Abstract
Neuroblastoma (NB) is the most common extracranial pediatric solid tumor with an undifferentiated status and generally poor prognosis, but the basis for these characteristics remains unknown. In this study, we show that upregulation of the Polycomb complex histone methytransferase EZH2, which limits differentiation in many tissues, is critical to maintain the undifferentiated state and poor prognostic status of NB by epigenetic repression of multiple tumor suppressor genes. We identified this role for EZH2 by examining the regulation of CASZ1, a recently identified NB tumor suppressor gene whose ectopic restoration inhibits NB cell growth and induces differentiation. Reducing EZH2 expression by RNAi-mediated knockdown or pharmacological inhibiton with 3-deazaneplanocin A (DZNep) increased CASZ1 expression, inhibited NB cell growth and induced neurite extension. Similarly, EZH2−/− mouse embryonic fibroblasts (MEFs) displayed 3-fold higher levels of CASZ1 mRNA compared to EZH2+/+ MEFs. In cells with increased expression of CASZ1, treatment with HDAC inhibitors decreased expression of EZH2 and the Polycomb complex component SUZ12. Under steady-state conditions H3K27me3 and PRC2 components bound to the CASZ1 gene were enriched, but this enrichment was decreased after HDAC inhibitor treatment. We determined that the tumor suppressors CLU, NGFR and RUNX3 were also directly repressed by EZH2 like CASZ1 in NB cells. Together, our findings establish that aberrant upregulation of EZH2 in NB cells silences several tumor suppressors, which contribute to the genesis and maintenance of the undifferentiated phenotype of NB tumors.
Keywords: CASZ1, neuroblastoma, EZH2, NGFR, CLU, RUNX3
Introduction
Neuroblastoma (NB) is the most common extracranial solid tumor in childhood, and accounts for 15% of all pediatric oncology deaths (1–3). Biological and genetic features have been used to stratify patients for risk and more intensive treatment. Biological features such as tumor histopathology or the Shimada classification system have revealed that undifferentiated or stroma-poor tumors have a worse prognosis than ganglioneuromas, which are mature, stroma rich tumors containing more differentiated ganglion cells. Patients with ganglioneuroblastomas, tumors that contain a spectrum of elements seen in both undifferentiated as well as well-differentiated tumors, have variable prognoses. Recent microarray analyses indicate the transcriptome of tumors from patients with poor prognoses is enriched in cell cycle related genes, while that of tumors from patients who have favorable prognoses tumors is enriched in differentiation-associated genes (4). The mechanisms by which the genetic alterations associated with neuroblastoma contribute to the undifferentiated state of NB are still unknown.
A number of genetic alterations are associated with NB and are thought to affect the differentiation potential of the pluripotent sympathetic neuroblasts from which neuroblastomas are thought to arise. Patients whose neuroblastoma tumors contain genomic MYCN amplification and allelic deletion of the short arm of chromosome 1 (Chr1p deletion) have the highest risk and worst prognoses (reviewed in (2)). Chr1p deletion is highly correlated with MYCN amplification (5). Loss of heterozygosity of 1p36 (1p36LOH) is found in 20%–40% cases and is independently associated with progression-free survival (6). The consistent loss of 1p36 brought attention to this region as the site of a putative NB tumor suppressor gene (2, 3). Genes in the 1p36 region such as CHD5 (7, 8), miR-34a (9), TP73 (10) and most recently CASZ1, a neuronal differentiation gene, have been shown to possess tumor suppressor activity (11).
CASZ1 is the human homologue of the drosophila zinc finger transcriptional factor Castor. Drosophila castor (cas) is specifically expressed in a subset of central nervous system (CNS) neuroblasts, and regulates late stage neurogenesis and neural fate determination (12). Liu et al report that the level of human CASZ1 expression increases upon the induction of differentiation in NB cells and mesenchymal cells (13). The restoration of CASZ1 expression in NB cells inhibits cell proliferation in vitro and decreases tumor growth in vivo (11). In an evaluation of primary NB tumors, the expression of CASZ1 is significantly decreased in aggressive NB compared with the favorable tumors (14, 15). The finding that no tumor-associated nucleotide mutation is found in the coding sequence of CASZ1 (15, 16) suggests that mechanisms such as epigenetic silencing may be involved in the decreased expression of CASZ1 in tumors of patients with unfavorable prognoses.
Major mechanisms of epigenetic silencing of gene expression include regulation of DNA methylation and the posttranslational modifications of histones. DNA methylation on the 1p36 region has been shown to mediate silencing of CHD5 in NB tumors cells (8). However, no consistent CpG methylation site in the 5′ proximal region or first intron of CASZ1 has been identified in either NB cell lines or primary tumors that differs from normal tissues (11, 15, 16). Thus it is unlikely that DNA methylation accounts for low CASZ1 expression in NB cells. The findings that the histone deacetylase inhibitors, depsipeptide (11) and trichostatin A (15) induce CASZ1 expression in NB cells, suggest that suppressive histone modifications inhibit CASZ1 gene expression.
Histone acetylation tightly associates with gene activation and the trimethylation of histone 3 on lysine 27 (H3K27me3) is a well-known histone mark associated with gene silencing. H3K27me3 is mediated by the methyltransferase EZH2, which is the enzymatically active component of the Polycomb Repressor Complex 2 (PRC2) (17). PRC2 contains three core subunits, enhancer of zeste 2 (EZH2), embryonic ectoderm development (EED) and suppressor of zeste 12 homolog (SUZ12) (reviewed in (18–20)). EZH2 is essential for stem cell identity and pluripotency (reviewed in (18–20)). PRC2 regulates a large set of developmental genes in embryonic stem cells, such as the HOX gene clusters, SOX, PAX and WNT gene families. In retinoic acid (RA) induced neural stem cell differentiation, EZH2 expression is decreased in differentiated neural cells, consistent with decreased binding of EZH2 to RA-inducible target genes (reviewed in (18). While PRC2 is released from genes (HOXA 1-5, ZIC1, CKM) expressed during the differentiation, it is also recruited to the certain genes (HOXA9-13, Neroug2, Olig2) that may be suppressed in specific cell lineages (reviewed in (19)). This dynamic recruitment and displacement of PRC2 together with the tissue specific transcriptional factors determines cell lineage (reviewed in (19)).
Over-expression of EZH2 is found in a number of different cancers and is associated with the progression of prostate (21, 22), breast (23), Ewing’s sarcoma (24) and glioblastoma (25). The oncogenic function of EZH2 is partially attributable to the ability of the PRC2 to localize to a number of well-known tumor suppressor genes, such as INK4A/B (26, 27), E-cadherin (28) and RUNX3 (29).
