stripped bar). These results suggest that the FOXO1 motif is responsible for the observed trans-activation of human Sell108 by FOXO1.
FOXO1 binds to the potential FOXO1 motif in vitro
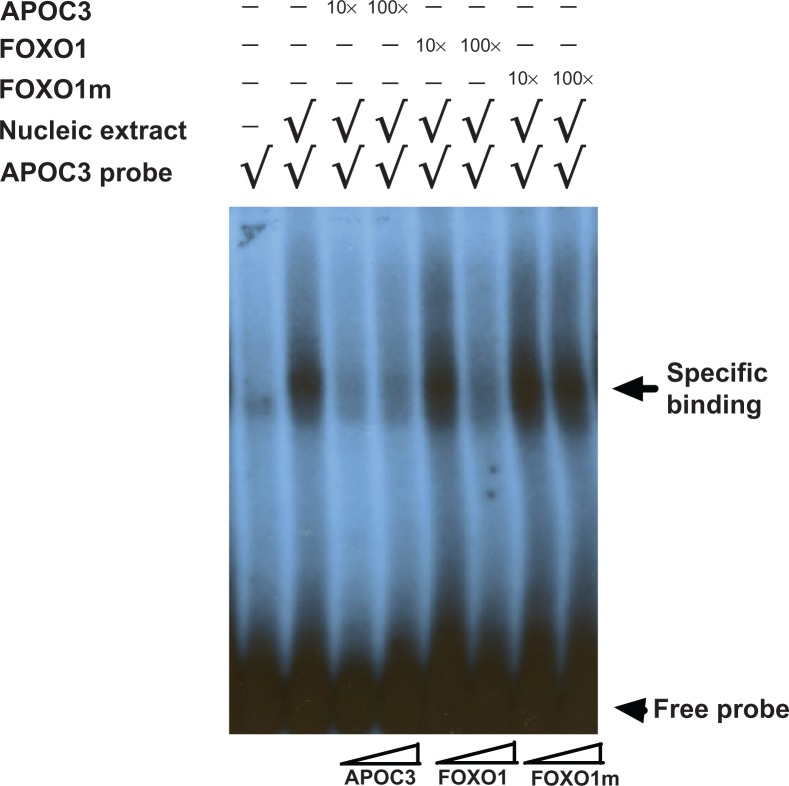
To confirm the potential FOXO1 motif in human Sell promoter binds to FOXO1, we performed the EMSA. A DNA probe containing the IRE (shown only sense oligo: 5′-CCTTTACTCCAAACACCCCCCA-3′) from apolipoprotein APOC3, whicht was well-characterized by others to bind to FOXO1 motif,29 was labeled with γ-32P-ATP (32P-APOC3). Indeed, when nuclear extract from HeLa cells over-expressing FOXO1-3A was incubated with γ-32P-APOC3, a moving retarded DNA-protein complex appeared as shown in Figure 7 lane 2, compared to the free probe in lane 1. This DNA-protein complex was competed out by 10× and 100× cold APOC3 probe (lane 3 and 4), confirming the specificity of the reported binding of FOXO1 to FOXO1 motif. Although we could not appreciate the competing by 10× cold FOXO1 probe from human Sell, the retarded DNA-protein complex did disappear by 100× cold probe (lane 5 and 6, FOXO1). In contrast, 10× and 100× cold FOXO1 probe carrying mutated FOXO1 motif (lane 7 and 8, FOXO1m) both failed to compete out the binding of APOC3 probe to FOXO1. Taken together, these results suggest that the sequence CCCTTTGG at −87/−80 in human Sell gene binds to FOXO1—although at lower affinity than that of the IRE characterized in APOC3 gene—and mediates the trans-activation of human Sell gene by FOXO1.
Figure 7.
FOXO1 binds to FOXO1 motif in vitro.
Notes: DNA probe containing IRE from APOC3 gene was labeled with γ-32P-ATP (lane 1), which was then incubated with nuclear extract from HeLa cells over-expressing FOXO1-3A (lane 2). The protein-DNA complex was then competed with either 10× and 100× cold APOC3 probe (lane 3 and 4, labeled as APOC3), or with 10× and 100× cold probe containing FOXO1 motif from human Sell (lane 5 and 6, labeled as FOXO1), or with 10× and 100× cold probe containing mutated FOXO1 motif from human Sell (lane 7 and 8, labeled as FOXO1m). Free probe was indicated with arrowhead and specific DNA-protein complex was indicated with arrow on the side.
