Abstract
Whether genetic factors account for differences in early kidney disease among blacks in a young healthy population is not well known. We evaluated the association of self-reported race and genetic African ancestry with elevated creatinine (≥1.3 mg/dl for men, ≥1.1 mg/dl for women) among 3,113 black and white participants in the Coronary Artery Risk Development in Young Adults (CARDIA) study, ages 38–50 years. We estimated individual African ancestry using 42 ancestry informative markers. Blacks were more likely to have elevated creatinine than whites, and this effect was more pronounced in men: adjusted odds ratio (AOR) for black versus white men = 7.03, 4.15–11.91; AOR for women = 2.40, 1.15–5.02. Higher African ancestry was independently associated with elevated creatinine among black men (AOR = 1.53,1.08–2.16 per SD increase in African ancestry), but not women. A graded increase in odds of elevated creatinine by African Ancestry was observed among black men compared with white men: AOR = 4.27 (2.26–10.06) for black men with 40–70% African ancestry; AOR = 8.09 (4.19–15.61) for black men with 70–80% African ancestry; AOR = 9.05 (4.81–17.02) for black men with >80% African ancestry. Genetic factors common to African ancestry may be associated with increased risk of early kidney dysfunction in a young, healthy population, particularly among black men.
Key Words: Ancestry, Creatinine, Race, Kidney
Introduction
Kidney disease and the progression to end-stage renal disease (ESRD) affect black Americans disproportionately [1,2,3]. In addition, differences between blacks and whites in prevalent kidney disease, progression and incident ESRD have been shown to be more pronounced among men compared to women [2,3,4]. Studies of ge- netic susceptibility to kidney disease have been mostly performed in white populations [5,6]. However, recent studies suggest a susceptibility gene for ESRD among nondiabetic African Americans: MYH9 [7,8,9]. However, polymorphisms in the MYH9 gene were not associated with estimated glomerular filtration rate in a population of hypertensive blacks and whites not enriched for kidney disease [10]. Thus, the extent to which genetic factors more commonly found among persons of African descent account, at least in part, for differences in early kidney disease risk among young healthy persons is still not well understood.
Genetic admixture analysis is a method to study potential genetic contributions to racial differences in complex traits such as kidney disease by estimating the percentage of a person's genome that is of a given ancestral origin [11,12,13,14]. Genetic admixture analysis uses the knowledge that individuals sampled worldwide can be classified into clusters that correspond to continental lines (Africans, Europeans/West Asians, East Asians, Pacific Islanders, and Native Americans) based on previously identified genetic markers to quantify the proportion of an individual's genome that is of a given ancestral origin (i.e. European, African) [15]. The technique uses ancestry informative markers (AIMs), which are single nucleotide polymorphisms (SNPs) with large frequency differences between the ancestral populations.
Blacks in the United States are a heterogeneous group, variably admixed with African and European ancestry [16]. Due to this heterogeneity, genetic admixture analysis offers a unique opportunity for studying the role of genetic factors within a single, admixed population, independent of social factors and comorbidities. Specifically, an association between genetic ancestry and a disease phenotype within an admixed group such as African Americans may be an indicator of genetic factors underlying differential expression among racial groups. Therefore, we designed this study to evaluate whether genetic African ancestry is independently associated with elevated serum creatinine among healthy, young men and women in the Coronary Artery Risk Development in Young Adults (CARDIA) study.
Methods
Participants
Study design details of CARDIA have been published previously [17]. Briefly, a cohort of young adults, age 18–30 at the time of enrollment, and balanced by race (black and white) and sex were recruited between 1985–1986 from four participating sites: Birmingham, Ala., Chicago, Ill., Minneapolis, Minn., and Oakland, Calif., USA. Follow-up examinations occurred at years 2, 5, 7, 10, 15 and 20. All black participants with stored DNA from the year 10 examination were genotyped for ancestry informative markers (n = 1,623). Of these, 1,240 returned for the year 20 examination and had a measured serum creatinine and were included in this analysis. All white participants with a measured serum creatinine from year 20 were also included (n = 1,873), for a total sample size of 3,113. All study protocols were approved by the appropriate institutional review boards.
