Abstract
Upon ATP binding, myosin motor protein is found in two alternative conformations, pre-recovery state M* and post-recovery state M**. The transition from one state to the other, known as the recovery stroke, plays a key role in the myosin functional cycle. Despite much recent research, the microscopic details of this transition remain elusive. A critical step in the recovery stroke is the rotation of the converter domain from “up” position in pre-recovery state to “down” position in post-recovery state that leads to the swing of the lever arm attached to it. In this work, we demonstrate that the two rotational states of the converter domain are determined by the interactions within a small structural motif in the force-generating region of the protein that can be accurately modeled on computers using atomic representation and explicit solvent. Our simulations show that the transition between the two states is controlled by a small helix (SH1) located next to the relay helix and relay loop. A small translation in the position of SH1 away from the relay helix is seen to trigger the transition from “up” state to “down” state. The transition is driven by a cluster of hydrophobic residues I687, F487 and F506 that make significant contributions to the stability of both states. The proposed mechanism agrees well with the available structural and mutational studies.
Keywords: myosin, recovery stroke, computer simulation, converter domain, replica-exchange
Introduction
Myosin II is a motor protein responsible for the contractile movement of muscles. Its functional cycle1 is driven by the hydrolysis of adenosine triphosphate (ATP) and consists of a number of conformational transitions alternating with actin binding/unbinding events. Key among these steps is the recovery stroke that takes the myosin molecule from the pre-recovery state M* that follows the force-generating power stroke to the post-recovery state M** that precedes the power stroke. The recovery stroke is a reversible transition2, 3 that occurs in ATP-bound state and can be controlled by temperature and pressure2.
While the end points of the recovery stroke, M* and M**, are available from crystallographic studies 4, the transition between them remains elusive. Numerous recent experimental2, 3, 5, 6 and theoretical7–18 studies have generated significant new insights into how the transition occurs but failed to provide the microscopic mechanism at the atomic level. Experimental approaches3, 6 typically lack sufficient resolution to provide the necessary detail. By measuring the distance distribution between select pairs of residues, for instance, recent paramagnetic resonance (EPR) and fluorescence energy transfer (FRET) studies3, 6, 19, 20, confirm that there are two alternative states in ATP-bound myosin but are unable to offer a suitable microscopic interpretation.
Theoretical methods, on the other hand, are challenged by the large size of the protein that contains more than 700 amino acid residues. Because the direct observation of the recovery stroke is not possible in simulations using all-atom protein models, various alternative approaches have been pursued 17, 21–28. One group of methods seeks to simplify the representation of the protein in an effort to make the problem more tractable. Elastic network studies21, 25, 27 and simulations employing reduced Go-style models23 are two notable recent contributions along this direction. While useful in predicting overall functional dynamics, these models lack microscopic resolution necessary for the recovery stroke. Another group of methods relies on the atomic architecture of the protein but applies techniques of accelerated dynamics such as transition path sampling. Although the recovery stroke transition is observed in these studies the reactive pathways are strongly influenced by the specifics of the employed method and thus remain approximate8, 13. The third group is comprised of approaches that also use atomic architecture but the transition between the reaction end points is simulated with the help of umbrella potentials applied along a pre-arranged reaction coordinate14, 16, 17. Much can be learned about the recovery stroke in these simulations but the proper choice of the reaction coordinate remains a significant weakness.
Crystallographic M* and M** states differ in two regions: 1) the ATP binding site and 2) the force-generating region, separated from the active site by close to 40Å. In the active site, a loop known as switch II can be either in the open state – M*, or the closed state – M**. The differences in the force-generating region concern: a) the relay helix which is straight in M* state and develops a kink in its C-terminal part starting at residue F487 in M** state, and b) the converter domain which undergoes a rotation upon the recovery stroke. Bound to the relay helix on one side, the converter domain connects myosin molecules with the myosin filaments via the neck domain, or the lever arm, on the other side; its rotation is the key step in the power stroke that leads to the lever arm swing during the muscle contraction.
Previous simulation studies by us18 and others8, 13 suggest that the switch II closing is the trigger of the M*-to-M** transition, preceding the conformational transition in the force-generating region. Focusing on that region, we showed in a previous paper29 through an exhaustive combinatorial search that the position of a short helix with the reactive thiol group near the relax helix, SH1, controls the conformation of the relay helix and the rotation of the converter domain. Drawing on that work29, we examine in this paper a small fragment from the force-generating region that contains SH1, converter domain, relay helix and relay loop and show in accurate replica-exchange simulations in explicit water that it experiences two-state folding. The rotation angles of the converter domain observed in our simulations are very similar to those seen in crystallographic M* and M** states, indicating that the designated fragment represents a functioning myosin motif. The small size of the fragment allows us, for the first time, to investigate the converter domain rotation responsible for the lever arm swing in atomic detail. Our simulations predict a critical role for SH1. A small displacement of this helix away from relay helix is seen to shift the population in the rotation angle toward predominantly M** state. The balance between the two states is controlled by the interactions among key residues, F487, F506 and I687 that lie at the interface between the relay helix, relay loop and SH1. The configuration of these three residues changes in response to the displacement of SH1 during the rotation, driven by hydrophobic interactions. The truncated myosin fragment introduced in this work is well suited for the investigations of mutations in the force-generating region of myosin, including mutations associated with the hypertrophic cardiomyopathy30–32.
Results
Force-generating region undergoes two-state folding
The truncated fragment of the myosin force-generating region designed in this work, which we refer to as the minimal recovery stroke model (MRSM), consists of the following structural elements: relay helix (RH) , relay loop (RL), converter domain (CD) and sulfhydryl (SH1) helix. Some parts of this model are constrained to their initial position while others are treated as fully flexible, as discussed in the “Methods” section in more detail. The key components, relay helix and relay loop, are not constrained. SH1 helix is allowed translational freedom, unlike in our previous study29.
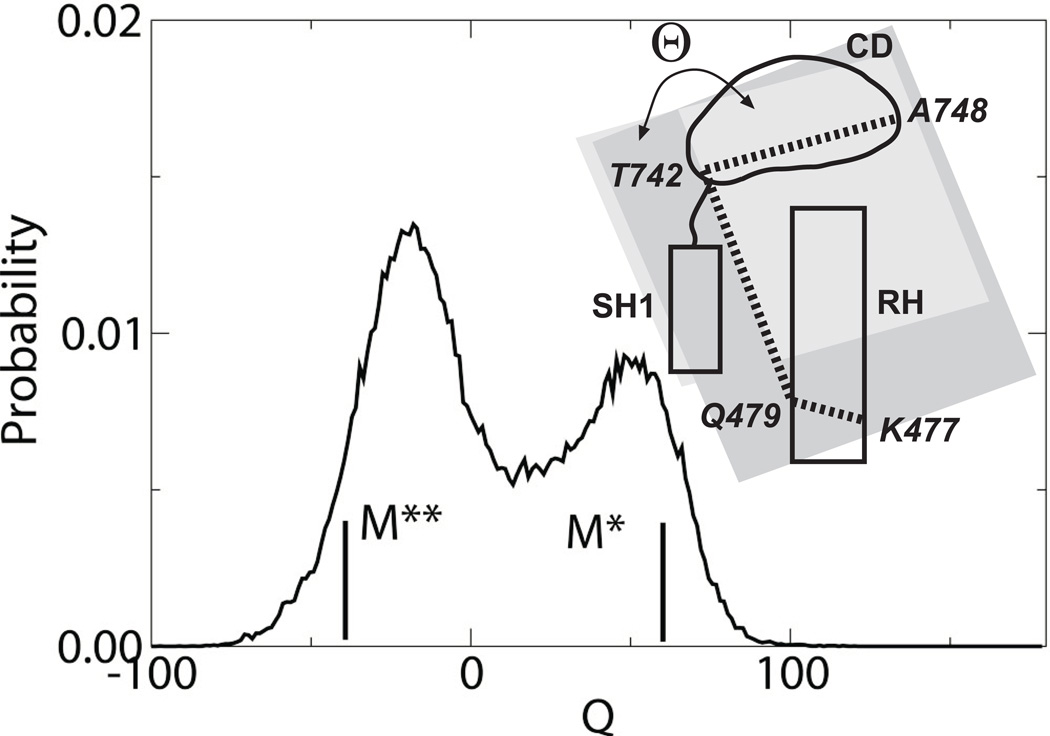
The structural states of the MRSM were characterized in accurate replica exchange simulations in explicit solvent. Figure 1 shows the observed distribution of the torsional anglei Θ formed by Cα atoms of residues K477, Q479, T742 and A748. The angle characterizes the orientation of the converter domain with respect to the N-terminal part of the relay helix. Its value is -38 degrees in M** state and 57 degrees in M* state. The distribution function has two maxima, indicating that the designed model is a two-state folder. Analysis of the locations of the maxima, -20 and 50 degrees, suggests that the two most populated states correspond to experimental M* and M** conformations.
