Abstract
Background:
Herbal drugs used to treat illness according to Ayurveda are often misidentified or adulterated with similar plant materials.
Objective:
To aid taxonomical identification, we used DNA barcoding to evaluate authentic and substitute samples of herb and phylogenetic relationship of four medicinal plants of family Asparagaceace and Asclepiadaceae.
Materials and Methods:
DNA extracted from dry root samples of two authentic and two substitutes of four specimens belonging to four species were subjected to polymerase chain reaction (PCR) and DNA sequencing. Primers for nuclear DNA (nu ITS2) and plastid DNA (matK and rpoC1) were used for PCR and sequence analysis was performed by Clustal W. The intraspecific variation and interspecific divergence were calculated using MEGA V 4.0.
Statistical Analysis:
Kimura's two parameter model, neighbor joining and bootstrapping methods were used in this work.
Results:
The result indicates the efficiency of amplification for ITS2 candidate DNA barcodes was 100% for four species tested. The average interspecific divergence is 0.12 and intraspecific variation was 0.232 in the case of two Asparagaceae species. In two Asclepiadaceae species, average interspecific divergence and intraspecific variation were 0.178 and 0.004 respectively.
Conclusions:
Our findings show that the ITS2 region can effectively discriminate Asparagus racemosus and Hemidesmus indicus from its substitute samples and hence can resolve species admixtures in raw samples. The ITS2 region may be used as one of the standard DNA barcodes to identify closely related species of family Asclepiadaceae but was noninformative for Asparagaceae species suggesting a need for the development of new markers for each family. More detailed studies involving more species and substitutes are warranted.
Keywords: Asparagus gonoclados, Asparagus racemosus, Decalepis hamiltonii, DNA barcoding, Hemidesmus indicus, ITS2 region
INTRODUCTION
Traditional herbal medicines are gaining popularity worldwide and about 80% of the world's population utilizes traditional medicines for well-being and healthcare.[1] The increased demand for botanical products is accomplished by an expanding industry but also accompanied by calls for assurance of quality, efficacy, and safety.[2] Studies on adulteration of herbal medicines or misidentification of plants have health consequences which may vary from life threatening and even to death.[3] Biological species are authenticated traditionally according to their morphological features. Morphological keys still are main basis of taxonomy. But, the traditional taxonomy study usually requires the expertise of an experienced professional taxonomist. In the case that the diagnostic morphological trait of a specimen is lacking, it becomes difficult even for the specialists to recognize a species correctly. Authentication using DNA barcodes overcomes these problems. The DNA barcode is a short fragment of the genome and species identification using DNA barcoding has been completed in several studies when morphological characteristics are absent. The DNA barcoding technique is a useful tool for taxonomists and is likely to usher the taxonomic research into a new era. In plants, several candidate DNA barcodes have attracted the attention of many researchers such as trnH-psbA, rbcL, matK, rpoB, rpoC1, ndhJ, accD, YCF5 in the plastid genome and rDNA ITS in the nuclear genome to classify the medicinal plants.[4,5] The advent of DNA barcoding to identify plant species in herbal medicine appears to be promising but remains to be fully evaluated.[6]
The species across families Asparagaceae and Asclepiadaceae possess important medicinal properties and proven healthcare benefits. Asparagus racemosus Willd. is a popular ayurvedic ingredient where the dried roots are used in different formulations. This plant has been described as a rasayana herb and has been extensively used as an adaptogen to increase the nonspecific resistance of organisms against a variety of stresses. In a recent literature, it has been examined for various pharmacological conditions such as antiinflammatory,[7] immune modulatory,[8] neuroprotective,[9] and several others. Asparagus gonoclados Baker is another plant of the same genera and is considered as a substitute to A. racemosus.[10] These two species are closely related and there is no marked difference in the flower and fruit structure, but there is difference in the leaf structure and in the distribution of the plant. The identification of the plant, especially the dry roots, becomes a difficult task and it can be easily adulterated with substitutes. Plant Hemidesmus indicus R. Br. is used to treat various medicinal conditions such as skin disease, diarrhea, fever, and rheumatism in Eastern part of India. The aromatic, woody, and deep roots are extremely important in Ayurveda and are used in various skin care formulations.[11] Due to scarcity of this plant in the wild in the recent past and the cumbersome extraction procedures, few other plants are now being used as substituents to H. indicus. The most common species used to substitute or adulterate H. indicus is Decalepish hamiltonii Wight and Arn. also a member of family Asclepiadaceae. Since the identification of these dried plants especially from roots is difficult, we explored utility of DNA barcoding to evaluate intraspecies specific variations and interspecies specific divergence in two species each of family Asparagaceace (A. racemous and A. gonocladus) and family Asclepiadaceae (H. indicus and D. hamiltonii).
