Abstract
Over the past decade, the importance of non-coding RNA such as microRNA has been established in numerous processes that drive human pathogenesis. These crucial molecular regulators modulate networks of target gene transcripts that, in turn, orchestrate cellular phenotypes such as cell survival, differentiation, proliferation, and metabolism among others and thus affect cardiopulmonary vascular disease conditions. Many of these same pathophenotypes figure prominently in the complex pathogenesis of pulmonary hypertension, an enigmatic vascular disorder characterized by a histological panvasculopathy and driven by disparate upstream triggers such as hypoxia, inflammation, and bone morphogenetic protein signaling. Yet, the importance of just a few microRNAs in pulmonary hypertension has been recognized, and we are only beginning to understand the integrative functions of these molecules in this disease. By combining systems biology with traditional experimental approaches, more direct insight into the pleiotropy of microRNA should not only further reveal the spectrum of molecular pathways that cause pulmonary hypertension, but also offer novel and much needed diagnostic and therapeutic strategies.
Keywords: hypoxia, microRNA, network biology, pulmonary hypertension
The manifestations of human disease were once thought to be driven solely by dysregulation of protein-coding genes and their molecular interactions with one another. In recent years, epigenetic alterations of the genome (e.g., DNA methylation) and the transcriptome (e.g., repression by non-coding RNA) have been recognized as powerful and alternative gene regulatory mechanisms that exponentially increase the complexity of how individualized pathophenotypes manifest. Specifically, since the first descriptions of their bona fide regulatory functions,[1,2] small non-coding RNA species termed microRNA (miRNA) have emerged as such essential regulators in a broad range of cellular adaptive processes, with an exceptionally rapid pace of discovery in this field (Fig. 1A).
Figure 1.
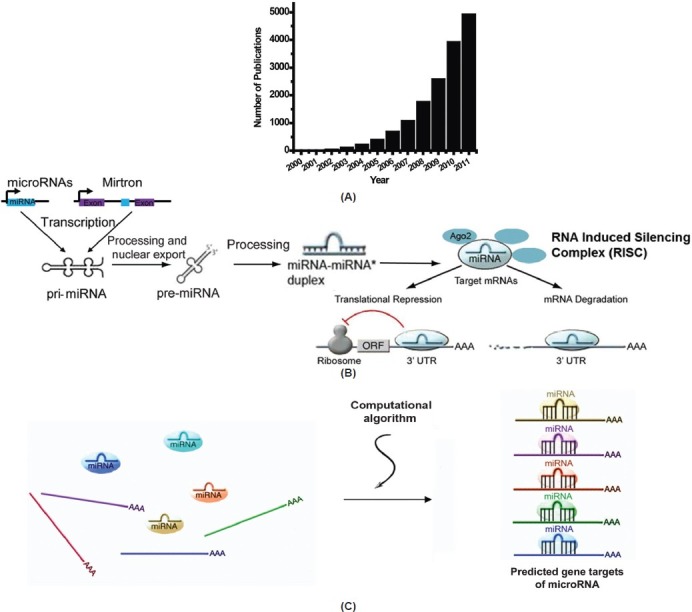
Overview of microRNA biology. (A) The number of publications related to miRNA biology has increased dramatically over the past 10 years, emphasizing the emerging importance of these small molecules in human health and disease. (B) MicroRNA biogenesis and mechanism of action. MiRNAs undergo several nuclear and cytosolic processing steps before maturation to a biologically active 19-24 nucleotide molecule containing the seed region. Through binding to the complementary sites in the 3? untranslated region (UTR) of their target genes, miRNA promote gene down-regulation via either translational repression or mRNA degradation. Despite their critical importance in diverse cellular processes, the functions of miRNA in PH remain mostly unclear. Adapted from[10,82] with permission. (C) Computational algorithms predict gene targets of miRNA. Based primarily on conserved Watson-Crick base pairing and other site-specific criteria, several computational algorithms (i.e. TargetScan and DIANA) are used to predict miRNA target genes.