Figure 1.
Probability distribution of the torsional angle Θ as seen in the simulations of MRSM. The angle, formed by Cα of residues K477, Q479, T742 and A748, characterizes the orientation of the converter domain relative to the relay helix. Values of crystallographic M* and M** states are shown as thick lines.
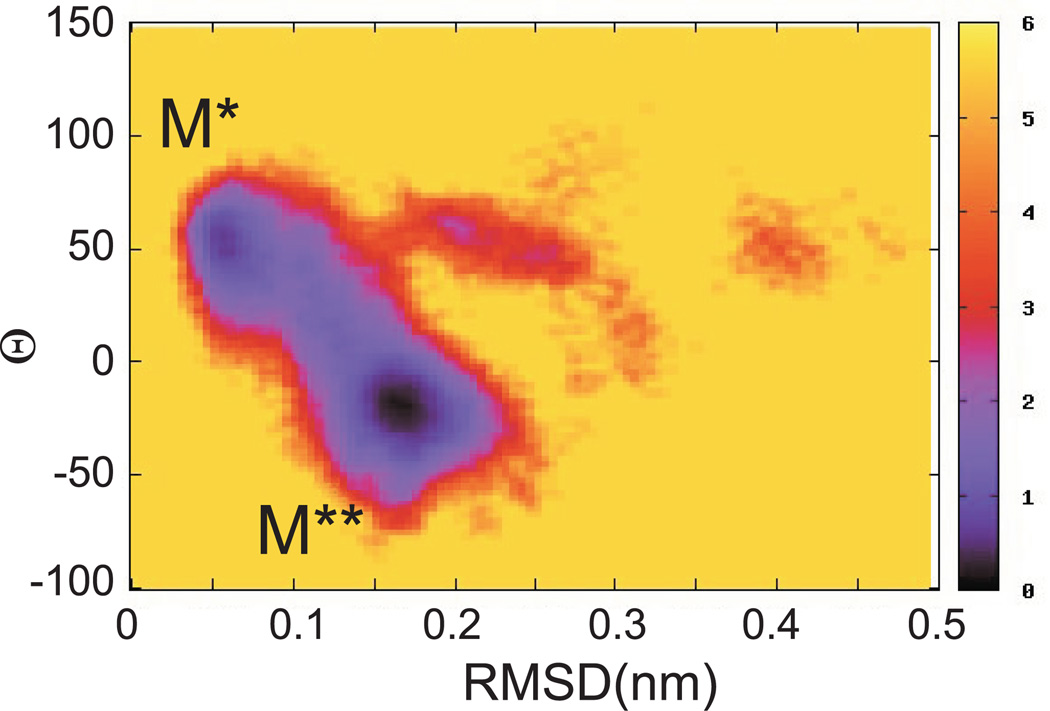
Further evidence that the observed states can be associated with M* and M** comes from the free energy map defined as a function of the root-mean square deviation (RMSD) over Cα atoms of the RH residues N479-E497 from the crystallographic M* conformation and the rotation angle Θ. Depicted in Figure 2, the map shows two minima located at RMSD~0.05nm, Θ~50 and RMSD~0.17nm, Θ~−20 demonstrating that RMSD and rotation angle are correlated. Conformations with Θ~50 in Figure 1, for instance, correspond to low RMSD, indicating straight relay helix. These states therefore can be associated with M* conformation. Similarly, conformations from the second minimum with bent helices can be assigned to M** state.
Figure 2.
Free energy map as a function of 1) RMSD over the relay helix from crystallographic M* conformation and 2) torsional angle Θ. Here and in other figures the energy is shown in units of kT, where k is the Boltzmann’s constant and T is the temperature. Two minima are seen with conformations associated with M* and M** states.
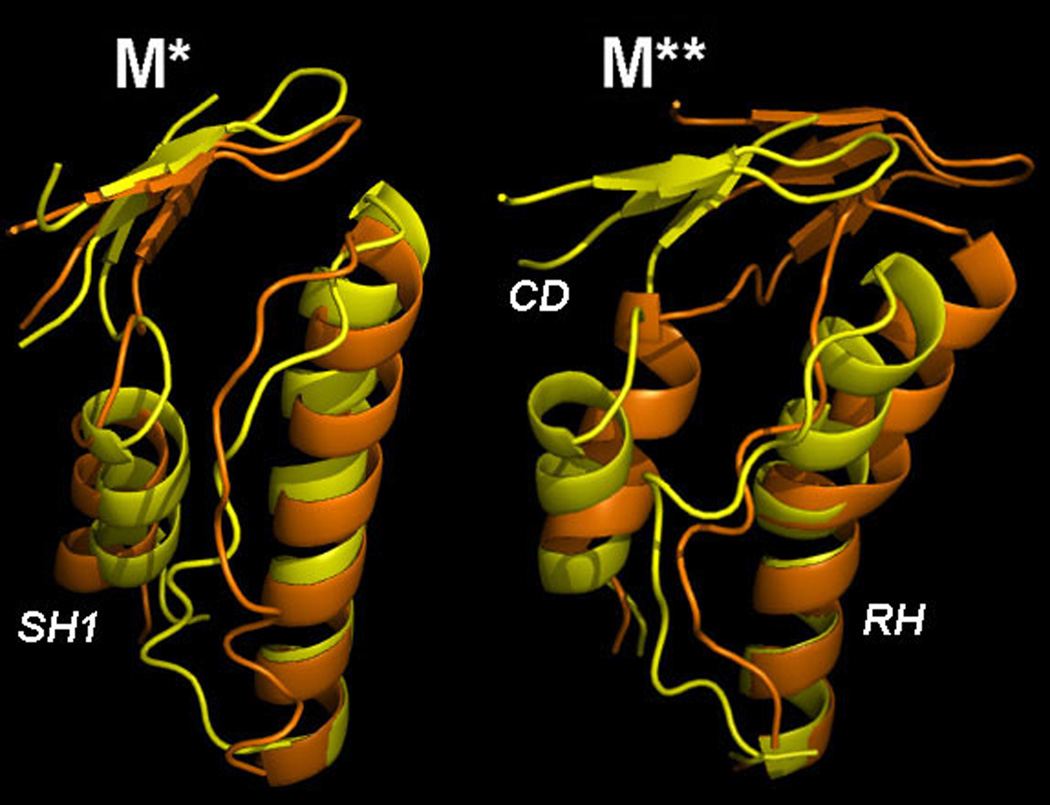
All conformations saved in our simulations were divided into two groups according to the torsional angle: those with Θ<10 correspond to M** and those with Θ>10, to M*. To find the most representative states, all conformations in each group were clustered according to the RH segment with the RMSD cutoff of 0.1nm. The clustering revealed a single predominant substate in each group, with ~60% population. These substates are shown in Figure 3 for M* and M** groups along with the corresponding crystallographic conformations. The computational and X-ray M* states are almost identical with small deviations seen in the C-terminal end of the relay helix and the position of SH1. Deviations in the same places but with greater magnitude are seen in M** conformation. The relay helix exhibits a kink at the same position in both computational and X-ray M** structures. However, the C-terminal part of the helix differs in the two structures, where it is more tilted toward SH1 helix in the computational conformation than in the experimental one.
Figure 3.
Most populated structures observed in our simulations for M* and M** ensembles, yellow, in comparison with the corresponding crystallographic structures, orange.
