Abstract
Lincomycin A is a potent antimicrobial agent noted for its unusual C1 methylmercapto-substituted eight-carbon sugar. Despite its long clinical history for the treatment of Grampositive infections, the biosynthesis of the C8-sugar, methylthiolincosamide (MTL), is poorly understood. Here, we report our studies of the two initial enzymatic steps in the MTL biosynthetic pathway leading to the identification of D-erythro-D-gluco-octose 8-phosphate as a key intermediate. Our experiments demonstrate that this intermediate is formed via a transaldol reaction catalyzed by LmbR using D-fructose 6-phosphate or D-sedoheptulose 7-phosphate as the C3 donor and D-ribose 5-phosphate as the C5 acceptor. Subsequent 1,2-isomerization catalyzed by LmbN converts the resulting 2-keto C8-sugar (octulose 8-phosphate) to octose 8-phosphate. These results provide, for the first time, in vitro evidence revealing the biosynthetic origin of the C8 backbone of MTL.
Many compounds important for the treatment and study of human disease have their origin in natural products. These compounds are frequently modified with carbohydrate appendages that are critical for their biological activities.1 By exploiting the biosynthetic machinery of these unusual sugars, it is possible to enhance or vary the biological characteristics of the parent molecules. To fully realize the therapeutic potential of such an approach, the biosynthetic pathways of these sugars must be characterized and the underlying chemistry understood at the mechanistic level.2 Despite recent advance made in deoxyhexoses biosynthesis research,3 our knowledge about how unusual octoses are constructed remains elusive.
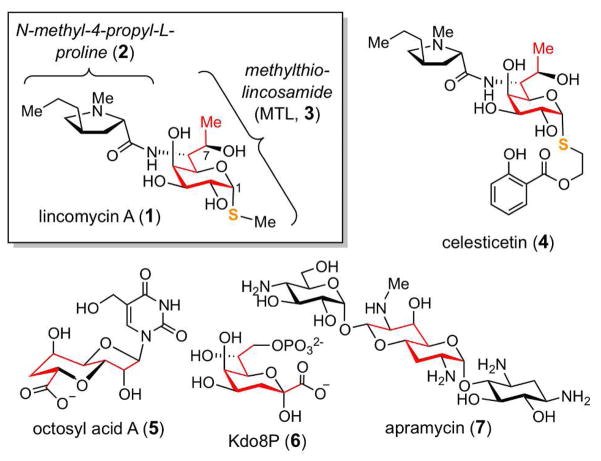
Lincomycin A (1), originally isolated from Streptomyces lincolnensis var. lincolnensis,4 is an octose-containing antimicrobial agent used for treating Gram-positive bacteria infections. The structure of 1 is composed of an N-methyl-4-propyl-L-proline moiety (2) and an unusual octose with a methylmercapto group at C-1, known as methylthiolincosamide (MTL, 3; Figure 1).5 The lincosamine component of this compound, acting as the structural mimic of the 3′-end of L-Pro-Met-tRNA and deacylated-tRNA, blocks microbial protein synthesis at the initial phase of the peptide elongation cycle.6 Thus far, only a handful natural products that contain eight-carbon sugar scaffolds have been identified. These include celesticetin (4), octosyl acid A (5), 2-keto-3-deoxy-D-manno-octulosonate 8-phosphate (Kdo8P, 6), and apramycin (7), in addition to 1 (Figure 1).7 Except for Kdo8P (6), which is a key structural component of lipopolysaccharides and has been established to be derived from D-arabinose 5-phosphate (A5P) and phospho-enolpyruvate (PEP) in an aldol-like reaction catalyzed by Kdo8P synthase,8 little is known about how the other C8 sugar scaffolds are biosynthesized. Herein, we report the in vitro functional characterization of two enzymes, LmbR and LmbN, involved in the early stage of MTL (3) biosynthesis. On the basis of the detailed investigation and stereochemical analysis of these two consecutive enzymatic reactions, we identified D-erythro-D-gluco- ocotose 8-phosphate (29) as a key intermediate in this pathway.
Figure 1.
