Abstract
Under genotoxic stress, activation of cell cycle checkpoint responses leads to cell cycle arrest, which allows cells to repair DNA damage before continuing to cycle. Drosophila larval epithelial sacs, called imaginal discs, are an excellent in vivo model system for studying radiation-induced cell cycle arrest. Larval imaginal discs go into cell cycle arrest after being subjected to low-dose irradiation, are subject to easy genetic manipulation, are not crucial for survival of the organism, and can be dissected easily for further molecular or cellular analysis. In this chapter, we describe methods for assessing low-dose irradiation-induced cell cycle arrest. Mitotic cells are identified by immunofluorescence staining for the mitotic marker phosphorylated histone H3 (phospho-histone H3 or pH3). When wandering third-instar control larvae, without transgene expression, are exposed to 500 rads of X-ray or γ-ray irradiation, the number of pH3-positive cells in wing imaginal discs is reduced from hundreds before irradiation to approximately 30 after irradiation, with an equal distribution between the anterior and posterior compartments (Yan et al., 2011, FASEB J). Using the GAL4/UAS system, RNAi, cDNA, or microRNA sponge transgenes can be expressed in the posterior compartment of the wing disc using drivers such as engrailed (en)-Gal4, while the anterior compartment serves as an internal control. This approach makes it possible to do genome-wide genetic screening for molecules involved in radiation-induced cell cycle arrest.
Keywords: Drosophila, larvae, imaginal discs, radiation, checkpoint, cell cycle arrest, mitosis
1. Introduction
The fruitfly Drosophila melanogaster was first introduced into the laboratory one hundred years ago by Thomas H. Morgan (2, 3) and has been used since then as a model organism for studies in genetics, animal development, and many other biological processes. The use of simple, yet elegant and powerful genetics tools in Drosophila has twice resulted in Nobel awards (4, 5). Today, we enjoy the insights that Drosophila has brought us regarding fundamental biological principles, and molecular mechanisms of human diseases. Each day we learn about new applications in biomedical research that are possible with this model organism.
Drosophila has been a great asset in studying cell cycle checkpoints (6). Many gene products involved in these checkpoints were first identified in Drosophila and were subsequently shown to have evolutionarily conserved functions in DNA damage responses and in cell cycle arrest in many other organisms including humans (7). It is likely that more genes which play important roles in cell cycle arrest will be discovered, since the Drosophila genome encodes many proteins and non-coding RNAs whose functions are still unknown.
Development of Drosophila larval wing epithelial sacs, called imaginal wing discs, has been studied intensively and comprises an excellent in vivo system for understanding organogenesis (8) and for investigating how different conserved signal transduction pathways function and interact during organ growth (9). During larval growth, a small population of wing progenitor cells proliferates rapidly, resulting in up to 50,000 cells at the late third-instar stage (10). Their rapid cellular division rate and their cell cycle arrest in response to low doses of radiation are among the advantages of using larval imaginal wing discs to study radiation-induced cellular responses (1). Furthermore, wing discs are dispensable for Drosophila survival, so they can be manipulated genetically with ease. For example, the Gal4/UAS system can be used to drive transgene expression in particular regions of discs (Figs. 1 and 2) (1, 11).
Fig. 1.