SH1 domain acts as a trigger of the recovery stroke
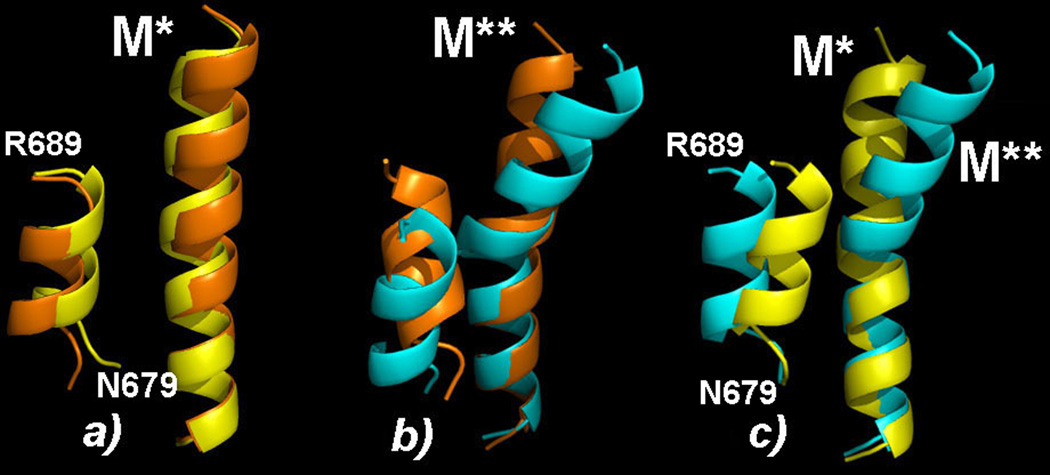
Similarly to the converter domain and the relay helix, SH1 helix occupies distinct configurations in M* and M** states. Figure 4 shows main clusters identified in our simulations together with the corresponding experimental structures. Compared to experiment, SH1 in the computational M* state (shown in Figure 4(a)) is located closer to the relay helix, especially in the N-terminal part, residue N679. This is most likely the consequence of SH1 not being attached to the remainder of the protein. In the computational M** state, SH1 is displaced downward and rotated so as to attain almost parallel orientation with RH. Figure 4(b) shows that the terminal N679 residue in this structure is closer to RH than in the experimental structure. The difference between the computational M* and M** states is shown in Figure 4(c). The N-terminal N679 residue has the same position in both structures, but the C-terminal R689 residue is closer to the relay helix in M* state by ~2Å. The comparison shows that a small displacement in the direction perpendicular to RH is the distinguishing characteristics of M** state.
Figure 4.
Orientation of SH1 helix in computational M* and M** clusters, shown in yellow and cyan respectively, in comparison with the corresponding crystallographic structures, shown in orange. In the computational M** state, SH1 is displaced by ˜2Å away from the relay helix.
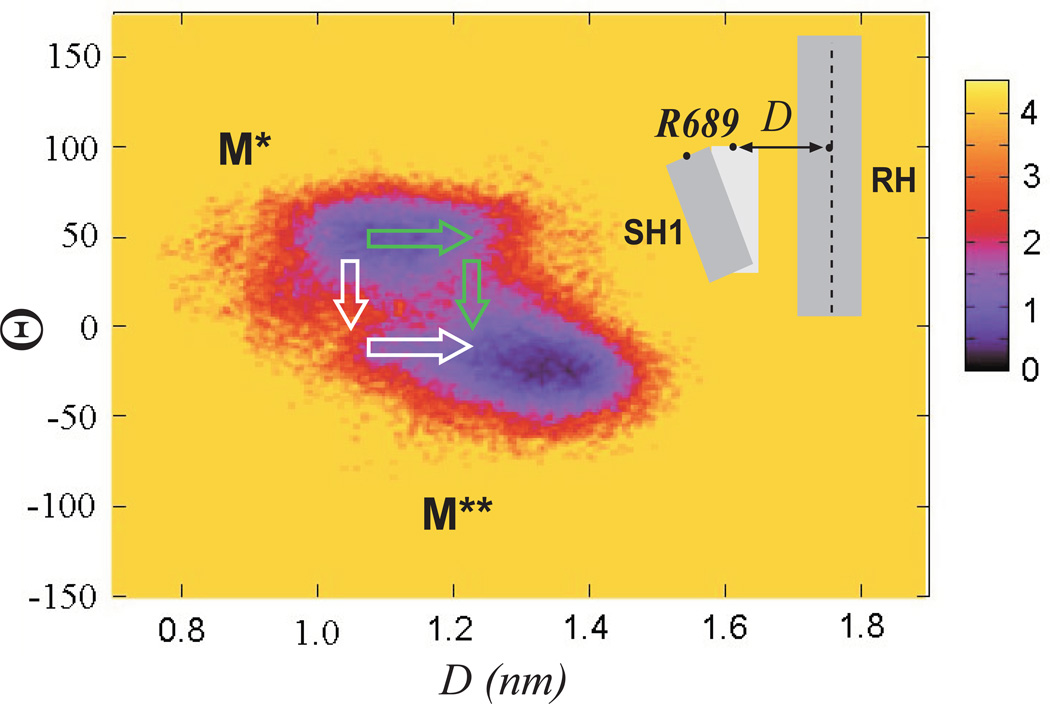
To determine how SH1 displacement is related to the rotation of the converter domain we use the free energy landscapes formalism, which is an accepted theoretical framework for understanding protein dynamics33. Figure 5 shows free energy map defined as a function of angle Θ and D, the distance from Cα of R689 to the long axis of RH (the line connecting Cα atoms of N475 and Y494). The map has two minima, indicating that the displacement and the angle are correlated. One minimum corresponds to M* state with D~1.1nm, Θ~50 and the other minimum is associated with M** state with D~1.3nm, Θ~−20. In addition to confirming the two-state character of our model, the map in Figure 5 sheds new light on the nature of the M*-to-M** transition, that is the recovery stroke. Starting initially from M* conformation, the transition in the 2D space of Θ and D variables can proceed by two scenarios. First, the rotation of the converter domain is followed by the displacement of SH1 helix. Second, the displacement of SH1 helix precedes the rotation of the converter domain. As seen in Figure 5, where these two scenarios are shown in white and green arrows respectively, the first pathway leads to a higher and broader free energy barrier, indicating a lower probability of observation. Based on these data, we conclude that SH1 helix most likely initiates the rotation of the converter domain and not vice versa. A more detailed transition mechanism can be obtained by using methods designed for generating reactive trajectories34–39 or for inferring dynamics from accelerated sampling approaches40, 41, 42
Figure 5.
Free energy map defined as a function of D, distance from Cα of R689 to RH, and the torsional angle Θ. Green arrows indicate the most likely scenario of M*-to M** transition, initiated by a displacement of SH1. White arrows illustrate an unlikely scenario where the converter domain rotates first and is followed by the displacement of SH1.
Hydrophobic interactions drive the converter domain rotation
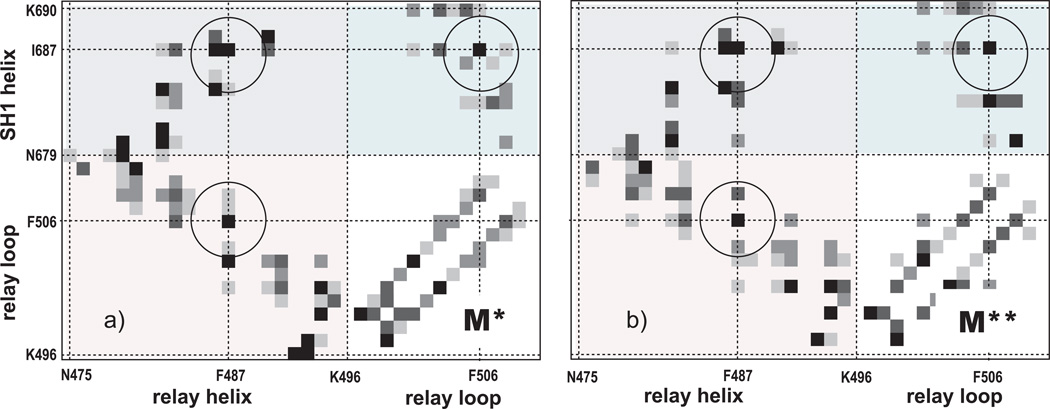
At the residue level, the transition between M* and M** states is driven by the interactions that occur among different parts of the model, RH, RL, SH1 and CD. Figure 6 shows contact probability maps for M* and M** ensembles. Ignoring the contacts between the N-terminal of SH1 and RH, which are probably artifacts, the other contacts shared between the two maps are F487-I687, F487-F506, and F506-I687, as highlighted in the figure. The three residues involved in these contacts, F487, F506 and I687 form a hydrophobic cluster that lies at the interface between RH, SH1 and RL. The intensity of other contacts varies between M* and M** states.