Discussion
PI3K/Akt/FOXO1 signaling pathway has been shown to participate in the homeostasis of immune system.30,31 Mice that were deficient in different PI3K isoforms or subunits showed various abnormalities, ranging from embryonic lethal issues to impairments in both T-cell and B-cell compartments.32 This contrasted with mice over-expressing a constitutive active PI3K variant, which showed increased T-cell viability and resistance to Fas-mediated apoptosis,33 suggesting a vital role of PI3K in normal development and functions of lymphocytes. During T-cell development, FOXO1 helped to maintain the levels of Sell expression that was indispensable for both Th1 polarization at the earlier stage and for T lymphocytes trafficking.27,34 Using transcriptional profiling, FOXO1 has been shown to up-regulate the expression of Sell, Klf2, and sphingosine-1-phosphate receptors (EDG1 and EDG6) that all participate in the regulation of lymphocyte trafficking.28 Deletion of the DNA-binding domain of FOXO1 eliminated its ability to regulate Sell expression.28 Furthermore, conditional knockout FOXO1 in T-cells resulted in CD62Llo surface phenotype T-cells that were hardly found in peripheral lymphoid compartment and relatively refractory to T-cell receptor stimulation.35 These results suggest the vital role of FOXO1 in regulating Sell expression. Characterization of human Sell promoter and mapping the regulatory elements for FOXO1 would be necessary to further address the roles of FOXO1 in the homeostasis of our immune system.
FOXO1 is a downstream target of PI3K/Akt signaling pathway,36 which has been shown to target its downstream genes involved in proliferation, apoptosis,37,38 control of oxidative stress, metabolism,39–41 and energy homeostasis.29,42,43 Upon activation, Akt phosphorylates FOXO1 and leads to its nuclear exclusion44 and increased proteosomal degradation,45,46 thus dampening its transcriptional regulation on targeted genes. The consensus sequence for FOXO1 binding was first characterized as (C/G)(A/T)AAA(C/A)A.47,48 Later FOXO1 was shown to bind to various forms of consensus sequences including at least two versions of IRE, TTGTTTAC,49 and T(G/A)TTT(T/G)(G/T),50 and a consensus sequence T(G/A)TT(G/T)(G/A)(C/T) from peroxisome proliferator-activated receptor-gamma.51 The FOXO1 core binding sequence in human Sell, 5′-CCCTTTGG-3′, bears high similarity to the IRE (Fig. 6A). To confirm the authenticity of the FOXO1 motif, we designed a competitive EMSA where oligonucleotides containing FOXO1 motif from Sell were used to compete the IRE-FOXO1 (DNA-Protein) complex that was well-characterized in APOC3 gene.29 As expected, the IRE-FOXO1 complex was completely disrupted by 100× wild-type FOXO1 oligos (Fig. 7, lane 6), but not by its mutant counterpart at the same concentration (Fig. 7, lane 8), suggesting the authenticity of this newly-identified FOXO1 motif.
We and others have shown that Sell gene can be transactivated by Klf226 and thus promote T cell quiescence and home to the lymph nodes.52 Interestingly, FOXO1 has been demonstrated to control the expression of both Sell and the transcription factor Klf2 in naïve T cells, deletion of which was sufficient to alter lymphocyte trafficking.27 These suggest that FOXO1 may transactivate Sell gene through at least two mechanisms, either binding to the FOXO1 motif directly or acting through a “FOXO1-Klf2-Sell” cascade-like reaction. Indeed, we observed that over-expression of FOXO1-3A increased the luciferase activity of the core promoter, Sell288 that contains both a Klf2 motif at 239/228 and the newly identified FOXO1 motif at 87/80, to more than 7 folds. This is in contrast to a 2-fold increase of the two shorter promoter constructs, Sell108 and Sell188, which contain only the newly identified FOXO1 motif (Fig. 5, solid bars). Of course, we cannot exclude the possibility that FOXO1 may up-regulate Sell expression through interaction with other bound transcription factors than Klf2.