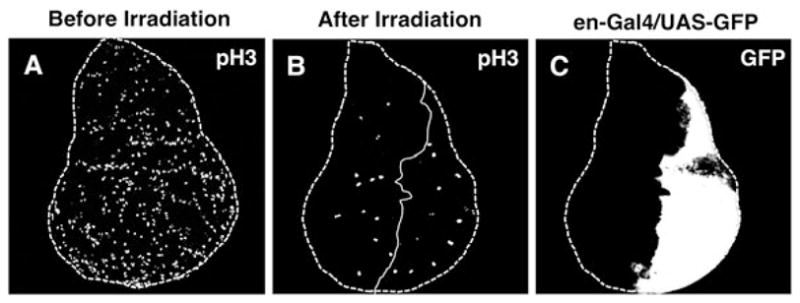
Low-dose radiation-induced cell cycle arrest. (a) A representative confocal image of a wild-type control, unirradiated imaginal wing disc immuno-stained with anti-pH3 and Alexa 546 (as the secondary antibody). Note >200 pH3-positive cells (white dots) in the disc. Imaginal wing discs are shown with anterior to the left and dorsal up. Dashed lines outline the wing discs. (b) Larvae were fixed 1 h post-irradiation (500 rads of X-rays or γ rays) and were immuno-stained with anti-pH3. Note that the number of pH3-positive cells is reduced to approximately 30, with a nearly equal distribution between the anterior and posterior compartments (about 15 in each). Solid lines demarcate the anterior–posterior boundaries. (c) en-Gal4 driven UAS-GFP expression in the posterior compartment. Note that the images in (b) and (c) come from the same disc.
Fig. 2.
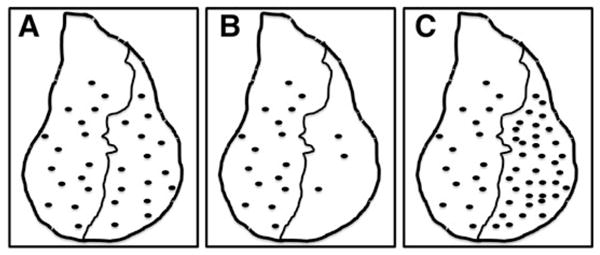
A schematic of different responses to radiation-induced cell cycle arrest. Following irradiation (500 rads of γ rays), the total number of pH3-positive cells is similar in the anterior and posterior compartments when only GFP was expressed in the posterior compartment (a), whereas fewer (b) and more (c) pH3-positive cells are found in the posterior compartments expressing particular UAS-cDNAs or UAS-RNAi trans-genes, respectively, indicating that such genes may control cell cycle arrest. Note that the anterior compartment of the discs serves as an internal control. It is possible to scale up such an assay for genome-wide genetic screening of molecules involved in the radiation-induced cell cycle arrest response.
Recently, UAS-cDNA transgenes from many fly laboratories and stock centers have become available at little or no cost, UAS-RNAi transgenes are available from the Vienna Drosophila RNAi Center (12), and an emerging Drosophila collection of UAS-microRNA sponge transgenes (13) is becoming available from Perrimon’s lab. The vast collection of UAS-RNAi trangenes has made it possible to do genome-wide genetic screening in order to study certain biological processes (14, 15). Such powerful tools also provide an opportunity to those who have not previously worked with Drosophila to begin using it in their research.
Here, we describe methods for studying low-dose radiation-induced cell cycle arrest. Our goal is to introduce these methods to researchers outside of as well as within the Drosophila community.