Examples of natural products containing a C8-sugar scaffold.
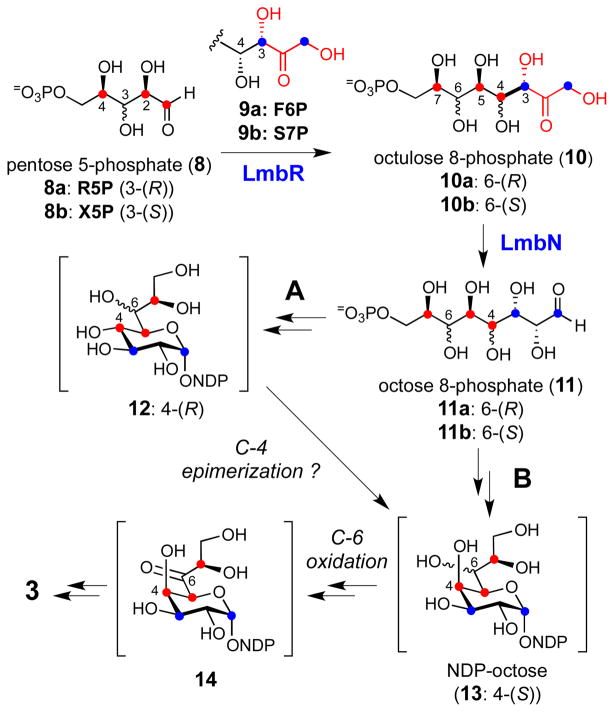
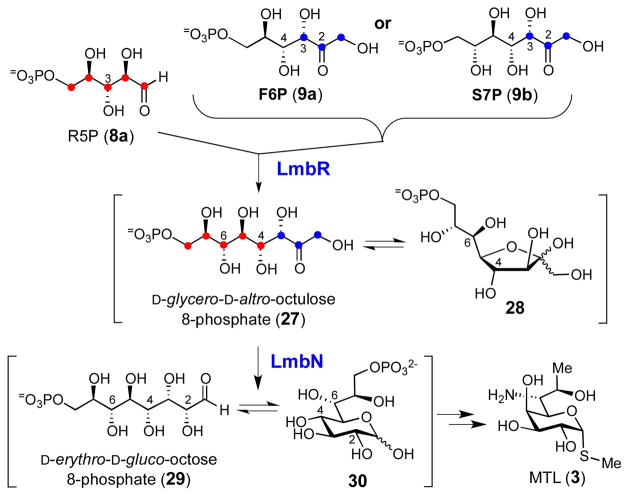
Early feeding experiments using 13C-labeled D-glucose showed that 3 may be assembled through the condensation of a pentose 5-phosphate (C5) and a C3 unit derived from the pentose phosphate pathway in a transaldolase-catalyzed reaction.9 Later, the biosynthetic gene cluster for 1 was isolated and sequenced,10 and a gene encoding a putative transaldolase, LmbR, was identified along with several genes homologous to those found in various NDP-deoxyhexose pathways.3 These results allowed us to propose a possible biosynthetic pathway for MTL (3), which consists of three key steps: i) the assembly of the C8 scaffold from a C5 acceptor and a C3 donor in a transaldol reaction catalyzed by LmbR, ii) the conversion of the resulting octulose 8-phosphate (10) to an NDP-activated octopyranose (NDP-octose, 13) through an octose phosphate intermediate (11), and iii) modification of the NDP-octose (13) by enzymes similar to those found in NDP-deoxyhexose biosynthetic pathways to yield MTL (3) as the end product (Scheme 1).11
Scheme 1.
Proposed biosynthetic pathway for MTL (3).