Conclusion
We provide evidence that FOXO1 can not only bind to and transactivate human Sell promoter directly, but may also upregulate Sell expression through a “FOXO1-Klf2-Sell” cascade-like reaction. This makes targeting FOXO1 a very efficient way to control Sell expression and thus an attractive drug target for therapeutic intervention.
Acknowledgements and Funding
This work was supported in large part by grant DK-57880 from the National Institutes of Health (to Klaus Ley) and by the funding for the key laboratory of the clinical pharmacology, Branch Hospital of Shanghai First People’s Hospital (to Yuefen Lou). Corresponding author is indebted to professor Klaus Ley for his scientific guidance and financial support.
Footnotes
Author Contributions
XD conceived and designed the experiments. YL, X Lu and XD performed the experiments. XD analyzed the data and wrote the manuscript. All authors reviewed and approved of the final manuscript.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
References
- 1.Whelan J. Selectin synthesis and inflammation. Trends Biochem Sci. 1996 Feb;21(2):65–9. [PubMed] [Google Scholar]
- 2.Verdrengh M, Erlandsson-Harris H, Tarkowski A. Role of selectins in experimental Staphylococcus aureus-induced arthritis. Eur J Immunol. 2000 Jun;30(6):1606–13. doi: 10.1002/1521-4141(200006)30:6<1606::AID-IMMU1606>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 3.Ley K, Tedder TF, Kansas GS. L-selectin can mediate leukocyte rolling in untreated mesenteric venules in vivo independent of E- or P-selectin. Blood. 1993 Sep 1;82(5):1632–8. [PubMed] [Google Scholar]
- 4.Wang CC, Biben C, Robb L, et al. Homeodomain factor Nkx2-3 controls regional expression of leukocyte homing coreceptor MAdCAM-1 in specialized endothelial cells of the viscera. Dev Biol. 2000 Aug 15;224(2):152–67. doi: 10.1006/dbio.2000.9749. [DOI] [PubMed] [Google Scholar]
- 5.Arbones ML, Ord DC, Ley K, et al. Lymphocyte homing and leukocyte rolling and migration are impaired in L-selectin-deficient mice. Immunity. 1994 Jul;1(4):247–60. doi: 10.1016/1074-7613(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 6.Galkina E, Florey O, Zarbock A, et al. T lymphocyte rolling and recruitment into peripheral lymph nodes is regulated by a saturable density of L-selectin (CD62L) Eur J Immunol. 2007 May;37(5):1243–53. doi: 10.1002/eji.200636481. [DOI] [PubMed] [Google Scholar]
- 7.Qian F, Hanahan D, Weissman IL. L-selectin can facilitate metastasis to lymph nodes in a transgenic mouse model of carcinogenesis. Proc Natl Acad Sci U S A. 2001 Mar 27;98(7):3976–81. doi: 10.1073/pnas.061633698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zoldhelyi P, Beck PJ, Bjercke RJ, et al. Inhibition of coronary thrombosis and local inflammation by a noncarbohydrate selectin inhibitor. Am J Physiol Heart Circ Physiol. 2000 Dec;279(6):H3065–75. doi: 10.1152/ajpheart.2000.279.6.H3065. [DOI] [PubMed] [Google Scholar]
- 9.Kretowski A, Kinalska I. L-selectin gene T668C mutation in type 1 diabetes patients and their first degree relatives. Immunol Lett. 2000 Nov 1;74(3):225–8. doi: 10.1016/s0165-2478(00)00259-5. [DOI] [PubMed] [Google Scholar]
- 10.Chen HY, Cui B, Wang S, et al. L-selectin gene polymorphisms in Graves’ disease. Clin Endocrinol (Oxf) 2007 Jul;67(1):145–51. doi: 10.1111/j.1365-2265.2007.02852.x. Epub Apr 25, 2007. [DOI] [PubMed] [Google Scholar]