Selection of Ancestry-Informative Markers and Genotype Analysis
Details on selection of markers and genotyping have been published previously [18]. Briefly, 42 autosomal SNP markers were selected on the basis of having large allele frequency differences (δvalues) between African and European populations and on being distantly spaced throughout the genome. A subset of 23 markers was selected from candidate genes involved in inflammation and thrombosis as part of the NHLBI's Program for Genomic Applications. Genotyping was performed using the TaqMan, SNPlex, and Illumina methods under standard conditions as previously described [18].
Based on this ancestral population data, we can calculate the probability for each person at every marker that their ancestry at that locus is from any particular ancestral population, using the assumption that the markers are independent. The proportion of African and European ancestry for each individual was estimated using a maximum likelihood method with the program FRAPPE (under a 2-population model) [12] and by the cluster analytic method of the program STRUCTURE [19]. Information on ancestral allele frequencies for white Europeans and Africans was obtained from HapMap from Yorubans and Utah Caucasians (CEPH), and FRAPPE and STRUCTURE estimates had high degrees of correlation [18].
Outcome: Elevated Serum Creatinine
Serum creatinine was measured by nephelometry and calibrated to National Institute of Standards and Technology (NIST) standards as recommended by The National Kidney Disease Education Program (NKDEP) Laboratory Working Group across all sites [20]. We defined an elevated creatinine as a serum creatinine of ≥1.3 mg/dl for men and ≥1.1 mg/dl for women at the year 20 exam. We chose not to use equations to estimate glomerular filtration rate (GFR) such as the Modification of Diet in Renal Disease (MDRD) equation [21] for several reasons: (1) these equations have not been validated in young blacks without chronic kidney disease, (2) these equations include interaction term for race which is the primary predictor of interest, and (3) since these interaction terms have not been derived in young adults without kidney disease, they are likely to inflate GFR estimates for young blacks. This is supported by prior data from CARDIA using 24-hour urine collections, where Jacobs et al. [22] found that the difference in creatinine excretion between blacks and whites was only 10%, compared to the current race adjustment factor of 21% in the MDRD equation [21]. In addition, we chose creatinine cut points rather than linear creatinine because prior work in CARDIA has shown that using linear creatinine is highly influenced by outliers [3]. We chose the cut points 1.3 mg/dl for men and 1.1 mg/dl for women after examination of the creatinine distributions between blacks and whites and because these represent documented clinically relevant cut points for kidney disease [3,23,24].
Secondary Predictors
Sociodemographic factors were assessed by questionnaire at all study visits. Race was determined by participant self-report at baseline. We used socioeconomic data from year 20 rather than baseline as a more accurate reflection of a person's socioeconomic status at the time of kidney function assessment. As a marker of socioeconomic status, we used income (categorized as USD <25,000, 25,000–<50,000, ≥50,000) and level of highest educational attainment (categorized as less than high school, high school graduate, some college, completed college, more than college).
We used year 20 measurements for most covariates. Participants were asked not to exercise on the day of the examination, and to fast for 12 h. Systolic and diastolic blood pressure were defined as the average of the second and third of three measurements taken at 1-min intervals after a 5-min rest. A diagnosis of hypertension was recorded if the participant ever had a systolic blood pressure ≥140 or diastolic blood pressure ≥90 or use of antihypertensive medications. Height and weight were used to calculate the body mass index (weight (kg) divided by height squared (m2)). Diabetes was defined by fasting glucose >126 mg/dl or use of diabetes medications. Smoking was defined as self-reported current tobacco use. History of cardiovascular disease (CVD) was defined by an adjudicated CVD CARDIA endpoint (myocardial infarction, congestive heart failure (CHF), or stroke) before the year 20 examination. LDL cholesterol (LDL-c) was derived using the Friedewald formula from fasting samples.. Plasma C-reactive protein (CRP) levels were assayed using BNII immunonephelometry (BNII Nephelometer 100 Analyzer, Dade Behring) on stored plasma from year 15 as it was unavailable for the year 20 examination.