Until now, the function of the PRC2 and EZH2 has not been evaluated in NB. In this study, we identify that NB patients with a poor prognoses have increased levels of EZH2 mRNA. Moreover we find that silencing of EZH2 leads to decreased H3K27me3 and increased expression of the NB tumor suppressor CASZ1, which is consistent with a model in which one allele of the CASZ1 may be lost by 1p LOH while remaining allele(s) are subject to epigenetic silencing by EZH2 mediated H3K27me3. Furthermore, we find that EZH2 silences a number of tumor suppressors, which control differentiation in NB such as CLU, RUNX3 and NGFR in NB cells. Finally we find that the genetic or pharmacologic inhibition of EZH2 inhibits NB cell growth and induces differentiation.
Material and Methods
Cell culture, transduction and Reagents
Neuroblastoma cell lines used in this study are listed in Supplemental Table 1. NB cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 2mM glutamine, and 100μg/ml of penicillin/streptomycin at 37°C in 5% CO2. The immortalized EZH2 −/− MEF cells and control MEF cells were constructed as described (30), and cultured in DMEM containing 10% fetal bovine serum, 2mM glutamine, and 100μg/ml of penicillin/streptomycin at 37°C in 5% CO2. For experiments, depsipeptide, 3-deazaneplanocin A (DZNep), and Z-VAD-FMK (R&D Systems Inc, Minneapolis, MN) were diluted to the indicated concentrations in cell culture media. SMS-KCNR cells were plated onto 100-mm dishes at a density of 5 million cells per dish. After 24 hrs, cells were infected by EZH2 or non-target shRNA lentiviral particles (Sigma-Aldrich Corp, Saint Louis, MO) following the manufacturer’s protocol. The cell viability was checked using the Cell-Titer Blue assay Kit or MTS assay (Promega, Madison, WI). Caspase 3/7 activities were evaluated using the Caspase-Glo 3/7 assay kit (Promega, Madison, WI).
RNA preparation and quantitative Real-time PCR
Cells were detached mechanically, washed with ice-cold PBS, and processed for RNA extraction using the RNeasy kit (Qiagen Inc., Chatsworth, CA). qRT-PCR was performed on ABI Prism 7000 (Applied Biosystems, Carlsbad, CA) to check the EZH2, β2MG, RUNX3, CLU, NGFR and CASZ1 expression using Maxima® Probe/ROX qPCR Master Mix or Maxima® SYBR Green/ROX qPCR Master Mix) (Fermentas Inc, Glen Burnie, MD). The samples were normalized to the levels of β-microglobulin (β2MG). The primer sets are listed in the Supplemental Table 2.
Protein assays
Cells were cultured and harvested as above, and nuclear protein extractions were prepared with NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific Inc., Rockford, IL) according to the manufacturer’s protocol. Immunologic detection of protein expression utilized the following antibodies: anti-EZH2 (BD, Franklin Lakes, NJ), anti-SUZ12, anti-EED (Abcam Inc, Cambridge, MA), anti-KDM6A (31), anti-H3K27me3, ant-H3K27Ac (Millipore Corp., Billerica, MA), and anti-H3 C-terminal (Active motif, Carlsbad, CA). Immunoreactivity was determined using the enhanced chemiluminescence method (Thermo Fisher Scientific Inc., Rockford, IL).
Chromatin immunoprecipitation assay
SMS-KCNR, SH-SY5Y and NGP cells were plated into 15cm dishes at a density of 5×106 cells per dish and treated with depsipeptide (2ng/ml). After 24 hours treatment, ChIP assays were carried out as described (30). For each ChIP assay, the following antibodies (2ug) were used; EZH2 (Active motif, Carlsbad, CA), SUZ12, EED (Abcam Inc, Cambridge, MA), H3K27me3, H3K27ac, RNA polymerase II (Millipore Corp., Billerica, MA) or IgG control (Santa Cruz Biotechnology, Santa Cruz, CA). ChIP enriched DNA and input DNA were subjected to qRT-PCR analysis with Maxima® SYBR Green/ROX qPCR Master Mix (Fermentas Inc, Glen Burnie, MD). Enrichment by ChIP assay on the specific genomic regions was assessed relative to the input DNA. The primer sets are listed in the Supplemental Table 3.
Xenograft model
SMS-KCNR cells were resuspended in Hanks balanced salt solution and Matrigel (Trevigen, Gaitherburg, MD) and 100ul cell suspension containing 2×106 cells was placed in the subcutaneous tissue of the right flanks of 4–5 week old nude mice (Taconic, Germantown, NY). Treatment was initiated when tumors reached approximately 100mm3. Cohorts of mice (7 mice/group) received DZNep (2.5mg/kg) or vehicle control (normal saline) twice daily, 3 days per week. The dimensions (length (L) and width (W)) of the tumors were assessed three times a week and the volume was calculated as (L×W2)/2. Student’s t test was used to compare the tumor volume between the DZNep and non-treatment groups and p values less than 0.05 were considered significant.