MATERIALS AND METHODS
Plant materials
H. indicus and D. hamiltonii roots were collected from a medicinal plants garden in Bangalore, and roots of A. gonoclados and A. racemosus were collected from Thirumala Hills (Andhra Pradesh) and Denkanikotta (Tamil Nadu). Botanical identification of all the four plants species was confirmed by in-house taxonomist (R&D, Natural Remedies, Bengaluru, India). Voucher specimens have been preserved with the authors at R&D of Natural Remedies. The voucher numbers for H.indicus, D. hamiltonii, A. gonoclados, and A. racemosus are Sariva I, Sariva II, AGR31, and AGR30 respectively. These samples hereafter referred to as reference material were used to develop DNA barcode. Roots were washed, chopped, and dried in sunshine.
Genomic DNA isolation, PCR amplification, and sequencing of DNA barcode
Dried roots were thoroughly rinsed in sterile distilled water and cut into small pieces. DNA extraction was performed according to the protocol of Warude et al.,.[12] Primer sequences for three candidate DNA barcodes (ITS2, rpoC1, matK) were taken from the previous study.[5] PCR amplification was performed in 25 μl reaction mixtures containing approximately 40 ng template DNA, 1 X PCR buffer, 1.5 mM MgCl2, 0.2 mM of each dNTP, 0.1 μM of each primers (Sigma Proligo and Sigma Life Science, India), and 1.0 U Taq DNA Polymerase (Invitrogen) in a thermal cycler (Eppendorf, Germany). The primer sequences and PCR reaction conditions used for PCR amplification are listed in Table 1. Purified PCR products were sequenced in both directions on ABI3130 sequencer (Applied Biosystem, USA).
Table 1.
Primers and reaction conditions used in the present study

Sequence alignment and genetic analysis
To estimate the quality of the generated DNA sequence traces, the original forward and reverse sequences of PCR products were assembled using sequence alignment tools (BLAST) from NCBI. The data analysis was done for all the four plant species of the two families, Asparagaceace and Asclepiadaceae for which the sequences are available in the GenBank and with the sequence generated in the present study to find the intraspecific divergence and interspecific variations. GenBank accession numbers of candidate DNA barcodes generated in the present study are H. indicus (JN712767), D. hamiltonii (JN712768), A. gonoclados (JN712766), and A. racemosus (JN712765), respectively. The GenBank accession numbers of candidate DNA barcodes of database taxa used in this study were A. densiflorus (HM357929), A. officinalis isolate A5 (HM35793), A. officinalis (GU474425), A. racemosus (GU474426), A. setaceus (GQ202238), A. filicinus (HM357928), and H. indicus (DQ916851).[13] Multiple sequence alignment was performed by Clustal W.[14] The basic sequence statistics including nucleotide frequencies and variability in different regions of the sequence were computed using the Kimura 2-parameter method by MEGA V 4.0 software. The sequence data were analyzed by neighbor joining and bootstrapping methods.[15–18]
RESULTS AND DISCUSSION
DNA barcodes must simultaneously contain enough variability to be used for species identification and adequate conservative regions for the universal primers.[19] Previous studies have suggested that ITS2 sequences are relatively easy to amplify using universal primers and that the ITS2 region possesses high interspecific divergence.[5] In this study, ITS2 DNA barcode candidate sequences in samples of Asparagaceace and Asclepiadaceae family tested were obtained through PCR amplification with universal primers followed by DNA sequencing and multiple sequence alignment. The success rate of PCR amplification of ITS2 sequences was 100% Figure 1 and alignment of sequences was performed with ease for all the four species studied in accordance with previously published studies.[20] Sequences were aligned using Clustal W and final alignment adjustments were made manually in order to remove artificial gaps. We assessed divergences within and between species using software MEGA V 4.0. In the species of Asparagaceae and Asclepiadaceae, average distances were computed based on pairwise comparisons of all sequences. For each species, of Asparagaceae average interspecific nucleotide differences was 0.198 and differences between individuals of the same species sequences was 0.233. The average interspecific percentage difference in sequence homology was 3.6% and the percentage difference between individuals of the same species is 5%. The distance between generated