Statistical Analyses
We studied participant characteristics by self-reported race. In order to extend previously published CARDIA findings from earlier exam years [3], we first studied the distributions of creatinine between blacks and whites by plotting the percent of participants of each race at each creatinine cut point by sex at year 20. We then studied differences in prevalent elevated creatinine by self-reported race in the entire CARDIA cohort at the year 20 visit. Using multivariable logistic regression, we estimated the odds of having elevated creatinine for blacks compared to whites within each sex group. We first adjusted for age, income and education. A second nested model adjusted for the above variables plus diabetes, hypertension, systolic blood pressure, cardiovascular disease, smoking, fasting glucose, LDL cholesterol, CRP. We also adjusted for clinical site. We used a nested model approach in order to identify potential mediators of the associations.
An individual's percentage of African ancestry was coded as a continuous variable for self-identified blacks. We then constructed models including only self-identified black participants to test whether African ancestry (per standard deviation increase of African ancestry) was independently associated with elevated creatinine among blacks. We used sequential multivariable logistic regression models as above. We performed a sensitivity analysis excluding those with ancestry <40% to avoid possible misclassification, as studies have rarely reported self-identified blacks in this range of African ancestry [16,25].
We then extended our analyses to evaluate whether quantification of African ancestry was associated with a gradient of risk in comparing black and white differences in prevalent elevated creatinine. We divided the cohort into five groups: self-identified whites who based on prior literature are assumed to have <5% African ancestry [25], and four groups of self-identified blacks divided as <40, 40–70, 70–80 and >80% African ancestry. We used these cut points of ancestry because only 19 black men and 28 black women had <40% ancestry and the three remaining groups roughly correspond to remaining tertiles of ancestry. We used sequential multivariable logistic regression models to test the association of African ancestry (by each group) with elevated creatinine, compared to whites as the referent group. All analyses were performed using SAS (version 9.1). Two-tailed p values <0.05 were considered significant.
Results
Participant Characteristics
The 3,113 self-identified black and white participants in CARDIA included in this analysis are described in table 1. Black participants had higher body mass index, higher systolic blood pressure, higher fasting glucose, and were more likely to have diabetes, hypertension and cardiovascular disease. Blacks also had lower income and a higher percentage with less than high school education.
Table 1.
Participant characteristics for 3,113 black and white participants in CARDIA at year 20 by self-reported race
| Characteristic | Black (n = 1,240) | White (n = 1,873) | p value |
|---|---|---|---|
| Age | 44.6 ± 3.8 | 45.7 ± 3.4 | <0.001 |
| Female | 757 (61.1) | 989 (52.8) | <0.001 |
| Income | <0.001 | ||
| USD <25,000 | 279 (22.5) | 148 (7.9) | |
| USD 25,000–49,999 | 338 (27.3) | 262 (14.0) | |
| USD ≥50,000 | 598 (48.2) | 1,447 (77.3) | |
| Education | <0.001 | ||
| Less than high school graduate | 80 (6.4) | 40 (2.1) | |
| High school graduate | 336 (27.1) | 242 (12.9) | |
| Some college | 440 (35.5) | 375 (20.0) | |
| College | 246 (19.8) | 581 (31.0) | |
| More than college | 130 (10.5) | 628 (33.5) | |
| LDL cholesterol | 108.3 ± 32.6 | 110.1 ± 30.9 | 0.14 |
| BMI | 31.4 ± 7.8 | 27.9 ± 6.5 | <0.001 |
| Systolic blood pressure | 120.5 ± 15.9 | 113.1 ± 13.2 | <0.001 |
| Fasting glucose | 99.6 ± 28.7 | 96.4 ± 22.0 | <0.001 |
| C-reactive protein | 4.0 ± 5.5 | 2.5 ± 4.5 | <0.001 |
| CVD | 16 (1.3) | 13 (0.7) | 0.09 |
| Diabetes | 169 (13.6) | 160 (8.5) | <0.001 |
| Hypertension | 369 (29.8) | 234 (12.5) | <0.001 |
| Smoker | 489 (39.5) | 762 (40.7) | 0.50 |
Figures in parentheses are percentage. LDL = Low-density lipoprotein;
CVD = cardiovascular disease.