Results
EZH2 expression is associated with NB outcome
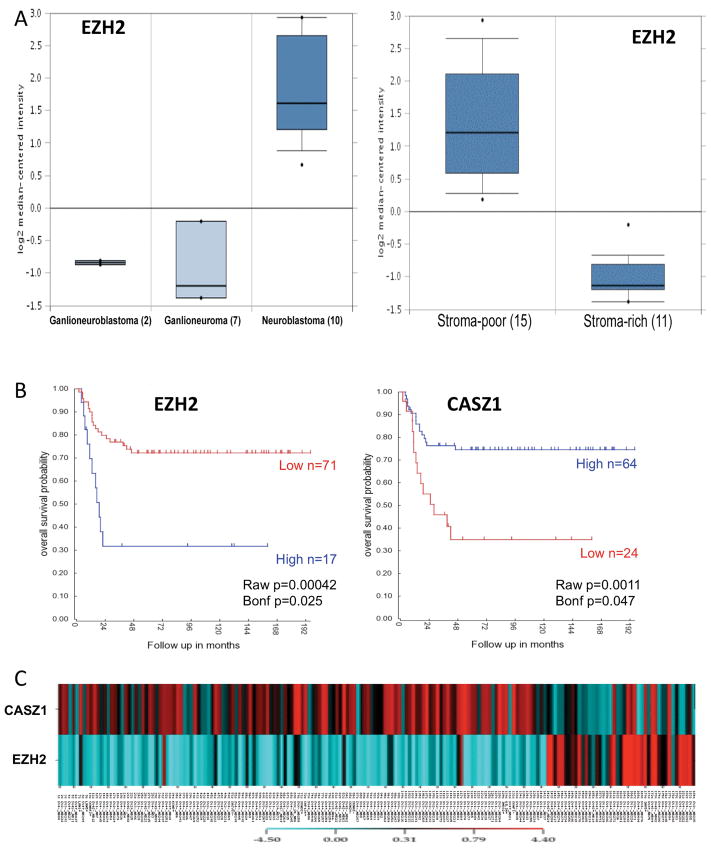
Since EZH2 is deregulated in a wide variety of cancers (21–23), we first analyzed the expression of EZH2 in NB patient tumor samples. According to the histopathological features, NB can be divided into Neuroblastoma (undifferentiated, schwannian stroma-poor, high risk), Gangioneuroblastoma and Ganglioneuroma (differentiated ganglion cells, schwannian stroma-rich, low-risk). Using the Oncomine microarray database (www.oncomine.com), we found that the expression level of EZH2 was higher in Neuroblastoma compared to Ganglioneuroblastoma and Ganglioneuroma (Fig. 1A, left panel) (32). Consistently EZH2 is expressed at a higher level in stroma-poor tumors compared to stroma-rich tumors (Fig. 1A, right panel). Patients, whose neuroblastoma tumors are stroma-poor or have an undifferentiated histopathology, have a worse prognosis (reviewed in (1, 3)). This suggests that EZH2 expression may be associated with NB prognosis. The Kaplan-Meier plots of overall survival, downloaded from the R2 microarray analysis and visualization platform (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi?&species=hs), demonstrated that high expression of EZH2 was associated with poor prognosis (p=0.00042 where the cut-off value was chosen by smallest p value), while high expression of CASZ1 was associated with good prognosis (p=0.0011, where the cut-off value was chosen by smallest p value) (Fig. 1B). Using another dataset (33) containing 168 clinical NB tumor samples and 13 NB cell lines we extracted the median-centered log-2 values for expression of CASZ1 and EZH2 which were clustered and color-coded using CIMminer software (Fig. 1C). The samples with high expression of EZH2 have low expression of CASZ1, while low expression of EZH2 was more frequently associated with high expression of CASZ1. The correlation between EZH2 and CASZ1 expression was statistically significant (ρ=−0.29, p=2.55×10−5) using the Spearman ranked correlation analysis. The findings that high levels of EZH2 expression and low levels of CASZ1 expression occur in the tumors of poor prognosis NB patients suggests that PRC2 components repress CASZ1 expression in NB cells.
Fig. 1. The level of EZH2 expression predicts the clinical outcome of NB patients.
A. EZH2 is highly expressed in more aggressive NB tumors comparing with the differentiated neuroblastic tumors, ganglioneuroblastoma and ganglioneuroma (Left panel, p=4.12×10−8), and high EZH2 expression is associated with stoma poor tumors (Right panel). The graph is downloaded from Oncomine microarray database.
B. Kaplan-Meier survival plots were downloaded from R2 microarray analysis and visualization platform. Patients with higher EZH2 and CASZ1 expression are highlighted in blue, while patients with lower EZH2 or CASZ1 expression are highlighted in red. (EZH2, p= 0.00042; CASZ1, p=0.0011).
C. EZH2 and CASZ1 expression is reverse correlated in NB clinical tumors and NB cell lines. The microarray expression data were downloaded from Oncogenomics Section Data center (http://pob.abcc.ncifcrf.gov/cgi-bin/JK). Log2 mean value is as the cut-off “0”, lower expression comparing with the mean value is negative number and colored blue, while higher is positive and colored red. The color codon is showed in color bar.
CASZ1 transcription is repressed by EZH2
To evaluate whether CASZ1 is a target of EZH2, lentiviral shRNAs were used to knock down EZH2 expression in SMS-KCNR cells. The expression of EZH2 decreased 3-fold in EZH2-shRNA transfected SMS-KCNR cells, while the mRNA expression of CASZ1 increased 2-fold (Fig. 2A). A pharmacologic inhibitor of EZH2, 3-Deazaneplanosin A (DZNep), which depletes EZH2 protein expression and is reported to relieve EZH2-mediated gene suppression (34), was used to further to determine whether CASZ1 is a target of EZH2. qRT-PCR showed that the mRNA expression of CASZ1 increased 2-fold after the DZNep treatment consistent with decreased EZH2 protein expression and a decrease in the global level of H3K27me3 (Fig. 2B). We also examined CASZ1 expression in immortalized EZH2−/− MEF cells (30). Expression of CASZ1 was increased 3-fold in EZH2−/− cells compared to EZH2+/+ MEF cells (Fig. 2C). These data indicate CASZ1 may be a target of EZH2.
Fig. 2. Genetic and pharmacologic inhibition of EZH2 expression leads to increase in CASZ1 mRNA expression.
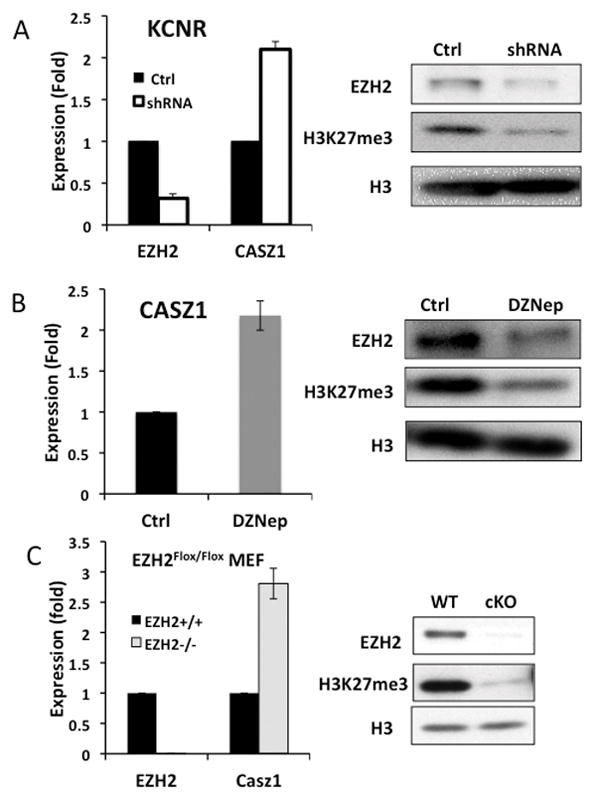
A. qRT-PCR measures the relative mRNA expression of EZH2 and CASZ1 in EZH2-shRNA-transfected KCNR (shRNA) comparing with non-target shRNA-transfected cells (Ctrl). The EZH2 protein expression and H3K27me3 status were detected by western blot. H3 served as loading control.