Because C5 and C7 of 3 are derived from C2 and C4 of the C5 acceptor and each has a (R)-configuration, both D-ribose 5-phosphate (R5P, 8a) and D-xylose 5-phosphate (X5P, 8b) could serve as the C5 precursor in the transaldolase-catalyzed reaction. Although the configuration of the 3-OH group of R5P is opposite to that of X5P, the stereochemical distinction becomes irrelevant as this stereogenic center is transformed to the C6 carbon in MTL (3) bearing an amino functionality. The substitution of a hydroxyl group with an amino moiety at this position is likely accomplished by a transamination reaction via the 6-keto intermediate (14). Thus, both R5P and X5P were considered as possible candidates for the C5 unit. Likewise, the C3 donor may be D-fructose 6-phosphate (F6P, 9a) or D-sedoheptulose 7-phosphate (S7P, 9b), both of which are known C3 precursors for the physiological transaldolase reaction in the pentose phosphate pathway.12 On the basis of the general stereochemical course established for most aldolase-catalyzed reactions, the resulting octulose 8-phosphate product (10) is expected to inherit the stereochemistry at the C3 and C4 positions from the corresponding chiral centers of the C3 donor.13 This would yield a C8 sugar intermediate having S and R configuration at C3 and C4, respectively. However, the predicted (R)-configuration of the C4 hydroxyl group of 10 is in contrast to the observed C4-(S)-configuration of the final product, 3. To account for this discrepancy, the participation of a putative epimerase (encoded by the lmbM gene) may be necessary to epimerize the C4 hydroxyl of 12 to afford 13 (Scheme 1, pathway A). Although rare, reactions catalyzed by aldolases giving the inversed stereochemistry are known.14 It is thus possible that the LmbR-catalyzed reaction may generate an octulose 8-phosphate intermediate with the C4-(S)-configuration, which can be transformed to 13 (via 11) without going through 12 (pathway B). In either case, LmbN, which displays moderate sequence identity with a S7P isomerase, GmhA (31% identity and 47% similarity to the Escherichia coli protein),15 is proposed to catalyze the C1–C2 isomerization of the LmbR product (10) to produce the corresponding octose 8-phosphate (11).
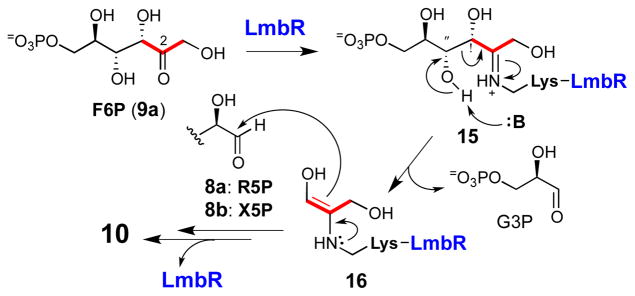
To verify the proposed transaldolase activity of LmbR, the recombinant LmbR with a C-terminal His6-tag was overexpressed in E. coli and purified to near homogeneity (see Figure S1 in the Supporting Information).16 A transaldolase-catalyzed reaction is known to be initiated by imine bond formation between an active site lysine of the enzyme and the 2-keto group of the ketosugar substrate (e.g., F6P (9a) or S7P (9b), see Scheme 2).12,17 To examine whether F6P (9a) is a competent C3 donor for the LmbR-catalyzed reaction, the purified C-His6-LmbR was incubated with F6P followed by sodium borohydride treatment. The recovered LmbR after incubation was subjected to MS analysis. In addition to the unmodified enzyme (calcd, 24952 Da; obsd 24952 Da), a set of MS signals corresponding to the reduced forms of the C-His6-LmbR–F6P conjugate (15: calcd, 25196 Da; obsd, 25193 Da) and C-His6-LmbR–dihydroxyacetone conjugate (16: calcd, 25026 Da; obsd, 25024 Da), were observed (see Supporting Information, Figure S2).16 The control reaction without adding F6P showed only the unmodified enzyme peak. Similar results were also noted when S7P (9b) was used in the incubation (see Supporting Information, Figure S3).16 These results indicate that both F6P and S7P are possible substrates of LmbR, and the reaction proceeds in a similar manner to other transaldolases involving the formation of an imine adduct (e.g., 15) followed by Cα–Cβ bond cleavage via a retro-aldol reaction (e.g., 15 → 16) to generate the C3 donor unit (16) that reacts with pentose 5-phosphate (8).
Scheme 2.
Proposed mechanism for LmbR-catalyzed reaction.