Self-Reported Race and Elevated Creatinine
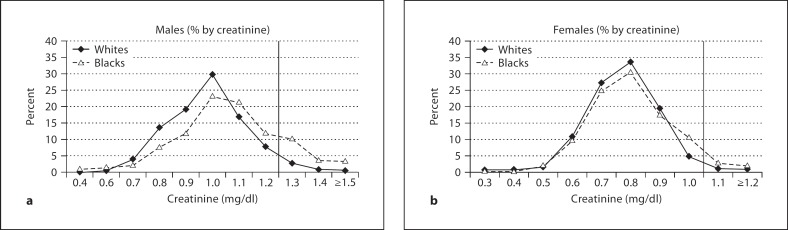
We plotted the proportion of participants in each creatinine group by race and sex and found differences at the tails of these distributions, with a higher proportion of black men with creatinine values ≥1.3 mg/dl compared to white men, and a higher proportion of black women with creatinine values ≥1.1 mg/dl compared to white women (fig. 1a, b). Based on examination of these distributions and our prespecified decision not to use the formulas, we chose these cut points for the analyses.
Fig. 1.
Distribution of serum creatinine (mg/dl) among blacks and whites in CARDIA by gender.
At year 20, there were significant race differences in the prevalence of elevated creatinine between blacks and whites. Compared to self-reported whites, black men had over 6-fold increased odds of elevated creatinine that was not attenuated by adjustment for sociodemographic characteristics or comorbidities. The magnitude of the association of race with elevated creatinine was lower among women in both unadjusted and adjusted analyses (table 2).
Table 2.
Association of self-reported black versus white race and elevated creatinine by sex
| Unadjusted | SES adjusteda | Adjustedb | |
|---|---|---|---|
| Men | 6.25 | 7.68 | 7.03 |
| (n = 1,367) | (4.01–9.76) | (4.71–12.53) | (4.15–11.91) |
| Women | 2.86 | 2.96 | 2.40 |
| (n = 1,746) | (1.59–5.12) | (1.56–5.62) | (1.15–5.02) |
Odds ratio (95% CI) for the association of self-reported race with elevated creatinine (reference white).
a Adjusted for age, income, education.
b Adjusted for age, income, education, smoking, systolic blood pressure, cardiovascular disease, diabetes, hypertension, BMI, fasting glucose, LDL cholesterol, C-reactive protein, and clinical site.
African Ancestry and Elevated Creatinine among Self-Identified Blacks
The distribution of African ancestry was similar for men and women. For men, the mean was 73% (SD 16%, median 76%) and it was 72% for women (SD 15%, median 74%). The standard error of ancestry estimates ranged between 3 and 13%. Among blacks, higher African ancestry (per SD) was independently associated with elevated creatinine among men in unadjusted models. Adjustment in sequential models for sociodemographic factors and comorbidities did not attenuate this effect in men. African ancestry was not associated elevated creatinine among self-identified black women (table 3).
Table 3.
Association of African ancestry and elevated creatinine by sex among self-identified blacks only in CARDIA
| Unadjusted | SES adjusteda | Adjustedb | |
|---|---|---|---|
| Men | 1.43 | 1.58 | 1.53 |
| (n = 483) | (1.09–1.90) | (1.16–2.15) | (1.08–2.16) |
| Women | 1.10 | 1.10 | 1.18 |
| (n = 757) | (0.77–1.59) | (0.75–1.61) | (0.76–1.83) |
Odds ratio (95% CI) for the association of African ancestry with elevated creatinine (per SD of African ancestry).
a Adjusted for age, income, education.
b Adjusted for age, income, education, smoking, systolic blood pressure, cardiovascular disease, diabetes, hypertension, BMI, fasting glucose, LDL cholesterol, C-reactive protein, and clinical site.
We conducted sensitivity analyses excluding participants with less than 40% African ancestry. This did not materially change our findings: among men, African ancestry (per SD) was associated with elevated creatinine in age and demographic adjusted models (AOR = 1.47, 95% CI 1.05–2.05 and AOR = 1.43, 95% CI 0.99–2.08 in fully adjusted models). There was no association of African ancestry and elevated creatinine among women (AOR = 0.89, 95% CI 0.67–1.19).