B. SMS-KCNR cells were treated with 0.5uM DZNep for 48 hours. The mRNA expression of CASZ1 was measured using qRT-PCR, and western blot examined protein expression as indicated.
C Relative mRNA expression of EZH2 and CASZ1 were measured by qRT-PCR in EZH2 knock out (EZH2−/−) and control MEF (EZH2+/+) cell lines, and western blot showed the proteins expression as indicated.
CASZ1 is a direct target of PRC2 complex
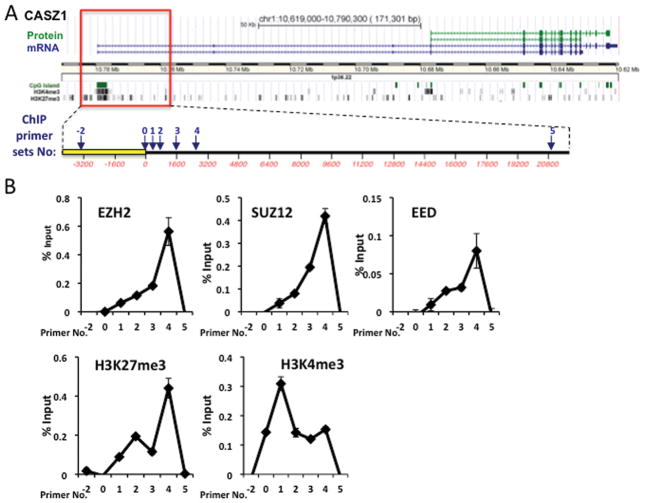
To determine whether CASZ1 is directly regulated by EZH2, we first scanned the CASZ1 gene using the UCSC genome browser, which showed that the CASZ1 transcriptional start site (TSS) region is enriched with H3K4me3 and H3K27me3 in embryonic stem cells (Fig. 3A). The enrichment of the bivalent mark of H3K4me3 and H3K27me3 is commonly found on silenced genes critical for development (35). Based on the location of H3K27me3 enrichment on the CASZ1 gene in ES cells, a series of PCR primer sets were designed to probe the PRC2 complex binding sites on the region of CASZ1 from 4,000 base pairs upstream to 20,000 base pairs downstream of its TSS (Fig. 3A). Under steady-state conditions, ChIP assays indicated that the three subunits of PRC2 all bound and were enriched around the CASZ1 TSS in SMS-KCNR cells (Fig. 3B). Moreover in the same region there was enrichment of H3K27me3 and H3K4me3, which is consistent with silencing or low expression of CASZ1 in the SMS-KCNR cells. The ChIP data reveals CASZ1 is subject to silencing chromatin modifications and is an EZH2 target in NB cells.
Fig. 3. PRC2 complex binds to the CASZ1 promoter.
A. Schematic representation of CASZ1 locus, transcripts, coded proteins, CpG islands and histone modifications, which was downloaded and modified from UCSC Genome Browser. The box frames the genomic section of CASZ1 expanded below. Numbered arrows indicate the genomic regions analyzed for PRC2 binding and histone modifications using ChIP assays.
B. ChIP-PCR assays revealed the binding of PRC2 components and the indicated histone marks, at CASZ1 locus in SMS-KCNR cells under steady-state conditions. The enrichment of examined histone modifications and the PRC2 components in the indicated regions were examined by qRT-PCR and plotted relative to input DNA.
HDAC inhibitor induces CASZ1 and represses PRC2
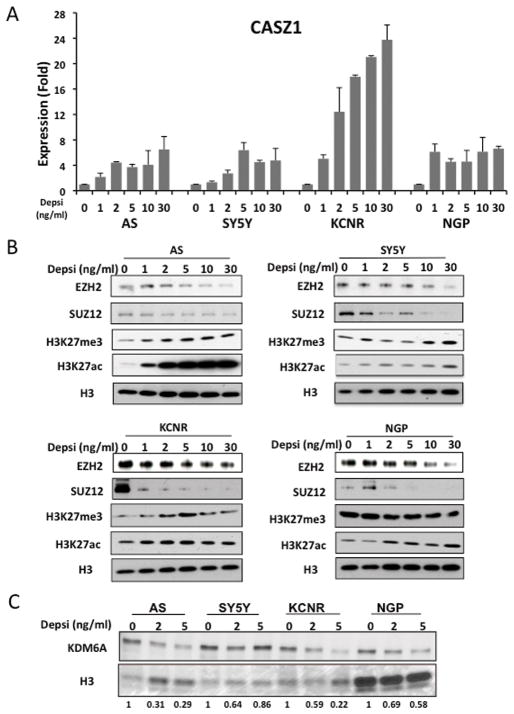
Since previous studies indicated that HDAC inhibitors could restore the expression of EZH2-repressed genes (21), we treated four different NB cell lines (Supplemental Table 1) with varying concentrations of depsipeptide, a Type 1 HDAC inhibitor (36). The four different NB cell lines contain characteristic genetic alterations found in NBs (Supplemental Table 1). After 24 hours treatment, CASZ1 mRNA expression increased 2–20 fold in all cell lines evaluated (Fig. 4A). The steady-state expression levels of PRC2 components, EZH2 and SUZ12 decreased in depsipeptide treated NB cells. While the global level of H3K27ac increased after depsipeptide treatment, the global level of H3K27me3 was unchanged or even increased in SK-N-AS and SH-SY5Y cells (Fig. 4B). We evaluated the H3K27me3 demethylases, KDM6A and KDM6B, in the depsipeptide treated cells and found that the steady-state protein expression of KDM6A decreased after depsipeptide treated NB cells (Fig. 4C). We failed to detect significant levels of KDM6B in NB cells (data not shown). This suggests that a compensating decrease in a H3K27 demethylase maintains the global levels of H3K27me3.
Fig. 4. HDACi depsipeptide induces CASZ1 and represses PRC2 complex.
A. NB cells were treated with different concentrations of depsipeptide for 24 hours. The relative CASZ1 expression is determined by qRT-PCR.
B. Western blot showed the protein expression of EZH2, SUZ12 and the status of global H3K27me3 and H3K27ac after treatment with depsipeptide (24hrs).
C. Western blot showed the protein expression of KDM6A after treatment with depsipeptide (24hrs). Desitometric analysis was performed using Image J. The relative expression of KDM6A (normalized to Ponceau S staining of Histone 3) in depsipeptide treated cells is normalized to the untreated control, normalized KDM6A values and expressed as relative density units (RDU).