MiRNA are expected to carry central regulatory roles in the progression of pulmonary vascular diseases such as pulmonary hypertension (PH). However, appreciation of their importance in PH is just emerging. Broadly, PH is a serious and at times fatal pulmonary vascular condition that afflicts a growing number of patients worldwide.[3,4] PH is characterized by an enigmatic and complex panvasculopathy, marked by the dysregulation of multiple cell types and molecular pathways. In the most severe forms of PH, lumen destruction is often present at bifurcation points distal to small pulmonary arteries (50-250 µm external diameter) caused by the presence of pathognomonic complex vascular lesions, or so-called plexogenic lesions.[5] This vascular pathology is triggered by seemingly disparate genetic and environmental stimuli, such as hypoxia, inflammation, and transforming growth factor/bone morphogenetic protein (TGF/BMP) signaling, among many others. The underpinnings of cross-talk between these pathways in the pulmonary vasculature are only beginning to be understood.[6,7] Nonetheless, such processes ultimately drive pulmonary arterial remodeling and pruning, increased pulmonary arterial pressures, right ventricular dysfunction, and sometimes death.
Owing to their pleiotropic vascular effects,[8] miRNA may function as factors that integratively link these pathogenic signaling pathways to PH. Accordingly, over the past 5 years, an increasing cadre of miRNA have been found to be dynamically regulated by disease triggers of PH (Tables 1 and 2). In turn, many of these miRNA carry especially important adaptive roles in vascular function, relevant for vasomotor tone, thrombosis, metabolism, cellular proliferation, and cellular survival.[9] Thus, although only a handful of miRNA have been definitively proven to modulate directly the development and/or progression of PH, we would expect a much greater number of miRNA to regulate various overlapping phenotypic features of this complex disease. Furthermore, because of the growing appreciation of the cooperative activities of multiple miRNA in complex human disease, the discovery of novel functions of miRNA in PH may be especially amenable to systems-wide bioinformatic approaches in combination with traditional experimentation. This review will provide an overview of miRNA biology relevant for pulmonary vascular function, and discuss those scientific approaches designed to demonstrate the importance of miRNA in pulmonary vascular dysregulation. Moreover, we will speculate on the importance of other miRNA in PH, and we will comment on the robust direction of future research of miRNA biology in this disease.
Table 1.
Compiled list of hypoxamirs with functions in PH that have been experimentally established or highly suspected[24,75]
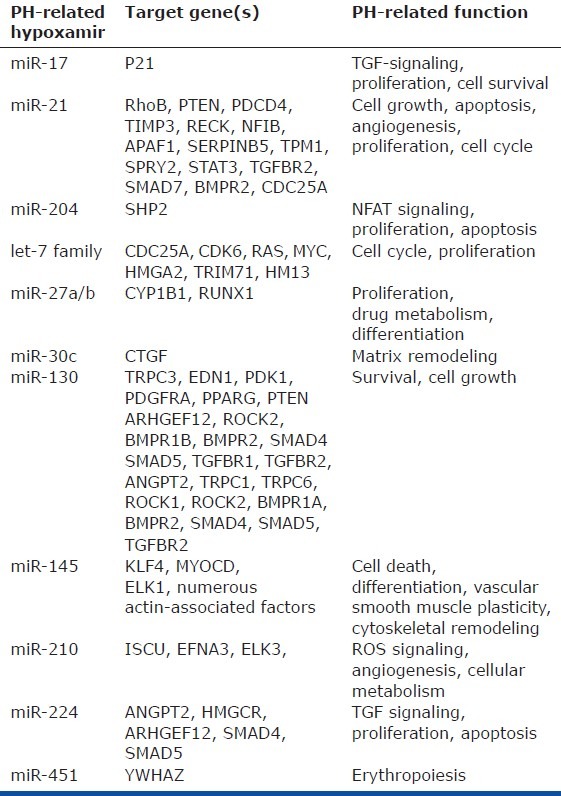
Table 2.