2. Materials
2.1. Culturing Flies
Fly food ingredients: 51 mL water, 69 g Agar (granulated), 90 g Brewer’s Yeast, 110 g molasses, 400 g malt extract, 400 g corn flour, 50 g soy flour, 31.3 mL propionic acid, 36.0 mL 20% Nipagin in 95% EtOH
Culture bottles (175 mL)
Culture vials (28.5 mL)
Fly stocks (see Note 1)
2.2. Collecting Larvae and Irradiation
Metal probes
Fly food vials
Apple juice plates (ingredients for 1 L): 750 mL water, 22.5 g Bacto Agar, 25.0 g sucrose (technical grade), 250 mL apple juice, 7.0 mL 20% Nipagin in 95% EtOH
An X-ray irradiator
A γ-ray irradiator
2.3. Dissecting Larvae and Fixing Larval Imaginal Discs
Dumont forceps #5
Glass dishes (see Note 2)
1× PBS buffer: 8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, 0.24 g KH2PO4 in 800 mL of distilled H2O. Adjust the pH to 7.4 with HCl. Add H2O to 1 L
A stereo microscope
4% Paraformaldehyde in 1× PBS
Ice buckets
2.4. Staining Larval Imaginal Discs
Primary antibodies: Rabbit anti-pH3 antibody (Upstate Biotechnology, used at 1:1000 in 5% normal goat serum/PBT) or mouse monoclonal anti-pH3 antibody (6G3, Cell Signalling, used at 1:2000 in 5% normal goat serum/PBT)
Secondary antibodies: Goat anti-rabbit or goat anti-mouse Alexa 488, 546, or 660 antibody (Invitrogen, used at 1:250 in 5% normal goat serum/PBT; see Note 3)
PBT (0.3% Triton X-100/1× PBS)
Normal goat serum
Sodium Azide
2.5. Dissecting and Mounting Larval Imaginal Discs
Same as Section 2.3 except using 1× PBT
100 μL and 1-mL Pasteur pipettes
Pipettemen
Glass slide coverslips 22 × 22 mm #1.5 (e.g., VWR)
Microscope glass slides 25 × 75 mm (e.g., VWR)
Lens paper (e.g., VWR)
Tissue paper (e.g., Kimwipes)
Vectashield mounting medium with DAPI (Victor)
Transparent nail polish
Slide holders (e.g., VWR)
2.6. Imaging Larval Imaginal Discs and Measuring Mitotic Cells
A confocal microcope (e.g., Leica SP2/SP5)
A fluorescence microscope (e.g., Zeiss Axio Imager M2m)
Imaging software (e.g., Leica Confocal or Photoshop)
3. Methods
3.1. Culturing Flies
Decide on the amount of fly food needed and proportionally increase or decrease each ingredient of the food recipe. Add the appropriate amount (60%) of hot water to a kettle. Close the lid and bring the water to a boil. Then turn down the heat to stop the water from boiling. Add agar a small amount at a time using a metal whisk. Mix well after each addition and then turn up the heat so that the ingredients in the kettle boil for 2 more minutes. Then slowly add the brewer’s yeast and mix well.
Dissolve the molasses and malt extract in 20% of the total volume of hot water. Stir into the agar/brewer’s yeast/water mixture.
Dissolve the corn flour and soy flour in 20% of the total volume of warm water. Pour into the kettle.
Cook the mix for 2 h with the lid on. Stir well.
Then reduce the heat further and cool for 1 h. Stir well.
When cooled to 80°C, add the propionic acid and Nipagin. Stir very well.
Pump food into vials and bottles (12 mL food for vials, 45 mL food for bottles). While dispensing, keep stirring occasionally to prevent food from thickening.
Cover vials and bottles with cheesecloth and let food solidify overnight before putting in cotton plugs. Place plastic bags over the cheesecloth if the humidity is low.
Wrap trays in plastic bags and store at 18°C. Label with the date the food was made.
Do crosses and transfer flies into fresh vials or bottles of fly food at least every 2–3 days) to prevent larvae from overcrowding (see Note 4). Depending on the number of flies, daily transfers may be necessary.
3.2. Collecting Larvae and Irradiation
Add agar and water to a 2-L flask. Add sucrose and apple juice to another flask. Stir well and then autoclave both flasks.
After autoclaving, cool the flasks down to about 50°C. Add the apple juice/sucrose solution to the agar solution slowly, to avoid the creation of bubbles.
Add the Nipagin solution and mix well without creating bubbles. Pour into 100 mm × 15 mm petri dishes. Wait until agar solidifies. Then store the plates at 4°C. Use a metal probe to pick wandering third-instar larvae (those that have just started crawling out of the food) and put them into fresh vials or onto apple juice plates (see Note 5) immediately before irradiation.
Carefully calculate the proper time required for low irradiation doses (250 or 500 rads of X-rays or γ rays), set the timer accordingly, put larvae into the irradiator, and turn it on (see Note 6).