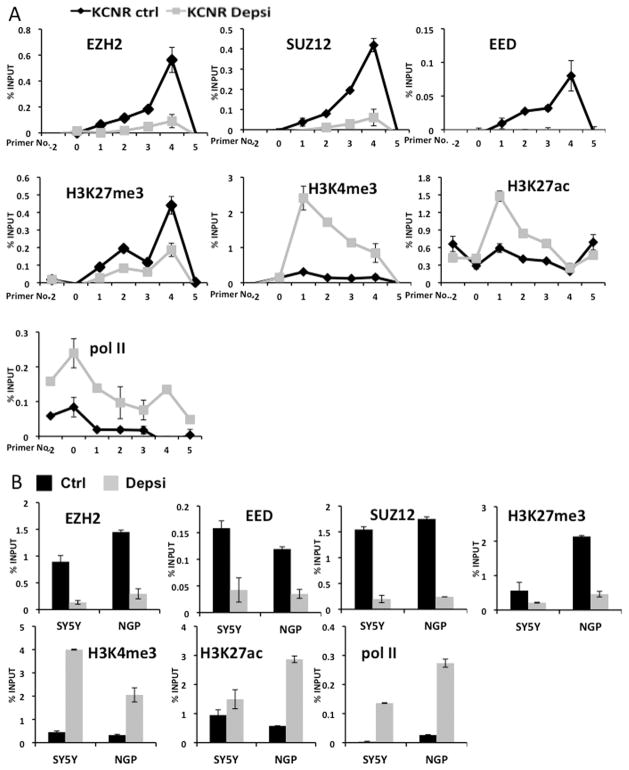
We used ChIP assays to examine the binding of PRC2 complex, RNA polymerase II and the status of H3 modifications on the CASZ1 gene after depsipeptide treatment. Following depsipeptide treatment, the binding of PRC2 complex to the CASZ1 TSS decreased, and the binding of RNA polymerase II increased (Fig. 5A). This is consistent with the increased CASZ1 expression after depsipeptide treatment (Fig. 4A). Consistent with the depsipeptide induced decrease in PRC2 binding there was also a decrease of H3K27me3 and an enrichment of H3K4me3 on the CASZ1 TSS (Fig. 5A). We further probed the binding of PRC2 complex, RNA polymerase II and the status of H3K27me3, H3K27ac and H3K4me3 in other NB cell lines (SH-SY5Y and NGP) following depsipeptide treatment (Fig. 5B). All core PRC2 components binding to CASZ1 decreased along with decreased enrichment of H3K27me3 after depsipeptide treatment. Consistent with the decreased occupancy of PRC2 components and H3K27me3, there was increased H3K4me3 and RNA polymerase II binding to the CASZ1 TSS. These data demonstrate that EZH2 directly represses CASZ1 in NB cells.
Fig. 5. ChIP analysis indicates that the binding of PRC2 complex is attenuated by HDACi depsipeptide treatment.
A. SMS-KCNR cells were treated with 2ng/ml depsipeptide for 24 hours. ChIP assays reveal the relative enrichment of indicated histone marks, RNA polymerase II (pol II) and the binding of PRC2 components at the CASZ1 locus in depsipeptide treated (KCNR Depsi, grey line) or steady-state or control KCNR cells (KCNR Ctrl, black line).
B. SH-SY5Y and NGP cells were treated with 2ng/ml depsipeptide for 24 hours (control, black; depsipeptide treated, grey). ChIP assays indicated the H3K27me3 status, PRC2 complex binding (primer No. 4) and H3K4me3, H3K27ac status and RNA polymerase II (pol II) binding (primer No. 1) on the CASZ1 promoter region.
EZH2 also represses other tumor suppressors in NB
We further explored whether other known tumor suppressors in NB were also repressed by EZH2. The expression of reported tumor suppressors, CLU (37), NGFR (reviewed in (3)), RUNX3 (29), CHD5 (7, 8), RASSF1A (38) and TP73 (reviewed in (10)) were examined in the shRNA-transfected SMS-KCNR cells. Decreasing EZH2 expression increased CLU, NGFR and RUNX3 expression (Fig. 6A), while the levels of CHD5, RASSF1A and TP73 expression did not change (data not shown). The expression of CLU, RUNX3 and NGFR were all increased after depsipeptide treatment (Fig. 6B). ChIP assays demonstrate the EZH2 binding and H3K27me3 enrichment on the promoter regions of these three genes were decreased, consistent with the respective increase in their expression (Fig. 6C). Our data indicate that EZH2 mediated repression contributes to the reduced expression of several genes in NB that have tumor suppressor activity or whose elevated expression is associated with a good prognosis.
Fig. 6. EZH2 also directly represses expression of NB tumor suppressor genes.
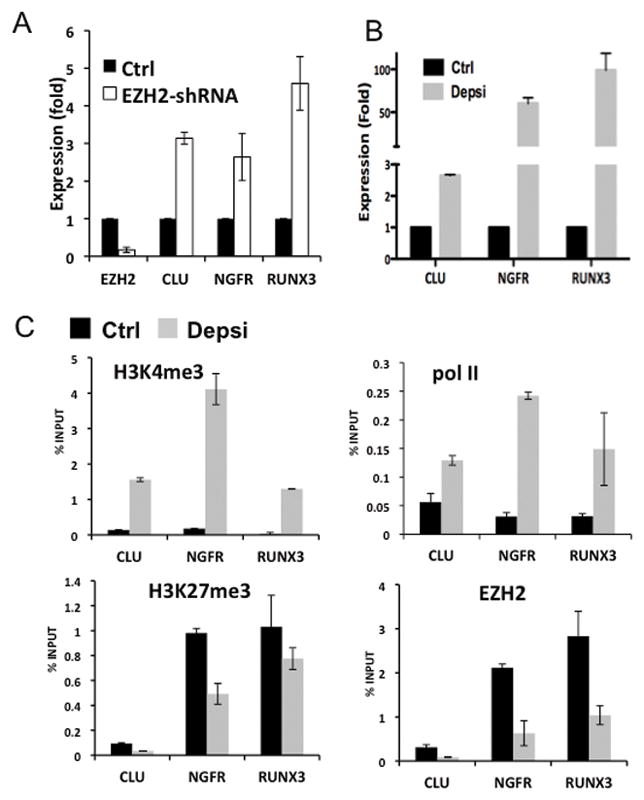
A. qRT-PCR measured the mRNA relative expression of EZH2, CLU, NGFR and RUNX3 in EZH2-shRNA-transfected KCNR (EZH2-shRNA) compared to non-target shRNA-transfected cells (Ctrl).