- 11.Steeber DA, Campbell MA, Basit A, Ley K, Tedder TF. Optimal selectin-mediated rolling of leukocytes during inflammation in vivo requires inter-cellular adhesion molecule-1 expression. Proc Natl Acad Sci U S A. 1998 Jun 23;95(13):7562–7. doi: 10.1073/pnas.95.13.7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Condon TP, Flournoy S, Sawyer GJ, Baker BF, Kishimoto TK, Bennett CF. ADAM17 but not ADAM10 mediates tumor necrosis factor-alpha and L-selectin shedding from leukocyte membranes. Antisense Nucleic Acid Drug Dev. 2001 Apr;11(2):107–16. doi: 10.1089/108729001750171353. [DOI] [PubMed] [Google Scholar]
- 13.Mascarell L, Truffa-Bachi P. T lymphocyte activation initiates the degradation of the CD62L encoding mRNA and increases the transcription of the corresponding gene. Immunol Lett. 2004 Jun 15;94(1–2):115–22. doi: 10.1016/j.imlet.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Sperandio M. Selectins and glycosyltransferases in leukocyte rolling in vivo. FEBS J. 2006 Oct;273(19):4377–89. doi: 10.1111/j.1742-4658.2006.05437.x. Epub Sep 5, 2006. [DOI] [PubMed] [Google Scholar]
- 15.Killock DJ, Ivetić A. The cytoplasmic domains of TNFalpha-converting enzyme (TACE/ADAM17) and L-selectin are regulated differently by p38 MAPK and PKC to promote ectodomain shedding. Biochem J. 2010 May 13;428(2):293–304. doi: 10.1042/BJ20091611. [DOI] [PubMed] [Google Scholar]
- 16.Le Gall SM, Bobé P, Reiss K, et al. ADAMs 10 and 17 represent differentially regulated components of a general shedding machinery for membrane proteins such as transforming growth factor alpha, L-selectin, and tumor necrosis factor alpha. Mol Biol Cell. 2009 Mar;20(6):1785–94. doi: 10.1091/mbc.E08-11-1135. Epub Jan 21, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walcheck B, Alexander SR, St. Hill CA, Matala E. ADAM-17-independent shedding of L-selectin. J Leukoc Biol. 2003 Sep;74(3):389–934. doi: 10.1189/jlb.0403141. [DOI] [PubMed] [Google Scholar]
- 18.Galkina E, Tanousis K, Preece G, et al. L-selectin shedding does not regulate constitutive T cell trafficking but controls the migration pathways of antigen-activated T lymphocytes. J Exp Med. 2003 Nov 3;198(9):1323–35. doi: 10.1084/jem.20030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Oliver P, Lancaster JR, Jr, et al. Reactive oxygen species mediate tumor necrosis factor alpha-converting, enzyme-dependent ectodomain shedding induced by phorbol myristate acetate. FASEB J. 2001 Feb;15(2):303–5. doi: 10.1096/fj.00-0371fje. Epub Dec 8, 2000. [DOI] [PubMed] [Google Scholar]
- 20.Kretowski A, Gillespie KM, Bingley PJ, Kinalska I. Soluble L-selectin levels in type I diabetes mellitus: a surrogate marker for disease activity? Immunology. 2000 Feb;99(2):320–5. doi: 10.1046/j.1365-2567.2000.00967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hafezi-Moghadam A, Ley K. Relevance of L-selectin shedding for leukocyte rolling in vivo. J Exp Med. 1999 Mar 15;189(6):939–48. doi: 10.1084/jem.189.6.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hafezi-Moghadam A, Thomas KL, Prorock AJ, Huo Y, Ley K. L-selectin shedding regulates leukocyte recruitment. J Exp Med. 2001 Apr 2;193(7):863–72. doi: 10.1084/jem.193.7.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans SS, Collea RP, Appenheimer MM, Gollnick SO. Interferon-alpha induces the expression of the L-selectin homing receptor in human B lymphoid cells. J Cell Biol. 1993 Dec;123(6 Pt 2):1889–98. doi: 10.1083/jcb.123.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tatewaki M, Yamaguchi K, Matsuoka M, et al. Constitutive overexpression of the L-selectin gene in fresh leukemic cells of adult T-cell leukemia that can be transactivated by human T-cell lymphotropic virus type 1 Tax. Blood. 1995 Oct 15;86(8):3109–17. [PubMed] [Google Scholar]