Self-Reported Race, African Ancestry and Elevated Creatinine among Blacks versus Whites
In order to understand the added value of genetically determined African ancestry to self-reported race alone, we divided our cohort into five groups (whites combined and self-identified blacks divided into four groups by African ancestry: <40, 40–70, 70–80 and >80%). We then studied the association of each African ancestry group and elevated creatinine compared to self-reported whites as the referent group among men and women. Among the men, there was a graded association of higher odds of elevated creatinine with increasing group of African ancestry. There were no cases of elevated creatinine among men with African ancestry <40%. Moreover, the odds of elevated creatinine were lower for those with ancestry <70% compared to 70–80 or >80% when each was compared to the white referent group. This effect was not seen among women (table 4).
Table 4.
Association of African ancestry groups versus whites and elevated creatinine
| Whites (combined) and blacks by African ancestry group | n | Unadjusted odds ratio (95% CI) | Adjusted1 odds ratio (95% CI) |
|---|---|---|---|
| Men | |||
| Self-reported | |||
| white (ref.) | 884 | 1.0 | 1.0 |
| African ancestry | |||
| <40% | 19 | * | * |
| 40–70% | 134 | 4.44 (2.36–8.36) | 4.27 (2.26–10.06) |
| 70–80% | 150 | 7.33 (4.21–12.74) | 8.09 (4.19–15.61) |
| >80% | 180 | 7.64 (4.52–12.91) | 9.05 (4.81–17.02) |
| Women | |||
| Self-reported | |||
| white (ref) | 989 | 1.0 | 1.0 |
| African ancestry | |||
| <40% | 28 | 2.12 (0.27–16.49) | 2.11 (0.25–18.11) |
| 40–70% | 110 | 3.04 (1.43–6.44) | 2.05 (0.79–5.32) |
| 70–80% | 252 | 2.12 (0.93–4.81) | 1.95 (0.73–5.23) |
| >80% | 239 | 3.56 (1.73–7.33) | 3.46 (1.41–8.50) |
* There were no black men with African ancestry <40% and elevated creatinine.
1 Adjusted for age, income, education, smoking, systolic blood pressure, cardiovascular disease, diabetes, hypertension, BMI, fasting glucose, LDL cholesterol, C-reactive protein, and clinical site.
Discussion
In a large and well-characterized cohort of young and middle-aged black and white adults, we found that self-reported black race was associated with higher odds of elevated creatinine compared to whites, particularly among men. When we used genetic admixture analysis, higher African ancestry was associated with higher odds of elevated creatinine among black men after adjustment for sociodemographic factors and comorbidities. Moreover, among men, across the two race groups, we found a graded increase in odds of elevated creatinine, with white men having the lowest risk, black men with lower African ancestry having intermediate risk, and black men with the highest African ancestry having the highest risk. Our results suggest that the use of genetically determined African ancestry may elucidate an important heterogeneity in the risk of kidney dysfunction among self-identified blacks that is not captured by self-reported race alone.
The association of African ancestry and elevated creatinine may be due to genetic factors, sociodemographic and environmental factors, or their interactions. Although familial aggregation of ESRD risk has been shown in both African American and white populations [26,27], a growing body of evidence has found African-specific polymorphisms associated with renal damage [28,29]. Most recently, two studies used admixture mapping to find an association of a gene thought to be involved in podocyte function (MYH9) with nondiabetic, proteinuric ESRD among African Americans [7,8]. However, this association was not seen with estimated glomerular filtration rate among blacks in a hypertensive population [10]. In the same study, there was a modest association with albuminuria, suggesting that the MYH9 polymorphisms may be more important for development of proteinuric disease. Our findings of an association of higher African ancestry with elevated creatinine in a young, healthy cohort are consistent with the possibility that there may be other genetic loci that may account for predisposition to kidney disease and reduced glomerular filtration rate due to a common genetic ancestry. Moreover, these positive findings suggest that early kidney disease (before ESRD) may be a complex trait amenable to genetic admixture mapping.