B. The relative CLU, NGFR and RUNX3 expression levels after treatment of NB cells with depsipeptide or control solvent were determined by qRT-PCR.
C. ChIP assays revealed the indicated histone marks, the binding of RNA polymerase II (pol II) and EZH2 at CLU, NGFR and RUNX3 promoters in depsipeptide treated (Depsi, grey) or non-treated KCNR cells (Ctrl, black).
Silencing of EZH2 decreases NB cell growth and induces neurite extensions
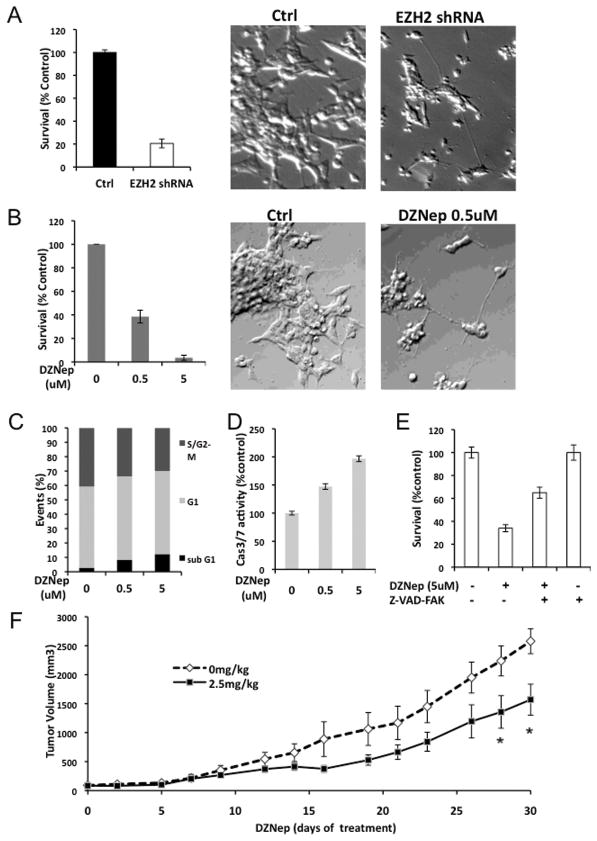
To investigate the biological function of EZH2 in NB, lentiviral shRNA were used to decrease EZH2 expression in SMS-KCNR cells. The growth of cells infected with the EZH2-shRNA was decreased to 20% after 3 days compared with control shRNA infected cells (Fig. 7A). Congruously NB cells displayed a concentration-dependent decrease in cell survival after 4-day DZNep treatment (Fig. 7B). There was also an increase in cells with neurite-like processes in the EZH2 shRNA infected cells (Fig. 7A) and similar morphologic changes were observed in DZNep treated cells (Fig. 7B).
Fig. 7. Decrease of EZH2 affects the cell growth and induces the neurite growth.
A. Cell survival in KCNR cells after 3-day infection with EZH2 or non-target shRNA lentivirus was assessed using a Cell-Titer Blue assay (left panel). The percentage of surviving cell was normalized by the absorbance value of the non-target shRNA infected cells (Control). Representative images (×200) of the non-target shRNA infected cells (Ctrl, middle panel), EZH2 shRNA infected cells (EZH2 shRNA, right panel).
B. KCNR cells were treated with different concentration of DZNep for 96 hours. MTS assay was used to detected cell survival (left panel). The percentage of surviving cells was normalized by the absorbance value of the non-treated cells. Representative images (×200) of the non-treated cells (Ctrl, middle panel) and 0.5uM DZNep treated cells (DZNep 0.5uM, right panel).
C. KCNR cells were treated with different concentration of DZNep for 96 hours. The cells were stained with propidium iodide and analyzed by flow cytometry. The data showed percentage of events in sub-G1, G1 and S/G2-M phase.
D. Caspase 3/7 activities were assessed after 48 hours with different concentration of DZNep in KCNR cells. The percentage of caspase 3/7 activities was graphed after normalization to non-treated cells.
E. KCNR cells were treated with 5uM DZNep in the absence or presence of 100uM pan caspase inhibitor, Z-VAD-FMK, for 72 hours. The percentage of surviving cells was graphed after normalization to untreated control.
F. Mice were treated with or without DZNep (2.5mg/kg) twice a day, 3 days per week for 4 weeks. The mean tumor volumes are plotted using the SEM (standard error of the mean). The time points with significant differences (p<0.05) were indicated with asterisk.
Cell cycle analysis of DZNep treated KCNR cells indicated a 4-fold increase in the subG1 phase of the cell cycle and a decrease in the growth fraction (S/G2-M) (Fig. 7C). To assess whether DZNep-induced cell death via a caspase-dependent apoptotic pathway, we evaluated caspase-3/7 activities. There was up to a 2-fold increase in caspase 3/7 activities in DZNep-treated NB cells (Fig. 7D). Incubation of cells with a pan caspase inhibitor (Z-VAD-FMK) partially blocked the DZNep mediated decrease in NB cell survival (Fig. 7E). These data indicate that the DZNep induced increases in cell death are partially due to induction of caspase-dependent apoptotic pathways. A preliminary study to explore whether DZNep inhibited NB xenograft tumor growth indicates a statistically significant (P<0.05) reduction in the growth of NB tumors in mice treated with DZNep (Fig. 7F).
Discussion
In this study, we find relatively high expression of EZH2 in more undifferentiated or stroma poor NB tumors and high EZH2 expression is detected in the tumors of patients with worse prognoses. We also find that the NB tumor suppressor genes, CASZ1, CLU, NGFR and RUNX3 are direct targets of H3K27me3 mediated gene silencing by EZH2. HDAC inhibition increases CASZ1 expression and decreases binding of PRC2 and the H3K27me3 enrichment on the CASZ1 promoter. Our findings suggest EZH2-mediated H3K27me3 is an important repressive mark of tumor suppressors in NB, and it also indicates that EZH2 cooperates with histone deacetylases for effective repression of its targets.
Over-expression of EZH2 is found in a number of cancers including those of prostate (21), breast (28), as well as Ewings sarcoma (24), glioblastoma (25) and melanoma (39, 40). Previous studies indicate that EZH2 is a prognostic biomarker in breast (23) and prostate cancer (22, 41). We do find that there is an association between EZH2 mRNA levels and NB prognosis but it is not as robust as previously described prognostic markers in NB. However, the finding of EZH2 association with tumor histology provides a clue as to the potential mechanism responsible for the variable differentiation status of NB tumors. This is consistent with the report that EZH2 blockade induces neural differentiation-associated genes in Ewings sarcoma (24), a tumor of undifferentiated mesenchymal origin.