ITS2 gene sequence of A. gonoclados and A. racemosus is 0.011 and the distance between generated sequence of A. gonoclados and database sequence of A. racemosus was 0.217. Further we observed that A. gonoclados is much closer to A. densiflorus (distance 0.012) than A. racemosus sequence from database [Tables 2 and 3]. In Asclepiadaceae species, gene sequences for ITS2 are available only for one species in the GenBank. The average interspecific nucleotide differences was 0.123 and differences between individuals of the same species sequences was 0.004. The average interspecific percentage difference in sequence homology was 13% and the percentage difference between individuals of the same species was 2% [Tables 4 and 5]. The distance between H. indicus and D. hamiltonii sequences of the present study was 0.1854 and the distance between the sequences of H. indicus from database was 0.008. Phylogenetic analyses were computed using the MEGA V 4.0 software. Phylogenetic inference was performed by the neighbor-joining method. Bootstrapping tests were conducted to examine the reliability of the interior branches and the validity of the trees. The optimal tree with the sum of branch length 0.805 was shown for Asparagaceae family species and the branch length was 0.629 for Asclepiadaceace family species. The percentage of replicate trees in which the associated taxa clustered together in the bootstrapping test (1000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The distances were computed and are in the units of the number of base substitutions per site. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated from the dataset. There were a total of 170 positions in the final dataset of Asparagaceae family species and of 232 positions in the final dataset of Asclepiadaceae family species [Figures 2 and 3]. The intraspecific percentage differences among H. Indicus species were less than the interspecific variations; hence the ITS2 region can be potentially used as a standard DNA barcode to identify medicinal plants of Asclepiadaceae family. But in the case of Asparagus species we found that the intraspecific percentage differences among the Asparagus species were greater than the interspecific variations, which suggest that the ITS2 gene is not an effective candidate barcode for Asparagaceae species.[21] All the samples were also tested using chloroplast-specific primers (rpoC1 and matK1) to amplify any contaminating chloroplast containing parts. No amplification products were obtained suggesting that the materials used primarily originated from the roots of the plants. Despite the potential benefits of DNA barcoding as a novel system designed to provide rapid and accurate species identifications by using short, standardized gene regions as internal species tags, but may at times barcoding may fail as sequences for some taxa are not as robust and broadly applicable.[22] In such cases new genomic markers may need to be adopted. In this study, DNA sequence alignment and sequence variation between species within the ITS2 region was examined for its usefulness as the candidate DNA barcode for species variation and species identification of Asparagaceace and Asclepiadaceae families. With 100% PCR amplification efficiency, straightforward sequence alignment, and ability to distinguish closely related species indicates that DNA sequences of the ITS2 region is an effective and powerful tool for identification of medicinal plants of Asclepiadaceae families in traditional Indian Ayurveda medicine and their contaminants. Although the ITS2 region of Asparagaceae species showed 100% PCR amplification efficiency, straightforward sequence alignment and had the ability to discriminate authentic Asparagus racemosus from its substitute sample, but the interspecific variation was low and intraspecific divergence was high which suggests that the ITS2 region is not an effective marker loci for Asparagaceae species. More studies are warranted with samples of most commonly available species and substitutes of these two families examined.
Figure 1.

PCR for the ITS2 region of root DNA samples of family Asparagaceae (Asparagus recemosus and Asparagus gonoclados) and Asclepiadaceae (Hemidesmus indicus and Decalepis hamiltonii) Detection of the ITS2 region of ~500 bp PCR product on a 1.5% agarose gel. Lane 1. Asparagus racemosus, Lane 2. Asparagus gonoclados, Lane 3. Hemidesmus indicus, Lane 4. Decalepis hamiltonii, Lane 5.100 bp ladder, Lane 6. Negative control
Table 2.