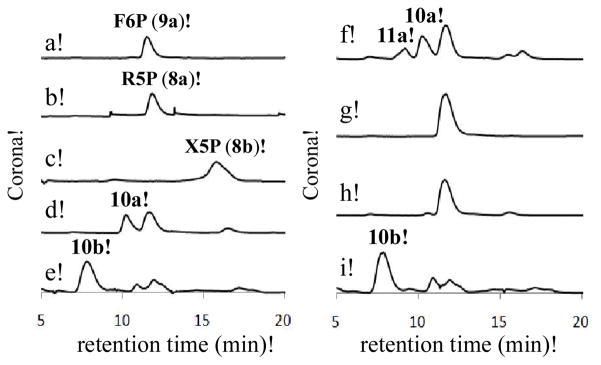
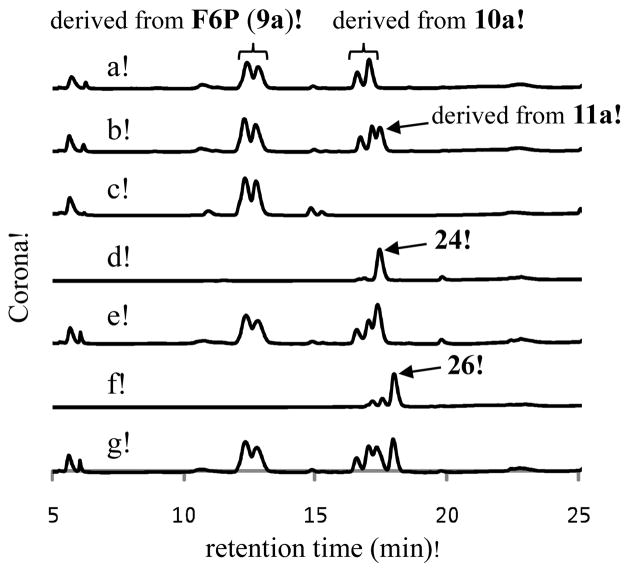
Next, we investigated the possible C5 acceptor substrate for the LmbR-catalyzed reaction. As discussed above, R5P (8a) and X5P (8b) are the two likely candidates. Hence, the purified C-His6-LmbR protein was incubated with F6P (9a) and R5P or X5P, and the reaction mixtures were analyzed using HPLC equipped with a Corona charged aerosol detector (CAD). When R5P (8a) was used in the incubation mixture, a new signal appeared (with a retention time ~10 min) and the substrate signals (retention time for F6P (9a): ~12 min, for R5P (8a): ~12 min) decreased in intensity (Figure 2, trace d). This new product peak, absent in the control reaction that excluded enzyme (trace g), was isolated and subjected to ESI-MS analysis. The molecular mass is consistent with that of the proposed octulose 8-phosphate product (10a; ESI− calcd for C8H16O11P− [M – H+], 319.0436; obsd, 319.0431). Similarly, a new signal (retention time ~ 8 min) was detected when X5P (8b) and F6P (9a) were incubated with the C-His6-LmbR protein (trace e). This new peak was also isolated and subjected to ESI-MS analysis. The molecular mass of the observed mass signal agrees with that of the proposed octulose 8-phosphate product (10b; ESI− calcd for C8H16O11P− [M – H+], 319.0, obsd, 319.1). These results clearly indicate that LmbR can accept both R5P (8a) and X5P (8b) as the C5 substrate. Conveniently, the C8 products derived from each C5 substrate have distinct HPLC retention times.
Figure 2.
The activity and substrate specificity assays for LmbR and LmbN. (a) F6P standard; (b) R5P standard; (c) X5P standard; (d) LmbR with R5P (10 mM) and F6P (10 mM); (e) LmbR with X5P (10 mM) and F6P (10 mM); (f) LmbR and LmbN with R5P (10 mM) and F6P (10 mM); (g) the control sample with F6P (10 mM) and R5P (10 mM), but no enzyme; (h) LmbN with F6P (10 mM) and R5P (10 mM); (i) LmbR and LmbN with X5P (10 mM) and F6P (10 mM).