In this study, we used the newly identified NB tumor suppressor CASZ1 to probe how EZH2 affects target genes and to shed light on mechanisms of histone methylations that regulate tumor suppressor genes expression in NB. Recently we have published the evidence supporting a tumor suppressor function of CASZ1 in NB (11). CASZ1 is a gene localized in the 1p36, which is a region frequently deleted in the various kinds of cancers. Over-expression of CASZ1 in NB cells decreases cell growth, cell migration, anchorage independent growth and NB tumor growth in xenograft models (11). In clinical NB tumor samples, low CASZ1 expression correlates with poor prognosis and high CASZ1 expression is found in the NB tumors with a differentiated histopathology. Since tumor suppressors also require a functional loss of expression, our finding that EZH2 represses CASZ1 expression in NB cell lines with 1pLOH (SMS-KCNR) as well as intact 1p (SH-SY5Y) provides a mechanism to account for the functional loss of CASZ1 expression. Using several model systems we provide evidence that CASZ1 is a direct target of EZH2. Moreover, we find that the tumor suppressors, CLU, RUNX3 and NGFR, are also repressed by EZH2 in NB. While EZH2 may also increase expression of genes associated with NB tumorigenesis, we did not find significant increases in MYCN, Cyclin D1 (CCND1), PHOX2B, or NTRK2 mRNA levels when EZH2 is over-expressed (C. Wang unpublished data). This suggests that EZH2 activity may represent a broad mechanism of suppression of genes already noted to be important in NB tumor biology.
There are numerous different mechanisms reported to regulate the EZH2 expression and function, which could lead to the over-expression or increased activity of EZH2 in NB. The genomic loss of microRNA-101, which inhibits EZH2 expression, leads to prostate tumor (42) and this genomic region may be depleted via 1pLOH in NB. Also, EZH2 expression is induced by E2F1 and repressed by activated Rb (43), and its activity is stimulated by CDK1/2 (44). Over-expression of cell cycle genes is found in NB tumors with the unfavorable prognoses (33) and over 75% of NB tumors have elevated levels of cyclin D (45). In lymphoma, MYC is reported to stimulate EZH2 expression by repression of miR-26a (46). Accordingly the deregulation of MYCN, which occurs in 20% NB patients (2) may be involved in the deregulation of EZH2. The EZH2 locus on 7q35-36 is amplified in 10–15% tumors (43) and gain of the entire chromosome 7 or 7q is detected in over 50% of NB tumors (47). Therefore, increased DNA copy number could also contribute to increased EZH2 expression in a subset of NB tumors. Preliminary evaluation of available microarray databases does not reveal a significant association of EZH2 mRNA with MYCN. However, qRT-PCR analyses are in progress to ascertain whether there is an association with other known prognostic markers in NB. The complexity of potential alterations leading to dysregulation of EZH2 warrants a more detailed analysis of EZH2 regulatory mechanisms in NB tumors.
Our future experiments are aimed at determining all EZH2 targets in NB cells using unbiased genomic strategies that include microarray analyses of NB cells with silencing of EZH2 using shRNA and ChIP-seq to map targets. While HDAC inhibitors are traditionally understood to reduce deacetylation, allowing unrestrained acetylation to open chromatin and thereby increase gene expression, studies that have looked at array data or at gene families have shown that approximately the same number of genes are repressed as are induced (48). It should be possible with these future studies to identify whether H3K27me3 (via EZH2 or another methytransferase) is involved in the repression of genes after depsipeptide treatment.
Recently, studies focusing on DZNep as an epigenetic cancer therapy drug have been reported in breast cancer (34), glioblastoma (49) and acute myeloid leukemia (50). Our studies show that inhibition of EZH2 by gene silencing or pharmacologic inhibition using DZNep leads to inhibition of NB cell growth and morphologic differentiation of NB cells. This suggests that inhibition of EZH2 function or expression may be an important drug targeting strategy for NB.
Supplementary Material
Acknowledgments
We thank Rogier Versteeg and Richard Volckmann (Academic Medical Center, Department of Human Genetics, Amsterdam, the Netherlands) for making the R2 bioinformatics database publically available. We thank Doo-Yi Oh, Stanley He and other lab members for their thoughtful review of the study and Lauren Marks for her support of the Cell & Molecular Biology Section. This research was supported and funded by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- 1.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–20. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.Oberthuer A, Theissen J, Westermann F, Hero B, Fischer M. Molecular characterization and classification of neuroblastoma. Future Oncol. 2009;5:625–39. doi: 10.2217/fon.09.41. [DOI] [PubMed] [Google Scholar]
- 3.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–16. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 4.Krasnoselsky AL, Whiteford CC, Wei JS, Bilke S, Westermann F, Chen QR, et al. Altered expression of cell cycle genes distinguishes aggressive neuroblastoma. Oncogene. 2005;24:1533–41. doi: 10.1038/sj.onc.1208341. [DOI] [PubMed] [Google Scholar]
- 5.Caron H, van Sluis P, de Kraker J, Bokkerink J, Egeler M, Laureys G, et al. Allelic loss of chromosome 1p as a predictor of unfavorable outcome in patients with neuroblastoma. N Engl J Med. 1996;334:225–30. doi: 10.1056/NEJM199601253340404. [DOI] [PubMed] [Google Scholar]
- 6.Attiyeh EF, London WB, Mosse YP, Wang Q, Winter C, Khazi D, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–53. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 7.Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–75. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 8.Fujita T, Igarashi J, Okawa ER, Gotoh T, Manne J, Kolla V, et al. CHD5, a tumor suppressor gene deleted from 1p36. 31 in neuroblastomas. J Natl Cancer Inst. 2008;100:940–9. doi: 10.1093/jnci/djn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei JS, Song YK, Durinck S, Chen QR, Cheuk AT, Tsang P, et al. The MYCN oncogene is a direct target of miR-34a. Oncogene. 2008;27:5204–13. doi: 10.1038/onc.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossi M, Sayan AE, Terrinoni A, Melino G, Knight RA. Mechanism of induction of apoptosis by p73 and its relevance to neuroblastoma biology. Ann N Y Acad Sci. 2004;1028:143–9. doi: 10.1196/annals.1322.015. [DOI] [PubMed] [Google Scholar]