Homology and distance matrix of Asparagus gonoclados plant ITS2 gene nucleotide sequences with ITS2 sequences of other Asparagus species
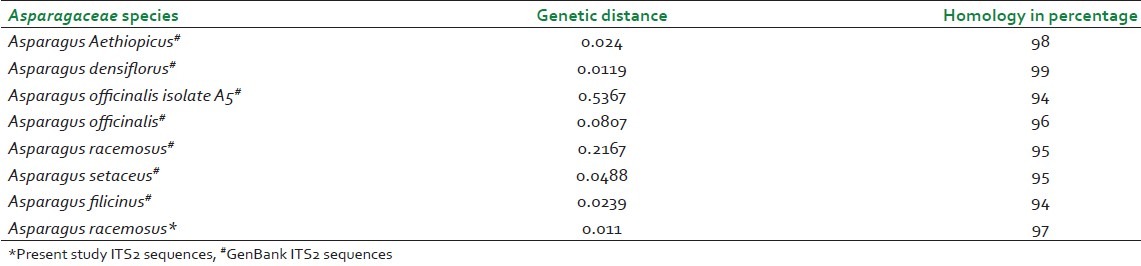
Table 3.
Estimated distance matrix for Asparagaceae species
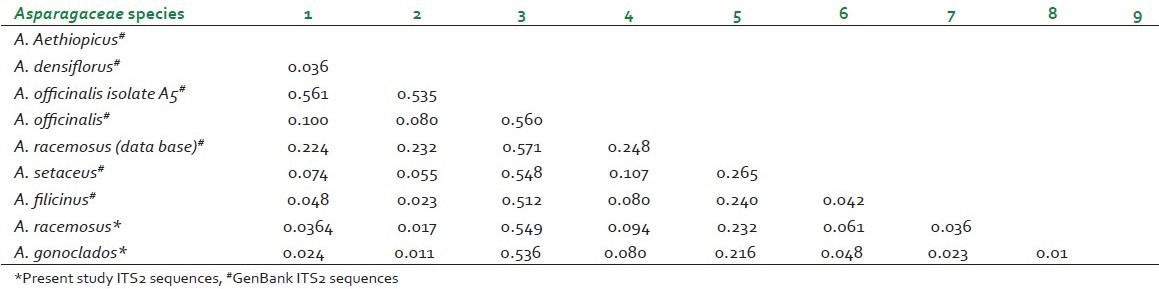
Table 4.
Homology and distance matrix of Decalepis hamiltonii ITS2 gene nucleotide sequences with ITS2 sequences of other species of the Asclepiadaceae family
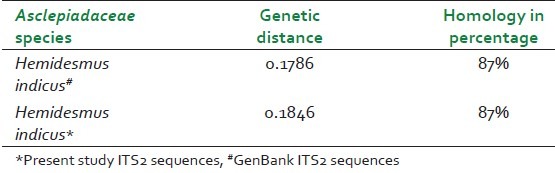
Table 5.
Estimation of distance matrix for Asparagaceae species
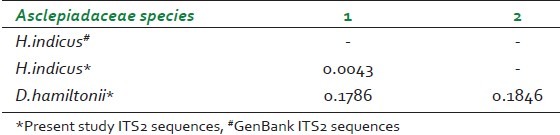
Figure 2.
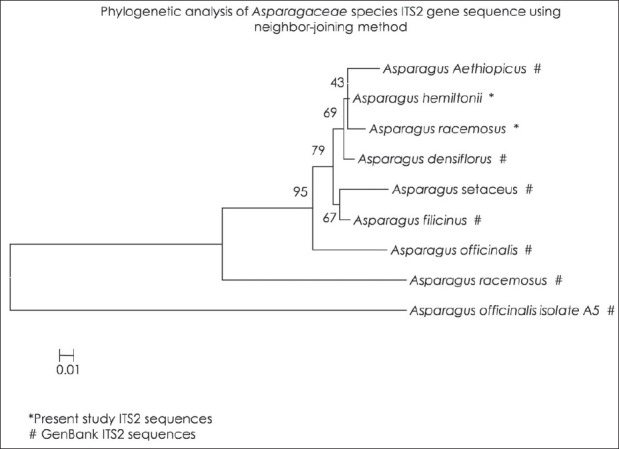
Phylogenetic analyses of the Asparagaceae species ITS2 gene sequence using the neighbor-joining method *Present study ITS2 sequences #GenBank ITS2 sequences
Figure 3.