Figure 6.
Contact probabilities of M* and M** ensembles observed in the reported simulations. A contact is considered formed between two residues if the shortest distance between any two atoms of their side chains is 6Å or less.
Table 1 lists the contacts (not including contacts involving glycines) that are newly formed or become stronger in each state respectively. Apart from electrostatic interaction D505-K690, which strongly favors M* state, the majority of all other contacts involve at least one hydrophobic residue. Residues predominantly involved in M** conformation are F506, L508 and I504. Additionally, this conformation is stabilized by the hydrophobic contact between I504 and F487. A similar interaction between F503 and F487, however, favors M* state, indicating that different contacts stabilize different states. As a consequence, the interplay among many hydrophobic interactions maintains the balance between M* and M** conformations.
Table 1.
| New/Stronger contacts | |
|---|---|
| M* | D505-K690, F503-K690, I687-F503, I687-Q483, T688-E490, F503-F487, Q483-E683 |
| M** | L508-E683, F506-E683, I687-I504, F487-E683, L508-Q479, L508-F487, F506-Q491, I504-E490, I504-Q491, F506-W501, I504-K690, I504-F487 |
List of contacts that stabilize M* and M** states
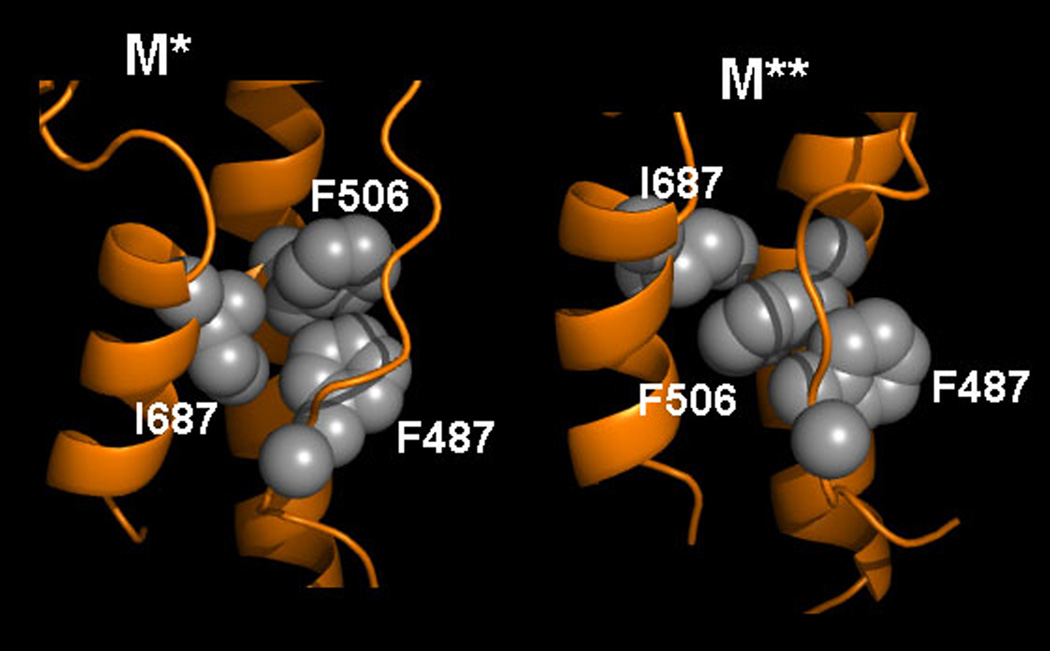
The arrangement of the critical hydrophobic residues F487, F506 and I687 in M* and M** conformations is shown in Figure 7, as determined by clustering. In M* states, the most common configuration is one where the side chains of all three residues form a triangle. In M** states, the side chains are arranged in a line, with F506 intercalating between I687 and F487. The comparison of the two states shows that the mostly hydrophobic interface between RH, SH1 and RL can occupy a number of alternative configurations, controlled by specific inter-residue contacts. Hydrophobic interactions thus seem to drive the M*-to-M** transition.
Figure 7.
Configuration of critical hydrophobic residues I687, F487 and F506 in M* and M** conformations.
Discussion
The force-generating region in the context of the full-length myosin
The MRSM model proposed in this work properly reproduces the two-state folding of the myosin recovery stroke. This is quite remarkable given the small size of the designed fragment that consists only of relay helix, relay loop and SH1 in the force-generating region of the protein. We showed in our previous study29 that these structural motifs are necessary to observe folding into two states. Here, we observe that they are also sufficient. It is clear, therefore, that the local interactions within a small fragment of the force-generating region of myosin II encode the functional dynamics of the entire protein.
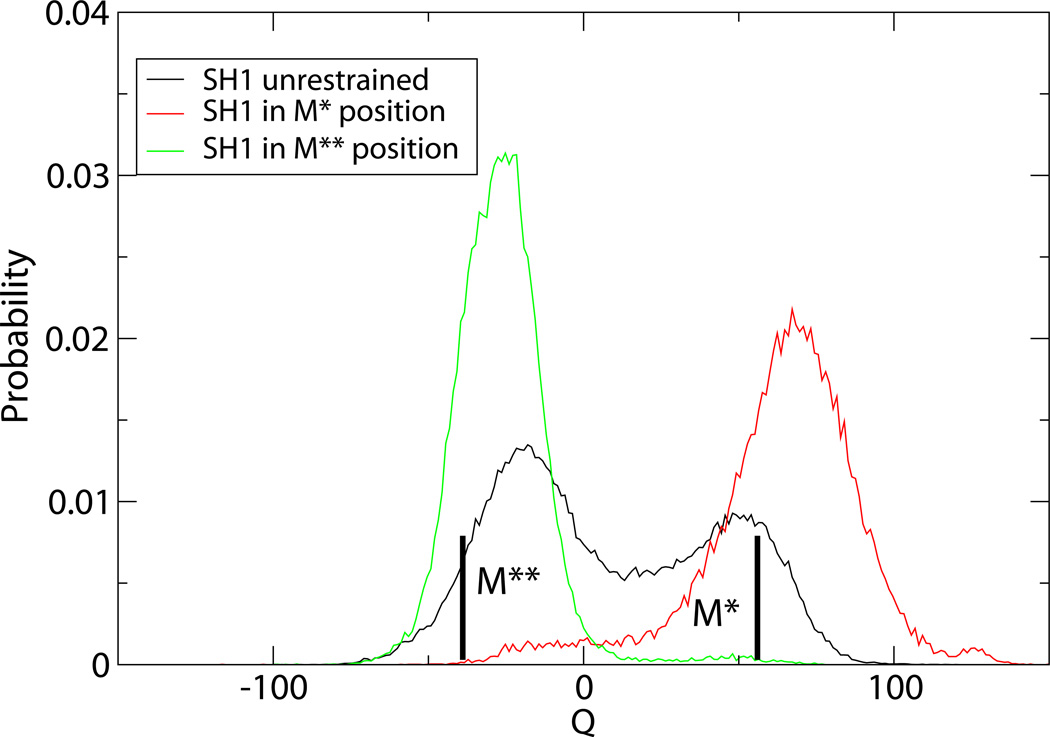
To function as a motor, the force-generating fragment needs to be manipulated externally. We showed that its free energy surface predisposes SH1 helix to act as a trigger of the converter domain rotation. A displacement of SH1 away from the relay helix creates a strong bias in the converter domain toward M** rotations. In available experimental structures, SH1 is seen to be shifted parallel to the relay helix in M** conformation, not perpendicular to it. We tested the effect of a parallel displacement by restraining SH1 to its position in crystallographic M* and M** states, as in our previous work29. Distribution functions obtained in these tests, shown in Figure 8, demonstrate that SH1 is a very efficient modulator of the conformational states of the converter domain: although both “up” and “down” rotations are seen for different SH1 positions, positioning as in M* or M** state creates a very strong affinity for that particular state. The parallel displacement is seen to have the same effect on the population of M** state as the perpendicular one. Additionally, the distribution functions with SH1 helix restrained come in better agreement with experiment.