It is important to recognize that sociodemographic and environmental factors may play an important role. That is, our findings may also be confounded by social and environmental factors not measured in this study. In addition, there may be important gene-environment interactions. Higher African ancestry may be a marker for factors such as individual poverty [30], neighborhood poverty, access to care, diet, social cohesion, discrimination, stress, or other environmental exposures. Higher African ancestry is known to be associated with darker skin color [31], and darker skin may, in turn, be associated with differing social experiences that may affect disease risk. However, our findings were independent of risk factors such as hypertension and diabetes that are likely affected by similar environmental characteristics. Moreover, our findings are robust after adjustment for income and education.
The observed gender differences in the association of genetic ancestry and elevated creatinine is noteworthy. Gender differences in racial associations with kidney disease prevalence, progression, and ESRD incidence have been previously reported in the epidemiology literature where differences are more striking among men [1,2,3,4]. Moreover, gender interactions in the association of genetic traits and complex phenotypes, including kidney disease, have been previously documented [28,29,32,33,34]. Several mechanisms have been suggested to explain these gender differences, including gene-gene or gene-environment interactions. One possible mechanism is that sex hormones may affect the expression of a polymorphism. In one well-studied example, estrogen levels modulate the expression of variants in a Ca2+ and voltage-dependent K+ channel involved in determining vascular tone [35], and polymorphisms in this gene have been linked to disruption of vascular tone, hypertension and left ventricular hypertrophy [36,37]. Thus, it is possible that, either directly or indirectly, sex hormone levels may affect kidney disease risk.
Our findings show that the use of genetic African ancestry captures heterogeneity in kidney dysfunction risk among black men that is not captured by self-reported race alone. This is supported by prior reports of the large heterogeneity in African ancestry among self-identified blacks across the United States [38,39]. Since prior reports have shown that self-report cannot adequately determine percent African ancestry among Blacks born in the United States [39], there may be additional information regarding risk using genetically determined ancestry.
Our study is limited by the lack of a gold standard measure of kidney function. Although blacks have been shown to have higher serum creatinine in some studies, and this may be due to differences in muscle mass and/or tubular secretion, our cut points are likely to represent kidney dysfunction in this young population, as has been shown in prior studies [23,24]. Our study is also limited by its cross-sectional nature. We are only able to study differences in prevalence of elevated creatinine and not progression of kidney disease, where racial differences may be greater. Moreover, we are limited by using education and income as markers of socioeconomic status, which may not reflect a person's entire social environment [40]. Our study is also limited by the use of a limited number of markers to determine genetic ancestry. This may lead to error in the estimates, which may have attenuated the effects we describe [13], and may have been a particular concern in the women where power was limited due to the small number with elevated creatinine.
In summary, higher African ancestry is associated with increased risk of elevated creatinine, particularly among black men. African Ancestry determination in black men elucidated a heterogeneity of risk not captured by race self-report alone. The observed associations of ancestry with elevated creatinine may be due to genetic or nongenetic effects or their interactions. Future studies should focus on elucidating those factors that may explain the observed associations and sex differences. Moreover, our study is important as these findings suggest that kidney disease prior to the onset of ESRD may be a complex trait amenable to admixture mapping.
References
- 1.Hsu CY, Lin F, Vittinghoff E, et al. Racial differences in the progression from chronic renal insufficiency to end-stage renal disease in the United States. J Am Soc Nephrol. 2003;14:2902–2907. doi: 10.1097/01.asn.0000091586.46532.b4. [DOI] [PubMed] [Google Scholar]