Having established the substrate specificity of LmbR, we decided to investigate the subsequent isomerization reaction catalyzed by LmbN using the LmbR reaction products, 10a and 10b, as substrates. The recombinant LmbN protein carrying a C-terminal His6-tag was overexpressed in E. coli (see Supporting Information, Figure S1),16 and the purified enzyme was incubated with the LmbR reaction mixture containing F6P (9a) and either R5P (8a) or X5P (8b). No apparent change of the HPLC trace for the reaction starting from X5P was noted (Figure 2, trace i), but a new peak with a retention time of ~9 min was observed for the reaction using R5P (trace f). This product was absent in a control reaction in which LmbR was left out (trace h). It is thus clear that the new product (11a) is derived from the octulose 8-phosphate (10a), and not from F6P or R5P in the reaction solution. This new compound was isolated and subjected to ESI-MS analysis. The molecular mass of the observed MS signal is consistent with that of the proposed octose 8-phosphate product (11a; ESI− calcd for C8H16O11P− [M – H+] 319.0436, obsd, 319.0431). The observed substrate specificity of the LmbN reaction provides strong evidence that the C5 precursor of MTL (3) is R5P (8a) instead of X5P (8b), even though both compounds can be processed by LmbR in vitro. These results also enable us to assign the (R)-configuration at C6 of the LmbR product (10a).
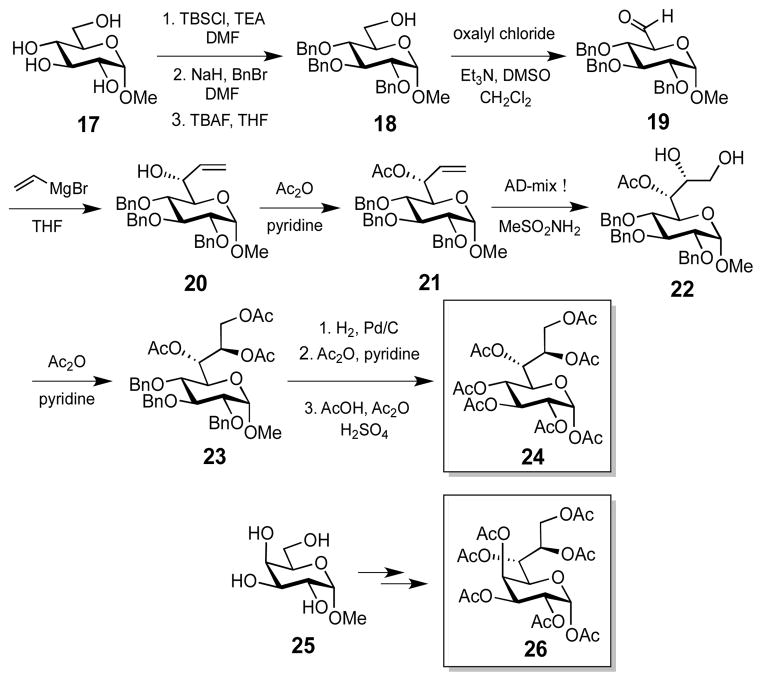
To fully characterize the LmbR and LmbN reactions, the stereochemistry at C4 of their products, 10a and 11a, must be determined (Scheme 1). Although a small amount of pure 11a could be isolated for MS analysis, it was difficult to secure sufficient amounts for NMR characterization due to the poor separation of 11a from 10a. Thus, we opted to chemically synthesize the peracetylated C4-(R)- and C4-(S)-octose standards (24 and 26, respectively (Scheme 3, also see the Supporting Information for details).16 For comparative analysis, the LmbR and LmbN products (10a and 11a, respectively) generated from the incubation with R5P (8a) and F6P (9a) were first treated with alkaline phosphatase, and the resulting dephosphorylated sugar compounds were subjected to the peracetylation conditions. The derivatized enzymatic product mixture along with the synthetic standards were then analyzed using HPLC.
Scheme 3.
Synthesis of (4R)- and (4S)-heptaacetyloctose standards.