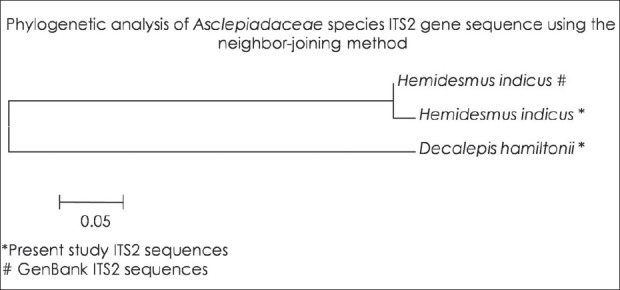
Phylogenetic analyses of the Asclepiadaceae species ITS2 gene sequence using the neighbor-joining method *Present study ITS2 sequences #GenBank ITS2 sequences
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Sucher NJ, Carles MC. Genome-based approaches to the authentication of medicinal plants. Planta Med. 2008;74:603–23. doi: 10.1055/s-2008-1074517. [DOI] [PubMed] [Google Scholar]
- 2.Shaw D, Leon C, Kolev S, Murray V. Traditional remedies and food supplements: A 5-year toxicological study (1991-1995) Drug Saf. 1997;17:342–56. doi: 10.2165/00002018-199717050-00006. [DOI] [PubMed] [Google Scholar]
- 3.Tewfik S. Authentication of medicinal plant material by DNA fingerprinting. World Rev Sci Tech Sustain Dev. 2008;5:151–60. [Google Scholar]
- 4.Song J, Yao H, Li Y, Li X, Lin Y, Liu C, et al. Authentication of the family Polygonaceae in Chinese pharmacopoeia by DNA barcoding technique. J Ethnopharmacol. 2009;124:434–9. doi: 10.1016/j.jep.2009.05.042. [DOI] [PubMed] [Google Scholar]
- 5.Chen S, Yao H, Han J, Liu C, Song J, Shi L, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One. 2010;5:e8613. doi: 10.1371/journal.pone.0008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joshi K, Chavan P, Warude D, Patwardhan B. Molecular markers in herbal drug discovery. Curr Sci. 2004;87:159–65. [Google Scholar]
- 7.Lee do Y, Choo BK, Yoon T, Cheon MS, Lee HW, Lee AY, et al. Anti-inflammatory effects of Asparagus cochinchinensis extract in acute and chronic cutaneous inflammation. J Ethnopharmacol. 2009;121:28–34. doi: 10.1016/j.jep.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Gautam M, Saha S, Bani S, Kaul A, Mishra S, Patil D, et al. Immunomodulatory activity of Asparagus racemosus on systemic Th1/Th2 immunity: implications for immunoadjuvant potential. J Ethnopharmacol. 2009;121:241–7. doi: 10.1016/j.jep.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 9.Parihar MS, Hemnani T. Experimental excitotoxicity provokes oxidative damage in mice brain and attenuation by extract of Asparagus racemosus. J Neural Transm. 2004;111:1–12. doi: 10.1007/s00702-003-0069-8. [DOI] [PubMed] [Google Scholar]
- 10.Madhavan V, Tijare RD, Mythreyi R, Gurudev MR, Yoganarasihman SN. Pharmacognostical studies on root tubers of Asparagus ganoclodos Baker- Alternate source for the Ayurvedic drug Shatavari. Indian J Nat Prod Res. 2010;1:57–62. [Google Scholar]
- 11.Aminuddin, Girach RD. Pluralistic folk uses of Hemidesmus indicus (L.) R. Br. from south eastern India. J Econ Taxon Bot. 1991;15:715–8. [Google Scholar]
- 12.Warude D, Chavan P, Joshi K, Patwardhan B. DNA Isolation from fresh and dry plant samples with highly acidic tissue extracts. Plant Mol Biol Rep. 2003;21:467. [Google Scholar]
- 13. [Last accessed on 2011 July 14]. Available from: http://www.ncbi.nlm.nih.gov/genbank .
- 14. [Last accessed on 2011 July 14]. Available from: http://www.mbio.ncsu.edu//bioedit .
- 15.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 17.Kimura MA. Simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Mol Biol Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 18.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 19.Gao T, Yao H, Song J, Zhu Y, Liu C, Chen S. Evaluating the feasibility of using candidate DNA barcodes in discriminating species of the large Asteraceae family. BMC Evol Biol. 2010;10:324. doi: 10.1186/1471-2148-10-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoeckle MY. Taxonomy, DNA, and the bar code of life. Biol Sci. 2003;53:2–3. [Google Scholar]
- 21.Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, et al. DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci U S A. 2008;105:2923–8. doi: 10.1073/pnas.0709936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hebert PD, Gregory TR. The promise of DNA barcoding for taxonomy. Syst Biol. 2005;54:852–9. doi: 10.1080/10635150500354886. [DOI] [PubMed] [Google Scholar]