- 2.System USRD. USRDS 2007 Annual Data Report. Bethesda, National Institutes of Health. 2007 [Google Scholar]
- 3.Stehman-Breen CO, Gillen D, Steffes M, et al. Racial differences in early-onset renal disease among young adults: the coronary artery risk development in young adults (CARDIA) study. J Am Soc Nephrol. 2003;14:2352–2357. doi: 10.1097/01.asn.0000083392.11042.14. [DOI] [PubMed] [Google Scholar]
- 4.Neugarten J, Acharya A, Silbiger SR. Effect of gender on the progression of nondiabetic renal disease: a meta-analysis. J Am Soc Nephrol. 2000;11:319–329. doi: 10.1681/ASN.V112319. [DOI] [PubMed] [Google Scholar]
- 5.Fox CS, Yang Q, Cupples LA, et al. Genomewide linkage analysis to serum creatinine, GFR, and creatinine clearance in a community-based population: the Framingham Heart Study. J Am Soc Nephrol. 2004;15:2457–2461. doi: 10.1097/01.ASN.0000135972.13396.6F. [DOI] [PubMed] [Google Scholar]
- 6.Kottgen A, Kao WH, Hwang SJ, et al. Genome-wide association study for renal traits in the Framingham Heart and Atherosclerosis Risk in Communities Studies. BMC Med Genet. 2008;9:49. doi: 10.1186/1471-2350-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kopp JB, Smith MW, Nelson GW, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40:1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kao WH, Klag MJ, Meoni LA, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freedman BI, Hicks PJ, Bostrom MA, et al. Polymorphisms in the non-muscle myosin heavy chain 9 gene (MYH9) are strongly associated with end-stage renal disease historically attributed to hypertension in African Americans. Kidney Int. 2009;75:736–745. doi: 10.1038/ki.2008.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freedman BI, Kopp JB, Winkler CA, et al. Polymorphisms in the nonmuscle myosin heavy chain 9 gene (MYH9) are associated with albuminuria in hypertensive African Americans: The HyperGEN Study. Am J Nephrol. 2009;29:626–632. doi: 10.1159/000194791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shriver MD, Kennedy GC, Parra EJ, et al. The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Hum Genomics. 2004;1:274–286. doi: 10.1186/1479-7364-1-4-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang H, Peng J, Wang P, et al. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 13.Rosenberg NA, Li LM, Ward R, et al. Informativeness of genetic markers for inference of ancestry. Am J Hum Genet. 2003;73:1402–1422. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakraborty R, Weiss KM. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc Natl Acad Sci USA. 1988;85:9119–9123. doi: 10.1073/pnas.85.23.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 16.Parra EJ, Kittles RA, Argyropoulos G, et al. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am J Phys Anthropol. 2001;114:18–29. doi: 10.1002/1096-8644(200101)114:1<18::AID-AJPA1002>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 17.Friedman GD, Cutter GR, Donahue RP, et al. CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol. 1988;41:1105–1116. doi: 10.1016/0895-4356(88)90080-7. [DOI] [PubMed] [Google Scholar]
- 18.Reiner AP, Carlson CS, Ziv E, et al. Genetic ancestry, population sub-structure, and cardiovascular disease-related traits among African-American participants in the CARDIA Study. Hum Genet. 2007;121:565–575. doi: 10.1007/s00439-007-0350-2. [DOI] [PubMed] [Google Scholar]
- 19.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myers GL, Miller WG, Coresh J, et al. Recommendations for improving serum creatinine measurement: a report from the Laboratory Working Group of the National Kidney Disease Education Program. Clin Chem. 2006;52:5–18. doi: 10.1373/clinchem.2005.0525144. [DOI] [PubMed] [Google Scholar]
- 21.Levey AS, Bosch JP, Lewis JB, et al. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med. 1999;130:461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs DR, Jr, Murtaugh MA, Steffes M, et al. Gender- and race-specific determination of albumin excretion rate using albumin-to-creatinine ratio in single, untimed urine specimens: the Coronary Artery Risk Development in Young Adults Study. Am J Epidemiol. 2002;155:1114–1119. doi: 10.1093/aje/155.12.1114. [DOI] [PubMed] [Google Scholar]