3.3. Dissecting Larvae and Fixing Larval Imaginal Discs
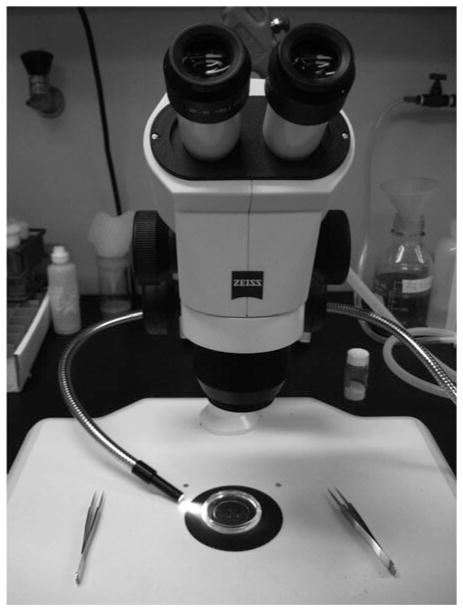
Set up the light source and stereo microscope properly before dissecting larvae (Fig. 3; see Note 7).
Exactly 1 h after irradiation, dissect larvae in 1× PBS with a pair of fine Dumont #5 forceps in each hand. Hold a larva with one pair of forceps and use the other pair to cut the larva about one-third of the way down from the mouth hook. Turn the anterior portion of the larva inside out by holding the head with one pair of forceps and pushing the mouth hook in with the other pair (see Note 8). Using the forceps, gently remove excess fat tissue, the salivary glands and the gut.
Immediately transfer the dissected, inside-out larval tissues into 4% paraformaldehyde in 1× PBS (in a 1.5-mL micro-tube) on ice. At least five larvae (ten imaginal wing discs) can be dissected and collected in the microtube. Then invert the microtube several times, making sure that all of the larvae are in the solution, and leave the tube at room temperature for 10 min, for fixation. Then remove the paraformaldehyde solution and rinse the larvae four times with 1× PBS.
Fig. 3.
Stereo microscope and light setup for larval dissections. The glass dish is under the microscope with the light source to the left side. Each hand will hold a pair of forceps during dissection.
3.4. Staining Larval Imaginal Discs
Incubate the fixed larvae, with wing imaginal discs attached to the body wall, with rabbit anti-phospho-histone H3 (pH3, used at 1:1000) in 5% normal goat serum/PBT overnight in a cold room at 4°C.
The next day, wash the larvae in PBT for 10 min three times at room temperature (see Note 9), and then incubate them with a fluorescent secondary antibody (e.g., goat anti-rabbit Alexa 546, used at 1:250) for 2 h at room temperature or overnight at 4°C (the latter is preferred).
Following the incubation with fluorescent antibody, wash the larvae in PBT for 10 min three times at room temperature and keep the larvae in PBT at 4°C before disc dissection and mounting.
3.5. Dissecting and Mounting Larval Imaginal Discs
Use the same setup as in Section 3.3, except that the dissections are done in PBT rather than in 1 × PBS.
Locate the wing imaginal discs, which are on either side of the body, attached to the trachea near the anterior spiracles (16). Gently remove the wing discs with forceps, without tearing or damaging them. Leave the dissected wing discs (at least 5–10) in PBT in the glass dishes used for dissection.
Clean glass microscope slides. Transfer 5–10 imaginal wing discs to a microscope slide using a 1-mL Pasteur pipette or a pipetteman. Carefully remove excess PBT by sucking it up with a pipette and then blotting with tissue paper.
Put 13 μL of Vectashield mounting medium onto the discs and cover them with a coverslip (avoid bubbles).
Seal the edges of the coverslip with transparent nail polish. Label the slides, put them into a slide holder, and store them at 4°C.
3.6. Imaging Larval Imaginal Discs and Measuring Mitotic Cells
Capture images of the discs using a confocal microscope (e.g., Leica SP2/SP5) or a fluorescence microscope (e.g., Zeiss Axio Imager M2m). Follow instructions in the user’s manual from the manufacturers of the microscope. For confocal images, use a 20× objective lens and a 1024×1024 scan format; set the pinhole diameter at 1 Airy unit.