Figure 8.
Distribution function of the torsional rotation angle obtained in the simulations of our model with SH1 a) unrestrained and b) restrained to its position in crystallographic M* and M** states.
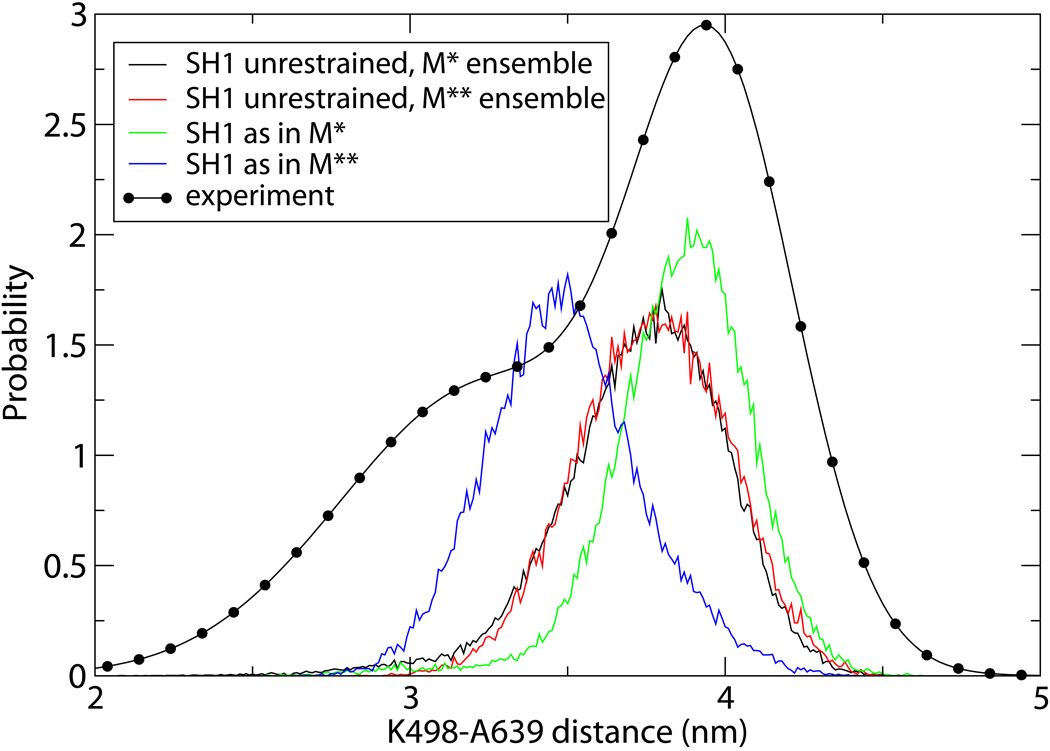
Additional evidence for the need to manipulate SH1 in the context of a functioning myosin comes from inter-residue distance distributions. Distances measured between specific residues in EPR and FRET experiments3, 6 show two maxima which are associated with M* and M** . As an illustration, we focus on the K498-A639 distance. Residue A639 is not part of our model, so we had to infer the distance from it to K498 by measuring the vector from K498 (atom Nζ) to N475 (atom Cα) and modeling the vector from N475 (atom Cα) to A639 (atom Cβ) as a Gaussian distribution with parameters (the mean and dispersion for all three components) extracted from short nanosecond-scale simulations of the full-length protein. The resulting distributions are shown in Figure 9 along with the MRSM results and the EPR data6. The maxima in simulations with free SH1 appear in the same place for both M* and M** ensembles (despite different rotation angles). But the simulations with restrained SH1 show two well separated maxima for M* and M** states, demonstrating that displacement of SH1 from outside produces better agreement with experiment (the quantitative comparison between theory and experiment is not justified since we did not model the experimental spin-label probes in our simulations). Similar improvement is observed for other distances, K498-D505 for instance, but is not as dramatic as for the K498-N475 pair.
Figure 9.
Distribution function of the distance between K498 and A639 obtained in this work in various simulations as explained in the legend. Symbols indicate EPR data6
Role of SH1 helix
SH1 was hypothesized43 to serve as a communication tool between the active site and the force-generating region on the basis of several lines of evidence: 1) Proximity of SH1 and SH2 (another helix with a sulfhydryl group that precedes SH1 along the sequence) to both the active site and the force-generating region, 2) Mutational studies showing that a) cross-linking of SH1-SH2 and b) substitutions in SH1/SH2 disrupt mechano-chemical coupling44, 3) X-ray structures suggesting that the pivotal point of converter domain rotation is in the vicinity of SH1 and SH243, 45, 4) Strong conservation of SH1/SH2 domains46. Our simulations are fully consistent with this hypothesis. The finding that SH1 needs to be manipulated from outside of the force-generating region makes it a prime candidate for the role of transmitting pertinent signals (switch I/II closing, ATP hydrolysis) from the active site.
SH1 was suggested to form a part of the minimal structural motif, the “little machine”, capable of capturing the complete motion of the converter domain during the recovery and power strokes47. Our simulations support this suggestion. The residues of the minimal motif, G691-P693 and F746, are found in the truncated myosin fragment identified in this work. Not only do we show that the motif captures the rotation of the converter domain, but we also explain how this happens.
A microscopic interpretation of how SH1 fulfils its communication role was offered by Fischer et al8, 9. Based on minimum energy paths connecting M* and M** states, the authors suggested that a piston-like movement of SH1 domain along the relay helix applies a mechanical force to the converter domain, causing it to rotate. Our simulations support a different scenario. We showed in this work that a small displacement in the perpendicular direction to the relay helix is able to induce the rotation. In a previous paper29, SH1 was seen to act by direct interactions with the relay helix, rather than by the mechanical force.
Mechanism of the lever arm swing in the recovery stroke
Moving SH1 away from the relay helix apparently creates a cavity in the hydrophobic cluster made of residues I687, F487 and F506 at the interface of RL, RH and SH1. The cavity is filled upon transition by the side chain of F487 which is pulled in-between I687 and F506 in M** conformation (see Figure 7), thereby creating a kink in the relay helix. The kink causes the rotation of the converter domain which is tightly coupled to the C-terminal part of the relay helix. Rather than mechanical8, 9, our model supports the statistical interpretation of the recovery stroke 13, 15, 16, 18, in which the restructuring of the hydrophobic cluster in response to the SH1 dislocation is the mechanism that drives the transition between M* and M** states.
Indirect support for this mechanism comes from mutational studies. We showed that residues F487, F506 and I687 are critical for the stability of both M* and M** states. Mutating any of them is expected to disrupt the structure of these states substantially, perhaps to the point of rendering the protein dysfunctional. This is exactly what happens in F487A substitution, which was shown48 to block motor activity in myosin II completely through the disruption of communication between the active site and the lever arm. The same effect was seen for F506A mutation, suggesting that it interferes with the folding of either M* or M**, or both 48. A later electron paramagnetic resonance study revealed49 that these states remain intact but with altered populations. Based on the analysis of crystallographic M* and M** states, hydrophobic interactions among a set of critical residues very similar to those identified in this work, N483, F487, F506, l508 and I687, were suggested to control the recovery stroke transition48.