- 25.Ord DC, Ernst TJ, Zhou LJ, et al. Structure of the gene encoding the human leukocyte adhesion molecule-1 (TQ1, Leu-8) of lymphocytes and neutrophils. J Biol Chem. 1990 May 15;265(14):7760–7. [PubMed] [Google Scholar]
- 26.Dang X, Raffler NA, Ley K. Transcriptional regulation of mouse L-selectin. Biochim Biophys Acta. 2009 Feb;1789(2):146–52. doi: 10.1016/j.bbagrm.2008.10.004. Epub Nov 11, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerdiles YM, Beisner DR, Tinoco R, et al. FOXO1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009 Feb;10(2):176–84. doi: 10.1038/ni.1689. Epub Jan 11, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fabre S, Carrette F, Chen J, et al. FOXO1 regulates L-Selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J Immunol. 2008 Sep 1;181(5):2980–9. doi: 10.4049/jimmunol.181.5.2980. [DOI] [PubMed] [Google Scholar]
- 29.Altomonte J, Cong L, Harbaran S, et al. FOXO1 mediates insulin action on apoC-III and triglyceride metabolism. J Clin Invest. 2004 Nov;114(10):1493–503. doi: 10.1172/JCI19992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dejean AS, Hedrick SM, Kerdiles YM. Highly specialized role of Forkhead box O transcription factors in the immune system. Antioxid Redox Signal. 2011 Feb 15;14(4):663–74. doi: 10.1089/ars.2010.3414. Epub Dec 13, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ouyang W, Li MO. Foxo: in command of T lymphocyte homeostasis tolerance. Trends Immunol. 2011 Jan;32(1):26–33. doi: 10.1016/j.it.2010.10.005. Epub Nov 23, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki T, Suzuki A, Sasaki J, Penninger JM. Phosphoinositide 3-kinases in immunity: lessons from knockout mice. J Biochem. 2002 Apr;131(4):495–501. doi: 10.1093/oxfordjournals.jbchem.a003126. [DOI] [PubMed] [Google Scholar]
- 33.Birkenkamp KU, Coffer PJ. FOXO transcription factors as regulators of immune homeostasis: molecules to die for? J Immunol. 2003 Aug 15;171(4):1623–9. doi: 10.4049/jimmunol.171.4.1623. [DOI] [PubMed] [Google Scholar]
- 34.Leenders H, Whiffield S, Benoist C, Mathis D. Role of the forkhead transcription family member, FKHR, in thymocyte differentiation. Eur J Immunol. 2000 Oct;30(10):2980–90. doi: 10.1002/1521-4141(200010)30:10<2980::AID-IMMU2980>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 35.Gubbels Bupp MR, Edwards B, Guo C, et al. T cells require FOXO1 to populate the peripheral lymphoid organs. Eur J Immunol. 2009 Nov;39(11):2991–9. doi: 10.1002/eji.200939427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kashii Y, Uchida M, Kirito K, et al. A member of Forkhead family transcription factor, FKHRL1, is one of the downstream molecules of phosphatidylinositol 3-kinase-Akt activation pathway in erythropoietin signal transduction. Blood. 2000 Aug 1;96(3):941–9. [PubMed] [Google Scholar]
- 37.Uddin S, Kottegoda S, Stigger D, Platanias LC, Wickrema A. Activation of the Akt/FKHRL1 pathway mediates the antiapoptotic effects of erythropoietin in primary human erythroid progenitors. Biochem Biophys Res Commun. 2000 Aug 18;275(1):16–9. doi: 10.1006/bbrc.2000.3266. [DOI] [PubMed] [Google Scholar]
- 38.Zheng WH, Kar S, Quirion R. Insulin-like growth factor-1-induced phosphorylation of the forkhead family transcription factor FKHRL1 is mediated by Akt kinase in PC12 cells. J Biol Chem. 2000 Dec 15;275(50):39152–8. doi: 10.1074/jbc.M002417200. [DOI] [PubMed] [Google Scholar]