Use Leica Confocal software or Photoshop to retrieve images. To convert color images to black and white, retrieve the images in Photoshop and select “grayscale” under the “Image > mode” menu (see Note 10).
Count the number of pH3 positive foci.
Fig. 4.
Vials, apple juice agar plates, and metal probe. Note that few larvae are in the vials and on the plates for illustration purposes; more larvae (>50) can be irradiated in a single vial or plate.
Fig. 5.
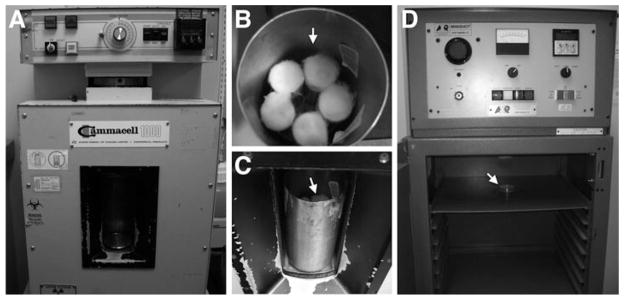
Irradiation equipment and setups. (a) A Cs-137 γ-ray irradiator. (b) A metal rotating cylinder is used to hold five vials (arrow) simultaneously for irradiation. (c) The cylinder with vials (arrow) is put into the irradiation chamber and is ready for irradiation. A shielding metal door is closed (not shown) during irradiation. (d) An X-ray irradiator with larvae on an apple juice agar plate (arrow) ready for irradiation.
Acknowledgments
We are grateful to Christine Summers in the Bohmann lab for providing recipes and protocols for making fly food and apple juice agar plates. We thank Dr L. Silver-Morse for critical reading of the manuscript. S.J.Y. is supported by a postdoctoral training grant (T32CA009363) from the National Institutes of Health. The Li lab is supported by grants from the National Institutes of Health, an American Cancer Society Research Scholar Grant, and a Leukemia & Lymphoma Society Research Scholar Grant to W.X.L.
Footnotes
Information about fly stocks is available through http://www.flybase.org.
Use glass dishes for dissecting larvae, since larval imaginal discs can stick to plastic. If plastic dishes are used, they must be the kind that are coated with glass on the inside.
Alexa Fluor 488 dye generally gives the brightest fluorescence and is therefore preferred if only a single primary antibody is used. When other fluorescence with a similar emission spectrum to Alexa Fluor 488 (e.g., GFP) is present, other fluorescent dyes (e.g., 546) can be used.
It is important to avoid larval overcrowding, as overcrowded larvae are developmentally delayed and smaller than uncrowded larvae. If necessary, use a spatula to transfer a small number of embryos or younger larvae (e.g., first instar larvae) to fresh fly food vials or bottles.
Apple juice plates are flat and can hold a rather large number of larvae. They are suitable for X-ray irradiation where equal distance from the radiation source is required (Figs. 4 and 5d) Normal fly food can be poured into plates for this purpose. However, transparent apple juice agar is more helpful in locating larvae for dissection. We use normal fly food vials to hold larvae for γ-ray irradiation since our γ-ray irradiator (GAMMACELL 1000) provides even penetrance of irradiation to a relatively large population of larvae, by rotating the vials. The GAMMACELL 1000 can irradiate five vials at a time (Fig. 5a–c).
Periodic calibration of the irradiators is important for consistency of the experimental data. Radiation safety training is also required for performing experiments involving irradiators.
To facilitate dissections, it is important to set up the microscope and light source properly. We find that it is easier to see the larvae and the tips of the forceps clearly using a 4 × objective zoom, since this setup provides adequate depth of field and high enough magnification. A 5 × zoom has higher magnification but less depth of field, which makes it more difficult to see both the larvae and forceps without constantly re-adjusting the focus. We also prefer having the light source at the left side instead of above the tissue; larval tissues are mostly transparent and can be seen more easily when lighted from the side. Light coming from above generates reflections, which make everything more difficult to see.