- 23.Rule AD, Bergstralh EJ, Melton LJ, 3rd, et al. Kidney stones and the risk for chronic kidney disease. Clin J Am Soc Nephrol. 2009;4:804–811. doi: 10.2215/CJN.05811108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nickolas TL, O'Rourke MJ, Yang J, et al. Sensitivity and specificity of a single emergency department measurement of urinary neutrophil gelatinase-associated lipocalin for diagnosing acute kidney injury. Ann Intern Med. 2008;148:810–819. doi: 10.7326/0003-4819-148-11-200806030-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang H, Jorgenson E, Gadde M, et al. Racial admixture and its impact on BMI and blood pressure in African and Mexican Americans. Hum Genet. 2006;119:624–633. doi: 10.1007/s00439-006-0175-4. [DOI] [PubMed] [Google Scholar]
- 26.Freedman BI, Volkova NV, Satko SG, et al. Population-based screening for family history of end-stage renal disease among incident dialysis patients. Am J Nephrol. 2005;25:529–535. doi: 10.1159/000088491. [DOI] [PubMed] [Google Scholar]
- 27.Lei HH, Perneger TV, Klag MJ, et al. Familial aggregation of renal disease in a population-based case-control study. J Am Soc Nephrol. 1998;9:1270–1276. doi: 10.1681/ASN.V971270. [DOI] [PubMed] [Google Scholar]
- 28.Keene KL, Mychaleckyj JC, Smith SG, et al. Comprehensive evaluation of the estrogen receptor alpha gene reveals further evidence for association with type 2 diabetes enriched for nephropathy in an African American population. Hum Genet. 2008;123:333–341. doi: 10.1007/s00439-008-0482-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gainer JV, Lipkowitz MS, Yu C, et al. Association of a CYP4A11 variant and blood pressure in black men. J Am Soc Nephrol. 2008 doi: 10.1681/ASN.2008010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peralta CA, Ziv E, Katz R, et al. African ancestry, socioeconomic status, and kidney function in elderly African Americans: a genetic admixture analysis. J Am Soc Nephrol. 2006 doi: 10.1681/ASN.2006050493. [DOI] [PubMed] [Google Scholar]
- 31.Shriver MD, Parra EJ, Dios S, et al. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet. 2003;112:387–399. doi: 10.1007/s00439-002-0896-y. [DOI] [PubMed] [Google Scholar]
- 32.Lieb W, Graf J, Gotz A, et al. Association of angiotensin-converting enzyme 2 (ACE2) gene polymorphisms with parameters of left ventricular hypertrophy in men: results of the MONICA Augsburg echocardiographic substudy. J Mol Med. 2006;84:88–96. doi: 10.1007/s00109-005-0718-5. [DOI] [PubMed] [Google Scholar]
- 33.Seibold MA, Wang B, Eng C, et al. An African-specific functional polymorphism in KCNMB1 shows sex-specific association with asthma severity. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao F, Zhang L, Wessel J, et al. Tyrosine hydroxylase, the rate-limiting enzyme in catecholamine biosynthesis: discovery of common human genetic variants governing transcription, autonomic activity, and blood pressure in vivo. Circulation. 2007;116:993–1006. doi: 10.1161/CIRCULATIONAHA.106.682302. [DOI] [PubMed] [Google Scholar]
- 35.Kundu P, Alioua A, Stefani E, et al. Regulation of mouse Slo gene expression: multiple promoters, transcription start sites, and genomic action of estrogen. J Biol Chem. 2007;282:27478–27492. doi: 10.1074/jbc.M704777200. [DOI] [PubMed] [Google Scholar]
- 36.Pluger S, Faulhaber J, Furstenau M, et al. Mice with disrupted BK channel beta1 subunit gene feature abnormal Ca(2+) spark/STOC coupling and elevated blood pressure. Circ Res. 2000;87:E53–E60. doi: 10.1161/01.res.87.11.e53. [DOI] [PubMed] [Google Scholar]
- 37.Brenner R, Perez GJ, Bonev AD, et al. Vasoregulation by the beta1 subunit of the calcium-activated potassium channel. Nature. 2000;407:870–876. doi: 10.1038/35038011. [DOI] [PubMed] [Google Scholar]
- 38.Ziv E, Burchard EG. Human population structure and genetic association studies. Pharmacogenomics. 2003;4:431–441. doi: 10.1517/phgs.4.4.431.22758. [DOI] [PubMed] [Google Scholar]
- 39.Yaeger R, Avila-Bront A, Abdul K, et al. Comparing genetic ancestry and self-described race in African Americans born in the United States and in Africa. Cancer Epidemiol Biomarkers Prev. 2008;17:1329–1338. doi: 10.1158/1055-9965.EPI-07-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merkin SS, Roux AV, Coresh J, et al. Individual and neighborhood socioeconomic status and progressive chronic kidney disease in an elderly population: the Cardiovascular Health Study. Soc Sci Med. 2007;65:809–821. doi: 10.1016/j.socscimed.2007.04.011. [DOI] [PubMed] [Google Scholar]