- 11.Liu Z, Yang X, Li Z, McMahon C, Sizer C, Barenboim-Stapleton L, et al. CASZ1, a candidate tumor-suppressor gene, suppresses neuroblastoma tumor growth through reprogramming gene expression. Cell Death Differ. 2011 doi: 10.1038/cdd.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellerick DM, Kassis JA, Zhang SD, Odenwald WF. castor encodes a novel zinc finger protein required for the development of a subset of CNS neurons in Drosophila. Neuron. 1992;9:789–803. doi: 10.1016/0896-6273(92)90234-5. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, Yang X, Tan F, Cullion K, Thiele CJ. Molecular cloning and characterization of human Castor, a novel human gene upregulated during cell differentiation. Biochem Biophys Res Commun. 2006;344:834–44. doi: 10.1016/j.bbrc.2006.03.207. [DOI] [PubMed] [Google Scholar]
- 14.Fransson S, Martinsson T, Ejeskar K. Neuroblastoma tumors with favorable and unfavorable outcomes: Significant differences in mRNA expression of genes mapped at 1p36. 2. Genes Chromosomes Cancer. 2007;46:45–52. doi: 10.1002/gcc.20387. [DOI] [PubMed] [Google Scholar]
- 15.Caren H, Fransson S, Ejeskar K, Kogner P, Martinsson T. Genetic and epigenetic changes in the common 1p36 deletion in neuroblastoma tumours. Br J Cancer. 2007;97:1416–24. doi: 10.1038/sj.bjc.6604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caren H, Ejeskar K, Fransson S, Hesson L, Latif F, Sjoberg RM, et al. A cluster of genes located in 1p36 are down-regulated in neuroblastomas with poor prognosis, but not due to CpG island methylation. Mol Cancer. 2005;4:10. doi: 10.1186/1476-4598-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 18.Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7:288–98. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–84. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 20.Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 22.Yu J, Rhodes DR, Tomlins SA, Cao X, Chen G, Mehra R, et al. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res. 2007;67:10657–63. doi: 10.1158/0008-5472.CAN-07-2498. [DOI] [PubMed] [Google Scholar]
- 23.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richter GH, Plehm S, Fasan A, Rossler S, Unland R, Bennani-Baiti IM, et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc Natl Acad Sci U S A. 2009;106:5324–9. doi: 10.1073/pnas.0810759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J, Son MJ, Woolard K, Donin NM, Li A, Cheng CH, et al. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell. 2008;13:69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–35. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–30. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Q, Yu J, Dhanasekaran SM, Kim JH, Mani RS, Tomlins SA, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–84. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–24. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Jin Q, Lee JE, Su IH, Ge K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc Natl Acad Sci U S A. 2010;107:7317–22. doi: 10.1073/pnas.1000031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A. 2007;104:18439–44. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albino D, Scaruffi P, Moretti S, Coco S, Truini M, Di Cristofano C, et al. Identification of low intratumoral gene expression heterogeneity in neuroblastic tumors by genome-wide expression analysis and game theory. Cancer. 2008;113:1412–22. doi: 10.1002/cncr.23720. [DOI] [PubMed] [Google Scholar]
- 33.Wei JS, Greer BT, Westermann F, Steinberg SM, Son CG, Chen QR, et al. Prediction of clinical outcome using gene expression profiling and artificial neural networks for patients with neuroblastoma. Cancer Res. 2004;64:6883–91. doi: 10.1158/0008-5472.CAN-04-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–63. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 36.Graham C, Tucker C, Creech J, Favours E, Billups CA, Liu T, et al. Evaluation of the antitumor efficacy, pharmacokinetics, and pharmacodynamics of the histone deacetylase inhibitor depsipeptide in childhood cancer models in vivo. Clin Cancer Res. 2006;12:223–34. doi: 10.1158/1078-0432.CCR-05-1225. [DOI] [PubMed] [Google Scholar]
- 37.Chayka O, Corvetta D, Dews M, Caccamo AE, Piotrowska I, Santilli G, et al. Clusterin, a haploinsufficient tumor suppressor gene in neuroblastomas. J Natl Cancer Inst. 2009;101:663–77. doi: 10.1093/jnci/djp063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Astuti D, Agathanggelou A, Honorio S, Dallol A, Martinsson T, Kogner P, et al. RASSF1A promoter region CpG island hypermethylation in phaeochromocytomas and neuroblastoma tumours. Oncogene. 2001;20:7573–7. doi: 10.1038/sj.onc.1204968. [DOI] [PubMed] [Google Scholar]
- 39.Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–73. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 40.McHugh JB, Fullen DR, Ma L, Kleer CG, Su LD. Expression of polycomb group protein EZH2 in nevi and melanoma. J Cutan Pathol. 2007;34:597–600. doi: 10.1111/j.1600-0560.2006.00678.x. [DOI] [PubMed] [Google Scholar]
- 41.Rhodes DR, Sanda MG, Otte AP, Chinnaiyan AM, Rubin MA. Multiplex biomarker approach for determining risk of prostate-specific antigen-defined recurrence of prostate cancer. J Natl Cancer Inst. 2003;95:661–8. doi: 10.1093/jnci/95.9.661. [DOI] [PubMed] [Google Scholar]
- 42.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–9. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–35. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen S, Bohrer LR, Rai AN, Pan Y, Gan L, Zhou X, et al. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat Cell Biol. 2010;12:1108–14. doi: 10.1038/ncb2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molenaar JJ, van Sluis P, Boon K, Versteeg R, Caron HN. Rearrangements and increased expression of cyclin D1 (CCND1) in neuroblastoma. Genes Chromosomes Cancer. 2003;36:242–9. doi: 10.1002/gcc.10166. [DOI] [PubMed] [Google Scholar]
- 46.Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, et al. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–12. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 47.Stallings RL, Howard J, Dunlop A, Mullarkey M, McDermott M, Breatnach F, et al. Are gains of chromosomal regions 7q and 11p important abnormalities in neuroblastoma? Cancer Genet Cytogenet. 2003;140:133–7. doi: 10.1016/s0165-4608(02)00681-7. [DOI] [PubMed] [Google Scholar]
- 48.Sasakawa Y, Naoe Y, Sogo N, Inoue T, Sasakawa T, Matsuo M, et al. Marker genes to predict sensitivity to FK228, a histone deacetylase inhibitor. Biochem Pharmacol. 2005;69:603–16. doi: 10.1016/j.bcp.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 49.Suva ML, Riggi N, Janiszewska M, Radovanovic I, Provero P, Stehle JC, et al. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009;69:9211–8. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- 50.Fiskus W, Wang Y, Sreekumar A, Buckley KM, Shi H, Jillella A, et al. Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood. 2009;114:2733–43. doi: 10.1182/blood-2009-03-213496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.