- 39.Tomizawa M, Kumar A, Perrot V, Nakae J, Accili D, Rechler MM. Insulin inhibits the activation of transcription by a C-terminal fragment of the forkhead transcription factor FKHR. A mechanism for insulin inhibition of insulin-like growth factor-binding protein-1 transcription. J Biol Chem. 2000 Mar 10;275(10):7289–95. doi: 10.1074/jbc.275.10.7289. [DOI] [PubMed] [Google Scholar]
- 40.Nakae J, Barr V, Accili D. Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. EMBO J. 2000 Mar 1;19(5):989–96. doi: 10.1093/emboj/19.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmoll D, Walker KS, Alessi DR, et al. Regulation of glucose-6- phosphatase gene expression by protein kinase Balpha and the forkhead transcription factor FKHR. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J Biol Chem. 2000 Nov 17;275(46):36324–33. doi: 10.1074/jbc.M003616200. [DOI] [PubMed] [Google Scholar]
- 42.Kim MS, Pak YK, Jang PG, et al. Role of hypothalamic FOXO1 in the regulation of food intake and energy homeostasis. Nat Neurosci. 2006 Jul;9(7):901–6. doi: 10.1038/nn1731. Epub Jun 18, 2006. [DOI] [PubMed] [Google Scholar]
- 43.Burgering BM, Medema RH. Decisions on life and death: FOXO Forkhead transcription factors are in command when PKB/Akt is off duty. J Leukoc Biol. 2003 Jun;73(6):689–701. doi: 10.1189/jlb.1202629. [DOI] [PubMed] [Google Scholar]
- 44.Jackson JG, Kreisberg JI, Koterba AP, Yee D, Brattain MG. Phosphorylation and nuclear exclusion of the forkhead transcription factor FKHR after epidermal growth factor treatment in human breast cancer cells. Oncogene. 2000 Sep 21;19(40):4574–81. doi: 10.1038/sj.onc.1203825. [DOI] [PubMed] [Google Scholar]
- 45.Aoki M, Jiang H, Vogt PK. Proteasomal degradation of the FOXO1 transcriptional regulator in cells transformed by the P3k and Akt oncoproteins. Proc Natl Acad Sci U S A. 2004 Sep 14;101(37):13613–7. doi: 10.1073/pnas.0405454101. Epub Sep 1, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsuzaki H, Daitoku H, Hatta M, Tanaka K, Fukamizu A. Insulin-induced phosphorylation of FKHR (FOXO1) targets to proteasomal degradation. Proc Natl Acad Sci U S A. 2003 Sep 30;100(20):11285–91120. doi: 10.1073/pnas.1934283100. Epub Sep 17, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J Biol Chem. 1999 Jun 11;274(24):17184–92. doi: 10.1074/jbc.274.24.17184. [DOI] [PubMed] [Google Scholar]
- 48.Ganjam GK, Dimova EY, Unterman TG, Kietzmann T. FOXO1 and HNF-4 are involved in regulation of hepatic glucokinase gene expression by resveratrol. J Biol Chem. 2001 Aug;29(Pt 4):552–8. doi: 10.1074/jbc.M109.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Brien RM, Streeper RS, Ayala JE, Stadelmaier BT, Hornbuckle LA. Insulin-regulated gene expression. Biochem Soc Trans. 2001 Aug;29(Pt 4):552–8. doi: 10.1042/bst0290552. [DOI] [PubMed] [Google Scholar]
- 50.Nakae J, Cao Y, Oki M, et al. Forkhead transcription factor FOXO1 in adipose tissue regulates energy storage and expenditure. Diabetes. 2008 Mar;57(3):563–76. doi: 10.2337/db07-0698. Epub Dec 27, 2007. [DOI] [PubMed] [Google Scholar]
- 51.Armoni M, Harel C, Karni S, et al. FOXO1 represses peroxisome proliferator-activated receptor-gamma1 and -gamma2 gene promoters in primary adipocytes. A novel paradigm to increase insulin sensitivity. J Biol Chem. 2006;281:19881–91. doi: 10.1074/jbc.M600320200. [DOI] [PubMed] [Google Scholar]
- 52.Bai A, Hu H, Yeung M, Chen J. Kruppel-like factor 2 controls T cell trafficking by activating L-selectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription. J Immunol. 2007 Jun 15;178(12):7632–9. doi: 10.4049/jimmunol.178.12.7632. [DOI] [PubMed] [Google Scholar]