Precisely how SH1 is controlled by the closing of switch II cannot be answered using the reported model alone. This issue remains to be addressed in simulations that include the whole protein, not just its fragments. Our model, however, is well suited for the studies of the effect of mutations in the force-generating region of myosin. The best candidates for such studies are F487A and F506A substitutions mentioned above and certain mutations, including E490D, E492K, R695L and L508R, associated with the hypertrophic cardiomyopathy30–32.
Methods
Design of the minimal recovery stroke model
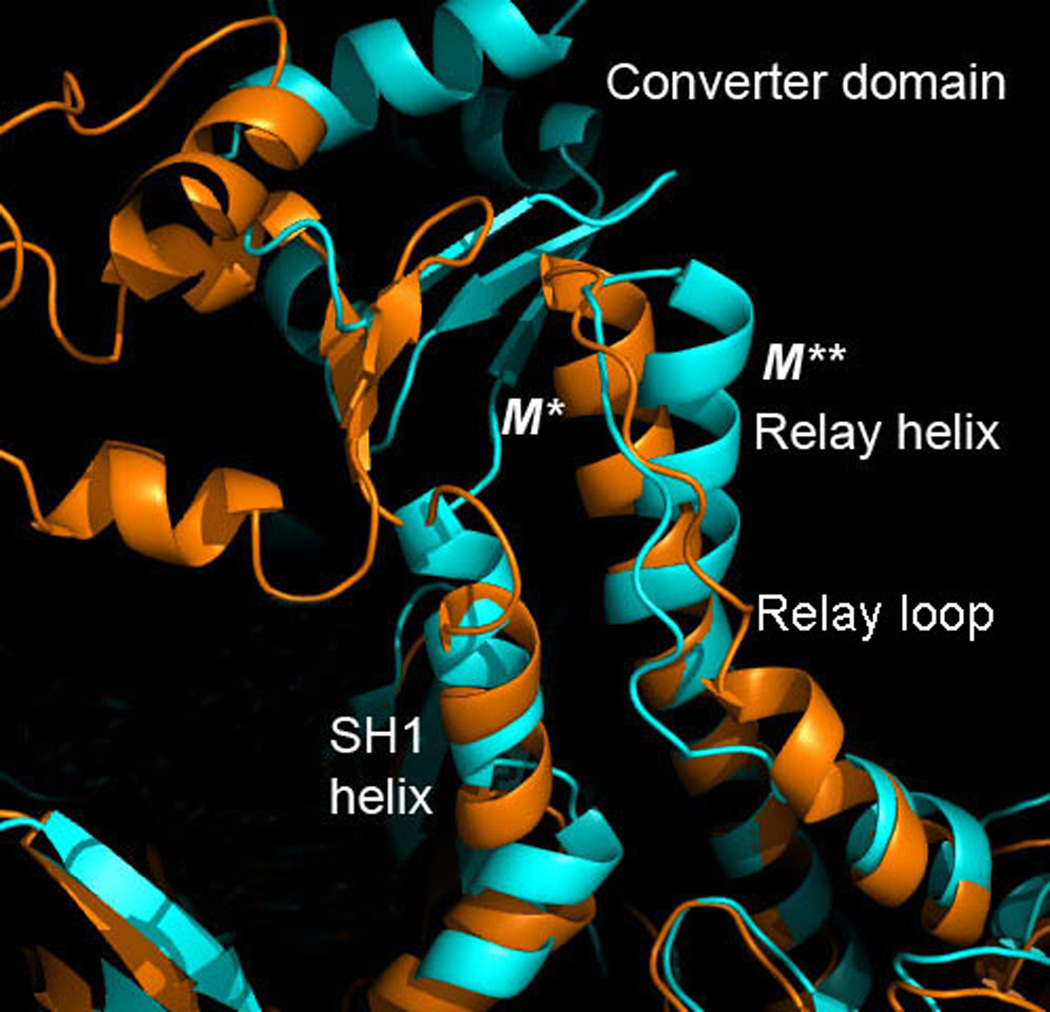
Figure 10 shows a close-up of crystallographic M* and M** conformations aligned along the N-terminal part of the relay helix. The kink is seen in M** conformation in the C-terminal end of RH that interacts with the converter domain, the relay loop linking the relay helix with the actin binding region of myosin, and SH1 helix. The N-terminal end of RH is identical in both M* and M** states. The conformation of the converter domain is the same as well, except for its orientation with respect to the relay helix. SH1 in M** state is shifted upwards by 4Å along the relay helix.
Figure 10.
Close up of M*, orange, and M**, cyan, conformations aligned along the N-terminal section of the relay helix. The kink visible in M** state occurs in the C-terminal part that interacts with relay loop, converter domain and SH1 helix. The latter is shifted by 4Å in M** relative to M*.
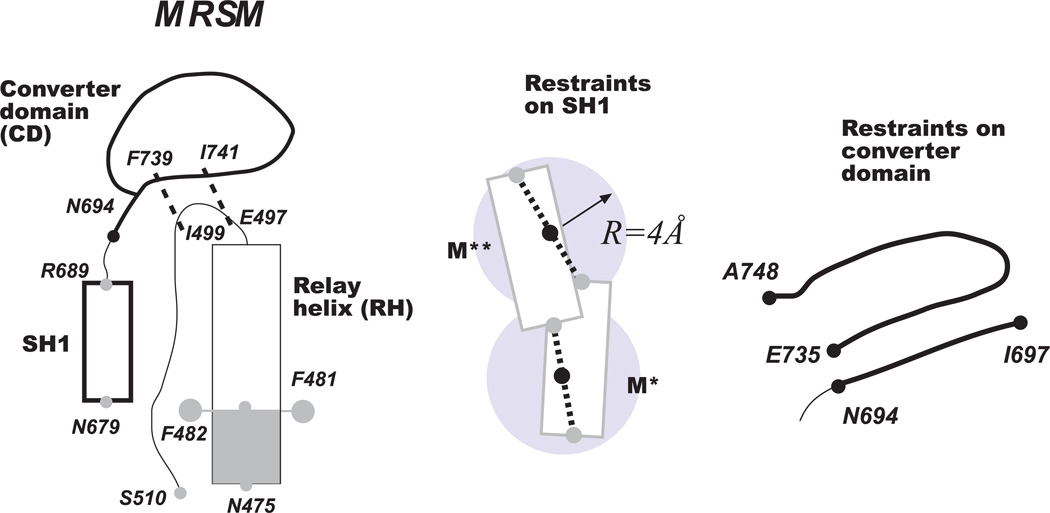
We consider a truncated version of myosin consisting of the fragments shown in Figure 10 and ask whether it exhibits an M*-to-M** transition similar to that seen experimentally. The model, illustrated in Figure 11, is designed to reflect the differences between M* and M** conformations discussed above. The N-terminal part of the relay helix is kept near its initial crystallographic position by harmonic restraints applied to Cα atoms of N475 to F482 and, additionally, to Cζ atoms of F481 and F482. The last residue of the relay loop, S510, occupies almost identical position in aligned M* and M** states, with the mutual distance less than 2Å. We therefore restrain Cα of this residue as well but with the force 10 times weaker than in the relay helix. The conformation of two segments, SH1 helix spanning N679 to R689, and converter domain that includes residues N694 to A748 is kept constant by pairwise inter-residue restraints applied to Cα atoms. The list of the employed restraints is provided in the “Supporting Information” section. All other parts of the model are completely flexible.
Figure 11.
The minimal recovery stroke model (MSRM) introduced in this work. The model consists of relay helix (RH), relay loop (RL), converter domain (CD) and SH1 helix. Shaded areas correspond to the parts restrained to their initial crystallographic positions. Parts shown in bold lines are made to maintain their conformation by inter-residue restraints. Thin lines show completely flexible parts. SH1 helix is kept near RH by two restraints acting on N679 and R689 that permit both M** and M* conformations. The converter domain contains two fragments, kept in contact with RH by two restraints.
The converter domain that consists of two segments, N694-I697 and E735-A748, is kept in contact with the relay helix by two restraints placed on F739 and I499, and I741 and E497, as shown in Figure 11. The restraints are designed to allow partial dissociation of the converter domain from the relay helix. Additionally, Cα of P693 was kept at least 15Å away from Cα of F482 to prevent the unphysical collapse of the converter domain onto the relay helix.