As shown in Figure 3, two sets of signals (retention times of ~13.5 min and ~17.5 min) arise from the LmbR reaction (trace a). The first set of two peaks match the signals observed for the control reaction, which generate pentaacetylated fructose as the product (trace c). This assignment is supported by MS analysis of the collected fraction containing these two peaks (ESI+, calcd for C14H19O9 + [M – AcO−] 331.1, found 331.2). The splitting peak pattern likely reflects the formation of two anomeric isomers (α and β) of the pentaacetylfructose product during chemical derivatization. Similarly, the second set of two peaks (retention time of ~17.5 min, trace a) can be attributed to the C-2 anomers of the octulofuranose heptaacetate (see 28) derived from the LmbR product 10a. The MS data of the isolated peaks are consistent with this assignment (ESI+, calcd for C20H27O13 + [M – AcO−] 475.1, found 475.2). When both LmbR and LmbN were used, a new peak with a retention time of ~18 min emerged (trace b). This peak was isolated and subjected to high resolution MS analysis. The results are in agreement with the proposed heptaacetyloctose product (ESI+, calcd for C22H30O15Na+ [M + Na]+ 557.1477, found 557.1478). Importantly, the retention time of this peak matches that of the C4-(R) standard 24 (trace d). Moreover, this product and 24 co-eluted when co-injected (trace e). In contrast, the C4-(S) isomer (26) eluted with a longer retention time (trace f) than the peracetylated LmbN product (see trace g).18 These results unambiguously show that the C4 position of the LmbR/N reaction products has a (R)-configuration (Scheme 1, pathway A).
Figure 3.
HPLC analysis of the acetylated LmbR and LmbN reaction products. (a) LmbR reaction using R5P (10 mM) and F6P (50 mM) followed by dephosphorylation and acetylation. (b) the LmbR and LmbN reactions using R5P (10 mM) and F6P (50 mM) followed by dephosphorylation and acetylation. (c) Control reaction using only F6P. (d) Synthetic standard 24. (e) Coinjection of the sample derived from LmbR and LmbN reaction (trace b), and the synthetic standard 24. (f) Synthetic standard 26. (g) Coinjection of the sample derived from LmbR and LmbN reaction, and the synthetic standard 26.
In summary, we performed in vitro functional characterization of LmbR and LmbN and determined the early intermediates of the MTL biosynthetic pathway. Our results clearly demonstrate that the products of LmbR and LmbN reactions are D-glycero-D-altro-octulose 8-phosphate (27/28) and D-erythro-D-gluco-ocotose 8-phosphate (29/30), respectively (Scheme 4). Our experiments also establish that the C8 sugar backbone of 3 is assembled using R5P (8a) as the C5 acceptor and F6P or S7P as the C3 donor in a transaldol reaction catalyzed by LmbR. Interestingly, the C4 stereochemistry of the LmbR and LmbN products (see 29) is different from that of MTL (3) rendering an C4 epimerization necessary in a later step of the pathway. Significantly, this work provides unambiguous evidence for the biosynthetic precursors of the octose skeleton in MTL, thereby defining a key feature in the overall pathway for this unusual thiooctose-containing natural product. This represents one of many pieces of the puzzle that make up thiosugar biosynthesis, which remains a largely unexplored subject. 19
Scheme 4.
Reactions catalyzed by LmbR and LmbN.
Supplementary Material
Acknowledgments
We thank Sei Hyun Choi for useful discussion of the chemical synthesis. This work was supported in part by grants from the National Institutes of Health (GM035906) and the Welch Foundation (F-1511).