Like other skills, dissecting larvae requires practice and patience. The first few tries can be frustrating. However, once hand-eye coordination is achieved, it will take only about 10–20 s to dissect a larva.
The antibody solutions can be reused at least three times if sodium azide is added to a final concentration of 0.02% (w/v), to prevent microbial contamination.
We find that, for quantification, black and white images are easiest on the eyes and are preferred for presentation and publication if a single antibody is being used.
References
- 1.Yan SJ, Lim SJ, Shi S, Dutta P, Li WX. Unphosphorylated STAT and heterochromatin protect genome stability. FASEB J. 2011;1:232–241. doi: 10.1096/fj.10-169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan TH. Sex limited inheritance in Drosophila. Science. 1910;32:120–122. doi: 10.1126/science.32.812.120. [DOI] [PubMed] [Google Scholar]
- 3.Bellen HJ, Tong C, Tsuda H. 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat Rev Neurosci. 2010;11:514–522. doi: 10.1038/nrn2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen B. Nobel committee rewards pioneers of development studies in fruitflies. Nature. 1995;377:465. doi: 10.1038/377465a0. [DOI] [PubMed] [Google Scholar]
- 5.Raju TN. The Nobel chronicles. 1933: Thomas Hunt Morgan (1866–1945) Lancet. 1999;353:157. doi: 10.1016/s0140-6736(05)76205-x. [DOI] [PubMed] [Google Scholar]
- 6.Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 7.Song YH. Drosophila melanogaster: a model for the study of DNA damage checkpoint response. Mol Cells. 2005;19:167–179. [PubMed] [Google Scholar]
- 8.Weatherbee SD, Carroll SB. Selector genes and limb identity in arthropods and vertebrates. Cell. 1999;97:283–286. doi: 10.1016/s0092-8674(00)80737-0. [DOI] [PubMed] [Google Scholar]
- 9.Yan SJ, Gu Y, Li WX, Fleming RJ. Multiple signaling pathways and a selector protein sequentially regulate Drosophila wing development. Development. 2004;131:285–298. doi: 10.1242/dev.00934. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Bellido A, Merriam JR. Parameters of the wing imaginal disc development of Drosophila melanogaster. Dev Biol. 1971;24:61–87. doi: 10.1016/0012-1606(71)90047-9. [DOI] [PubMed] [Google Scholar]
- 11.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 12.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 13.Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pospisilik JA, Schramek D, Schnidar H, Cronin SJ, Nehme NT, Zhang X, Knauf C, Cani PD, Aumayr K, Todoric J, Bayer M, Haschemi A, Puviindran V, Tar K, Orthofer M, Neely GG, Dietzl G, Manoukian A, Funovics M, Prager G, Wagner O, Ferrandon D, Aberger F, Hui CC, Esterbauer H, Penninger JM. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown versus white adipose cell fate. Cell. 2010;140:148–160. doi: 10.1016/j.cell.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 15.Neely GG, Kuba K, Cammarato A, Isobe K, Amann S, Zhang L, Murata M, Elmen L, Gupta V, Arora S, Sarangi R, Dan D, Fujisawa S, Usami T, Xia CP, Keene AC, Alayari NN, Yamakawa H, Elling U, Berger C, Novatchkova M, Koglgruber R, Fukuda K, Nishina H, Isobe M, Pospisilik JA, Imai Y, Pfeufer A, Hicks AA, Pramstaller PP, Subramaniam S, Kimura A, Ocorr K, Bodmer R, Penninger JM. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell. 2010;141:142–153. doi: 10.1016/j.cell.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bodenstein D. The postembryonic development of Drosophila. In: Demerec M, editor. Biology of Drosophila. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 275–367. Facsimile edn. [Google Scholar]