The position of SH1 helix was subject to two restraints applied to Cα of N679 and R689, designed to keep it at an interaction distance from RH. The anchor points for the restraint potentials were chosen at the midpoint of the line connecting positions of the corresponding residues in M* and M** conformations, as shown in Figure 11. The residues were allowed to deviate by 4Å before the restraining potentials were switched on, allowing for a wide ensemble of unbiased conformations including M* and M**. To probe the effect of SH1 positioned as in crystallographic structures, two sets of anchor points without permitted free deviations were considered at the positions of M* and M** conformations, in analogy to our previous work29, where all Cα atoms of SH1 helix were restrained.
Finally, in order to prevent SH1 from rotating around its own axis and forming an interface with RH not seen in crystallographic structures, an angular restraint was applied to G684, I685, F481 and F482. The angle formed by Cα atoms of these residues was kept near zero, the value seen in both M* and M** states.
With the restraints mimicking the neglected parts of the myosin, we refer to the model shown in Figure 11 as the minimal recovery stroke model (MRSM). More computational details are provided in the “Supporting Information” section.
Computational details
All myosin fragments considered in this work were modeled in full atomic detail using OPLS/AA50 force field in combination with TIP3P model51 of water. Neutralizing ACE and NH2 groups were placed at N- and C-termini of all three fragments to avoid interference from terminal charged groups. Initial coordinates were downloaded from PDB data bank52 for Dictyostelium myosin, structures 1w9k for M* state and 1w9l for M** state, and truncated according to the size of the model.
All simulations were performed using GROMACS53 software set. The chemical bonds in water molecules were held constant by the SETTLE54 algorithm. The bonds involving hydrogen atoms in the protein were constrained according to the LINCS55 algorithm. Crystallographic structures for each model were placed in a cubic box of 6.2nm across, containing 7490 water molecules, and equilibrated with the heavy atoms constrained to their initial positions. The replica-exchange algorithm56 was used to conduct the simulations using Nose-Hoover thermostat57 with a 0.5ps time constant to maintain constant temperature. A total of 60 replicas were considered, spaced equidistantly in inverse temperature between end-points of 300 and 550K. All data were analyzed at T=300K. A single cut-off of 1nm was used for the van der Waals interactions, with the neighbor lists updated every 10 time steps. Smooth-particle mesh Ewald (PME) method58 was used to treat electrostatic interactions. The time step was set at 2fs in all simulations.
Supplementary Material
Acknowledgment
We would like to thank Yuri Nesmelov for his comments on the early version of the manuscript and for providing his EPR data. We gratefully acknowledge the support of the National Institutes of Health, grant R01GM083600-04. The computational time was allocated at UNC Charlotte Cobra cluster supported by NIH grant 1S10RR026514-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lymn RW, Taylor EW. Mechanism of Adenosine Triphosphate Hydrolysis by Actomyosin. Biochemistry. 1971;10(25):4617–4624. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]
- 2.Malnasi-Csizmadia A, Pearson DS, Kovacs M, Woolley RJ, Geeves MA, Bagshaw CR. Kinetic resolution of a conformational transition and the ATP hydrolysis step using relaxation methods with a Dictyostelium myosin II mutant containing a single tryptophan residue. Biochemistry. 2001;40(42):12727–12737. doi: 10.1021/bi010963q. [DOI] [PubMed] [Google Scholar]
- 3.Agafonov RV, Nesmelov YE, Titus MA, Thomas DD. Muscle and nonmuscle myosins probed by a spin label at equivalent sites in the force-generating domain. P Natl Acad Sci USA. 2008;105(36):13397–13402. doi: 10.1073/pnas.0801342105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sweeney HL, Houdusse A. Structural and Functional Insights into the Myosin Motor Mechanism. Annual Review of Biophysics, Vol 39. 2010;39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- 5.Wray J, Urbanke C. A fluorescence temperature-jump study of conformational transitions in myosin subfragment 1. Biochemical Journal. 2001;358:165–173. doi: 10.1042/0264-6021:3580165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agafonov RV, Negrashov IV, Tkachev YV, Blakely SE, Titus MA, Thomas DD, Nesmelov YE. Structural dynamics of the myosin relay helix by time-resolved EPR and FRET. P Natl Acad Sci USA. 2009;106(51):21625–21630. doi: 10.1073/pnas.0909757106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mesentean S, Koppole S, Smith JC, Fischer S. The principal motions involved in the coupling mechanism of the recovery stroke of the myosin motor. J Mol Biol. 2007;367(2):591–602. doi: 10.1016/j.jmb.2006.12.058. [DOI] [PubMed] [Google Scholar]
- 8.Fischer S, Windshugel B, Horak D, Holmes KC, Smith JC. Structural mechanism of the recovery stroke in the myosin molecular motor. P Natl Acad Sci USA. 2005;102(19):6873–6878. doi: 10.1073/pnas.0408784102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koppole S, Smith JC, Fischer S. The structural coupling between ATPase activation and recovery stroke in the myosin II motor. Structure. 2007;15(7):825–837. doi: 10.1016/j.str.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Koppole S, Smith JC, Fischer S. Simulations of the myosin II motor reveal a nucleotide-state sensing element that controls the recovery stroke. J Mol Biol. 2006;361(3):604–616. doi: 10.1016/j.jmb.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 11.Gyimesi M, Kintses B, Bodor A, Perczel A, Fischer S, Bagshaw CR, Malnasi-Csizmadia A. The mechanism of the reverse recovery step, phosphate release, and actin activation of Dictyostelium myosin II. Journal of Biological Chemistry. 2008;283(13):8153–8163. doi: 10.1074/jbc.M708863200. [DOI] [PubMed] [Google Scholar]
- 12.Yamanaka K, Okimoto N, Neya S, Hata M, Hoshino T. Behavior of water molecules in ATPase pocket of myosin. Journal of Molecular Structure-Theochem. 2006;758(2–3):97–105. [Google Scholar]
- 13.Elber R, West A. Atomically detailed simulation of the recovery stroke in myosin by Milestoning. P Natl Acad Sci USA. 2010;107(11):5001–5005. doi: 10.1073/pnas.0909636107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Yu HB, Cui Q. Extensive conformational transitions are required to turn on ATP hydrolysis in myosin. J Mol Biol. 2008;381(5):1407–1420. doi: 10.1016/j.jmb.2008.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu HB, Ma L, Yang Y, Cui Q. Mechanochemical coupling in the myosin motor domain. I. Insights from equilibrium active-site simulations. Plos Comput Biol. 2007;3(2):199–213. doi: 10.1371/journal.pcbi.0030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris MJ, Woo HJ. Energetics of subdomain movements and fluorescence probe solvation environment change in ATP-bound myosin. European Biophysics Journal with Biophysics Letters. 2008;38(1):1–12. doi: 10.1007/s00249-008-0347-3. [DOI] [PubMed] [Google Scholar]
- 17.Woo HJ. Exploration of the conformational space of myosin recovery stroke via molecular dynamics. Biophysical Chemistry. 2007;125(1):127–137. doi: 10.1016/j.bpc.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Baumketner A, Nesmelov YE. Early stages of the recovery stroke in Dictyostelium discoideum myosin II studied by molecular dynamics simulations. Protein Sci. 2011;20:2013–2022. doi: 10.1002/pro.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nesmelov YE, Agafonov RV, Burr AR, Weber RT, Thomas DD. Structure and dynamics of the force-generating domain of myosin probed by multifrequency electron paramagnetic resonance. Biophys J. 2008;95(1):247–256. doi: 10.1529/biophysj.107.124305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nesmelov YE, Agafonov RV, Negrashov IV, Blakely SE, Titus MA, Thomas DD. Structural kinetics of myosin by transient time-resolved FRET. Proc Natl Acad Sci U S A. 2011;108(5):1891–1896. doi: 10.1073/pnas.1012320108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tekpinar M, Zheng WJ. Predicting order of conformational changes during protein conformational transitions using an interpolated elastic network model. Proteins. 2010;78(11):2469–2481. doi: 10.1002/prot.22755. [DOI] [PubMed] [Google Scholar]