Footnotes
Supporting Information. Experimental details, ESI-MS spectra, additional HPLC trace, and Mosher ester analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Thorson JS, Hosted TJ, Jiang JQ, Biggins JB, Ahlert J. Curr Org Chem. 2001;5:139–167. [Google Scholar]; (b) Kren V, Martinkova L. Curr Med Chem. 2001;8:1303–1328. doi: 10.2174/0929867013372193. [DOI] [PubMed] [Google Scholar]; (e) Chen X. ACS Chem Biol. 2011;6:14–17. doi: 10.1021/cb100375y. [DOI] [PubMed] [Google Scholar]
- 2.(a) Mendez C, Salas JA. Trends Biotechnol. 2001;19:449–456. doi: 10.1016/s0167-7799(01)01765-6. [DOI] [PubMed] [Google Scholar]; (b) Pelzer S, Vente A, Bechthold A. Curr Opin Drug Discov Devel. 2005;8:228–238. [PubMed] [Google Scholar]; (c) Griffith BR, Langenhan JM, Thorson JS. Curr Opin Biotechnol. 2005;16:622–630. doi: 10.1016/j.copbio.2005.10.002. [DOI] [PubMed] [Google Scholar]; (d) Fu X, Albermann C, Jiang J, Liao J, Zhang C, Thorson JS. Nat Biotechnol. 2003;21:1467–1469. doi: 10.1038/nbt909. [DOI] [PubMed] [Google Scholar]; (e) Melancon CE, Thibodeaux CJ, Liu H-w. ACS Chem Biol. 2006;1:499–504. doi: 10.1021/cb600365q. [DOI] [PubMed] [Google Scholar]; (f) Thibodeaux CJ, Liu H-w. Pure Appl Chem. 2007;79:785–799. [Google Scholar]; (g) Mendez C, Luzhetskyy A, Bechthold A, Salas JA. Curr Top Med Chem. 2008;8:710–724. doi: 10.2174/156802608784221532. [DOI] [PubMed] [Google Scholar]; (h) Williams GJ, Gantt RW, Thorson JS. Curr Opin Chem Biol. 2008;12:556–564. doi: 10.1016/j.cbpa.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Walsh CT, Fischbach MA. J Am Chem Soc. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) He X, Agnihotri G, Liu H-w. Chem Rev. 2000;100:4615–4661. doi: 10.1021/cr9902998. [DOI] [PubMed] [Google Scholar]; (b) He X, Liu H-w. Annu Rev Biochem. 2002;71:701–754. doi: 10.1146/annurev.biochem.71.110601.135339. [DOI] [PubMed] [Google Scholar]; (c) Thibodeaux CJ, Melancon CE, Liu H-w. Nature. 2007;446:1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]; (d) Thibodeaux CJ, Melancon CE, Liu H-w. Angew Chem, Int Ed. 2008;47:9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herr RR, Bergy ME. Antimicrob Agents Chemother. 1962:560–564. [Google Scholar]
- 5.Hoeksema H, Bannister B, Birkenmeyer RD, Kagan F, Magerlein BJ, Mackellar FA, Schroeder W, Slomp G, Herr RR. J Am Chem Soc. 1964;86:4223–4224. [Google Scholar]
- 6.(a) Lewis C, Clapp HW, Grady JE. Antimicrob Agents Chemother. 1963:570–582. [Google Scholar]; (b) Fitzhugh AL. Bioorg Med Chem Lett. 1998;8:87–92. doi: 10.1016/s0960-894x(97)10196-2. [DOI] [PubMed] [Google Scholar]; (c) Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]; (d) Poehlsgaard J, Douthwaite S. Nat Rev Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- 7.(a) Hoeksema H. J Am Chem Soc. 1964;86:4224–4225. [Google Scholar]; (b) Oconnor S, Lam LKT, Jones ND, Chaney MO. J Org Chem. 1976;41:2087–2092. doi: 10.1021/jo00874a003. [DOI] [PubMed] [Google Scholar]; (c) Isono K, Crain PF, Mccloskey JA. J Am Chem Soc. 1975;97:943–945. [Google Scholar]; (d) Raetz CRH. Annu Rev Biochem. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- 8.(a) Kaustov L, Kababya S, Du S, Baasov T, Gropper S, Shoham Y, Schmidt A. J Am Chem Soc. 2000;122:2649–2650. [Google Scholar]; (b) Cipolla L, Gabrielli L, Bini D, Russo L, Shaikh N. Nat Prod Rep. 2010;27:1618–1629. doi: 10.1039/c004750n. [DOI] [PubMed] [Google Scholar]
- 9.Brahme NM, Gonzalez JE, Mizsak S, Rolls JR, Hessler EJ, Hurley LH. J Am Chem Soc. 1984;106:7878–7883. [Google Scholar]
- 10.(a) Peschke U, Schmidt H, Zhang HZ, Piepersberg W. Mol Microbiol. 1995;16:1137–1156. doi: 10.1111/j.1365-2958.1995.tb02338.x. [DOI] [PubMed] [Google Scholar]; (b) Koberska M, Kopecky J, Olsovska J, Jelinkova M, Ulanova D, Man P, Flieger M, Janata J. Folia Microbiol. 2008;53:395–401. doi: 10.1007/s12223-008-0060-8. [DOI] [PubMed] [Google Scholar]
- 11.(a) Spížek J, Řezanka T. Appl Microbiol Biotechnol. 2004;63:510–519. doi: 10.1007/s00253-003-1431-3. [DOI] [PubMed] [Google Scholar]; (b) Piepersberg W, Distler J. In: Biotechnology. 2. Rehm HJ, Reed G, editors. Vol. 7. Wiley-VCH; Weinheim: 1997. pp. 397–488. [Google Scholar]; (c) Piepersberg W. In: Novel frontiers in the production of compounds for biomedical use. van Broekhoven A, Shapiro F, Anne J, editors. Vol. 1. Kluwer Academic; Dordrecht: 2001. pp. 161–167. [Google Scholar]; (d) Spížek J, Novotná J, Řezanka T. Adv Appl Microbiol. 2004;56:121–154. doi: 10.1016/S0065-2164(04)56004-5. [DOI] [PubMed] [Google Scholar]
- 12.Samland AK, Sprenger GA. Int J Biochem Cell Biol. 2009;41:1482–1494. doi: 10.1016/j.biocel.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 13.(a) Takayama S, McGarvey GJ, Wong CH. Annu Rev Microbiol. 1997;51:285–310. doi: 10.1146/annurev.micro.51.1.285. [DOI] [PubMed] [Google Scholar]; (b) Samland AK, Sprenger GA. Appl Microbiol Biotechnol. 2006;71:253–264. doi: 10.1007/s00253-006-0422-6. [DOI] [PubMed] [Google Scholar]; (c) Samland AK, Rale M, Sprenger GA, Fessner WD. Chembiochem. 2011;12:1454–1474. doi: 10.1002/cbic.201100072. [DOI] [PubMed] [Google Scholar]
- 14.Fitz W, Schwark JR, Wong CH. J Org Chem. 1995;60:3663–3670. [Google Scholar]
- 15.(a) Brooke JS, Valvano MA. J Bacteriol. 1996;178:3339–3341. doi: 10.1128/jb.178.11.3339-3341.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kneidinger B, Graninger M, Puchberger M, Kosma P, Messner P. J Biol Chem. 2001;276:20935–20944. doi: 10.1074/jbc.M100378200. [DOI] [PubMed] [Google Scholar]; (c) Taylor PL, Blakely KM, De Leon GP, Walker JR, McArthur F, Evdokimova E, Zhang K, Valvano MA, Wright GD, Junop MS. J Biol Chem. 2008;283:2835–2845. doi: 10.1074/jbc.M706163200. [DOI] [PubMed] [Google Scholar]
- 16.See the Supporting Information for details.
- 17.Jia J, Schorken U, Lindqvist Y, Sprenger GA, Schneider G. Protein Sci. 1997;6:119–124. doi: 10.1002/pro.5560060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.GmhA, the counterpart of LmbN in the NDP-heptose biosynthetic pathway, catalyzes the C1–C2 isomerization of S7P to give the corresponding heptose with a C2-(S)-configuration.15b,15c Interestingly, no isomerized product was observed when S7P was treated with LmbN (see Figure S4).16 This result together with the fact that LmbN does not process 10b indicates that LmbN has a relatively strict substrate specificity.
- 19.(a) Sasaki E, Ogasawara Y, Liu HW. J Am Chem Soc. 2010;132:7405–7417. doi: 10.1021/ja1014037. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sasaki E, Liu HW. J Am Chem Soc. 2010;132:15544–15546. doi: 10.1021/ja108061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.