- 22.Taylor WR, Katsimitsoulia Z. A coarse-grained molecular model for actin-myosin simulation. Journal of Molecular Graphics & Modelling. 2010;29(2):266–279. doi: 10.1016/j.jmgm.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Takagi F, Kikuchi M. Structural change and nucleotide dissociation of myosin motor domain: Dual G(o)over-bar model simulation. Biophys J. 2007;93(11):3820–3827. doi: 10.1529/biophysj.106.103796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cecchini M, Alexeev Y, Karplus M. Pi Release from Myosin: A Simulation Analysis of Possible Pathways. Structure. 2010;18(4):458–470. doi: 10.1016/j.str.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cecchini M, Houdusse A, Karplus M. Allosteric Communication in Myosin V: From Small Conformational Changes to Large Directed Movements. Plos Comput Biol. 2008;4(8) doi: 10.1371/journal.pcbi.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ovchinnikov V, Trout BL, Karplus M. Mechanical Coupling in Myosin V: A Simulation Study. J Mol Biol. 2010;395(4):815–833. doi: 10.1016/j.jmb.2009.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng W. Multiscale modeling of structural dynamics underlying force generation and product release in actomyosin complex. Proteins: Structure, Function, and Bioinformatics. 2010;78(3):638–660. doi: 10.1002/prot.22594. [DOI] [PubMed] [Google Scholar]
- 28.Tehver R, Thirumalai D. Rigor to Post-Rigor Transition in Myosin V: Link between the Dynamics and the Supporting Architecture. Structure. 2010;18(4):471–481. doi: 10.1016/j.str.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 29.Baumketner A. Interactions between relay helix and Src homology 1 domain helix (SH1) drive the converter domain rotation during the recovery stroke of myosin II Proteins. Structure, Function, and Bioinformatics. 2012 doi: 10.1002/prot.24051. at press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson P, McKenna W, Bristow M, Maisch B, Mautner B, OConnell J, Olsen E, Thiene G, Goodwin J, Gyarfas I, Martin I, Nordet P. Report of the 1995 World Health Organization International Society and Federation of Cardiology Task Force on the Definition and Classification of Cardiomyopathies. Circulation. 1996;93(5):841–842. doi: 10.1161/01.cir.93.5.841. [DOI] [PubMed] [Google Scholar]
- 31.Maron BJ. Hypertrophic Cardiomyopathy. JAMA: The Journal of the American Medical Association. 2002;287(10):1308–1320. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- 32.Van Driest SL, Ommen SR, Vasile VC, Will ML, Tajik AJ, Gersh BJ, Ackerman MJ. Comprehensive mutational analysis of myosin binding protein C in 389 unrelated patients with hypertrophic cardiomyopathy. Journal of the American College of Cardiology. 2004;43(5):165a–165a. doi: 10.1016/j.jacc.2004.04.039. [DOI] [PubMed] [Google Scholar]
- 33.Zhuravlev PI, Papoian GA. Protein functional landscapes, dynamics, allostery : a tortuous path towards a universal theoretical framework. Q Rev Biophys. 2010;43(3):295–332. doi: 10.1017/S0033583510000119. [DOI] [PubMed] [Google Scholar]
- 34.Elber R, Karplus M. A Method for Determining Reaction Paths in Large Molecules - Application to Myoglobin. Chemical Physics Letters. 1987;139(5):375–380. [Google Scholar]
- 35.Faradjian AK, Elber R. Computing time scales from reaction coordinates by milestoning. J Chem Phys. 2004;120(23):10880–10889. doi: 10.1063/1.1738640. [DOI] [PubMed] [Google Scholar]
- 36.Bolhuis PG, Chandler D, Dellago C, Geissler PL. Transition path sampling: Throwing ropes over rough mountain passes, in the dark. Annu Rev Phys Chem. 2002;53:291–318. doi: 10.1146/annurev.physchem.53.082301.113146. [DOI] [PubMed] [Google Scholar]
- 37.van Erp TS, Moroni D, Bolhuis PG. A novel path sampling method for the calculation of rate constants. J Chem Phys. 2003;118(17):7762–7774. [Google Scholar]
- 38.Henkelman G, Jonsson H. Improved tangent estimate in the nudged elastic band method for finding minimum energy paths and saddle points. J Chem Phys. 2000;113(22):9978–9985. [Google Scholar]
- 39.Maragliano L, Fischer A, Vanden-Eijnden E, Ciccotti G. String method in collective variables: Minimum free energy paths and isocommittor surfaces. J Chem Phys. 2006;125(2) doi: 10.1063/1.2212942. [DOI] [PubMed] [Google Scholar]
- 40.Chodera JD, Swope WC, Noe F, Prinz JH, Shirts MR, Pande VS. Dynamical reweighting: Improved estimates of dynamical properties from simulations at multiple temperatures. J Chem Phys. 2011;134(24) doi: 10.1063/1.3592152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muff S, Caflisch A. ETNA: Equilibrium Transitions Network and Arrhenius Equation for Extracting Folding Kinetics from REMD Simulations. J Phys Chem B. 2009;113(10):3218–3226. doi: 10.1021/jp807261h. [DOI] [PubMed] [Google Scholar]
- 42.Laio A, Gervasio FL. Metadynamics: a method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Rep Prog Phys. 2008;71(12) [Google Scholar]
- 43.Rayment I, Rypniewski WR, Schmidtbase K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. 3-Dimensional Structure of Myosin Subfragment-1 - a Molecular Motor. Science. 1993;261(5117):50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 44.Ruppel KM, Spudich JA. Structure-function analysis of the motor domain of myosin. Annual Review of Cell and Developmental Biology. 1996;12:543–573. doi: 10.1146/annurev.cellbio.12.1.543. [DOI] [PubMed] [Google Scholar]
- 45.Houdusse A, Kalbokis VN, Himmel D, Szent-Gyorgyi AG, Cohen C. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: A novel conformation of the myosin head. Cell. 1999;97(4):459–470. doi: 10.1016/s0092-8674(00)80756-4. [DOI] [PubMed] [Google Scholar]
- 46.Weiss A, Schiaffino S, Leinwand LA. Comparative sequence analysis of the complete human sarcomeric myosin heavy chain family: Implications for functional diversity. J Mol Biol. 1999;290(1):61–75. doi: 10.1006/jmbi.1999.2865. [DOI] [PubMed] [Google Scholar]
- 47.Ohki T, Mikhailenko SV, Morales MF, Onishi H, Mochizuki N. Transmission of force and displacement within the myosin molecule. Biochemistry. 2004;43(43):13707–13714. doi: 10.1021/bi048954f. [DOI] [PubMed] [Google Scholar]
- 48.Tsiavaliaris G, Fujita-Becker S, Batra R, Levitsky DI, Kull FJ, Geeves MA, Manstein DJ. Mutations in the relay loop region result in dominant-negative inhibition of myosin II function. Dictyostelium EMBO Rep. 2002;3:1099–1105. doi: 10.1093/embo-reports/kvf214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agafonov RV, Negrashov IV, Blakely SE, Titus Margaret A, Nesmelov YE, Thomas DD. Structural Basis for Uncoupling of Force Generation in the F506A Dictyostelium Myosin Revealed by Time-Resolved EPR and FRET. Biophys J. 2010;98(3) Supplement 1:143a–143a. [Google Scholar]
- 50.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 51.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 52.Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. Protein Data Bank - Computer-Based Archival File for Macromolecular Structures. J Mol Biol. 1977;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 53.Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, flexible, and free. Journal of Computational Chemistry. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 54.Miyamoto S, Kollman PA. SETTLE - an analytical version of the SHAKE and RATTLE algorithm for rigid water models. Journal of Computational Chemistry. 1992;13(8):952–962. [Google Scholar]
- 55.Hess B, Bekker H, Berendsen HJC, Fraaije J. LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry. 1997;18(12):1463–1472. [Google Scholar]
- 56.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 57.Nose S. Constant Temperature Molecular-Dynamics Methods. Progress of Theoretical Physics Supplement. 1991;(103):1–46. [Google Scholar]
- 58.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103(19):8577–8593. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.