Compiled list of miRNA related to TGF/BMP signaling, inflammatory stimuli, and hypoxia many of which lack substantial experimental support regarding their actions in PH[75]

MIRNA BIOGENESIS AND ESTABLISHED FUNCTIONS IN CARDIOVASCULAR BIOLOGY
At present, over 1400 distinct miRNA species have been predicted to be encoded by the human genome.[10] In total, between 50% and 60% of all mammalian messenger RNA (mRNA) transcripts are estimated to be subject to miRNA regulation[11,12] highlighting their role as ubiquitous biological regulators of gene expression. Processed, mature forms of miRNA molecules exist as double-stranded non-coding RNA species between 19 and 24 nucleotides in length. Within each miRNA exists a “seed sequence” which is an important determinant for complementary strand target sequence binding, typically positioned at the 3′ untranslated region (3′ UTR) of the target mRNA transcript. This miRNA-mRNA interaction results in the downregulation of the target gene transcript, via either translational repression or transcript degradation (Fig. 1B). As a result of the importance of conserved Watson-Crick binding for such target recognition, several well-validated computational algorithms have been developed (i.e., Targetscan 5 [Conserved][13] and DIANA,[14] among many others) to predict mRNA targets of specific miRNA (Fig. 1C). Although they suffer from a degree of false-positive prediction, these algorithms have been invaluable in rapidly identifying the biological roles of miRNA based on their predicted target pool.
Consequently, multiple miRNA have been implicated in cardiomyocyte biology such as those involved in myocardial fibrosis, infarction, and hypertrophy (Fig. 2A) as well as in vascular biology, such as those involved in atherogenesis, vasomotor tone and hypertension, and neointimal formation (Fig. 2B), among many others.[9,15] Notably, these studies have mostly focused on the study of the systemic rather than pulmonary vasculature. However, they nonetheless suggest the requirement of miRNA in preserving overall vascular function and integrity, thus supporting their presumed importance in the pulmonary vessel, as well. More specifically, it is already established that complex molecular dysregulation of multiple signaling pathways underlies the pathogenesis of PH, including TGF/BMP,[16] RhoA/Rho-kinase,[17] serotonin,[18] Notch,[19] nitric oxide/cyclic guanosine monophosphate (cGMP),[20] endothelin,[21] and Ang1/Tie2[22] pathways, among others. Importantly, a number of miRNA target genes have been predicted to be highly enriched within these PH-associated pathways, suggesting extensive miRNA-regulated control that awaits experimental validation. In fact, by cross-referencing a known module of PH-relevant genes in these pathways with the miRNA targets predicted by the TargetScan 5 (Conserved) algorithm, approximately 85% of all conserved miRNA are predicted to target at least one PH-relevant gene.[23] Such regulatory relationships certainly suggest the pervasive importance of miRNA in PH, yet they also reflect their extremely complicated and sometimes overlapping, synergistic, or redundant actions.
Figure 2.
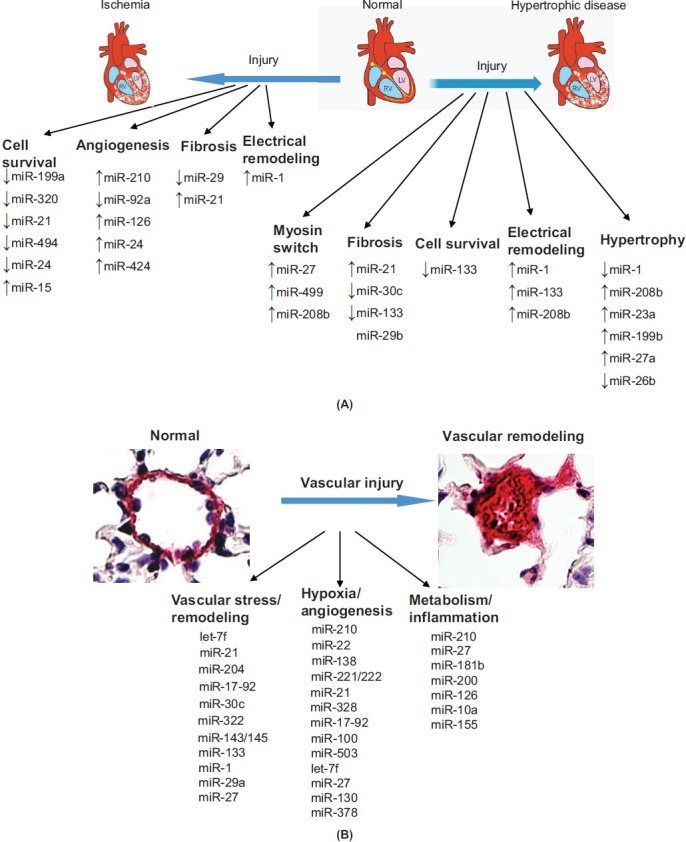
MiRNA regulate cardiovascular functions. (A) Control of cardiomyocyte function by microRNA. The molecular pathologic events that occur in cardiomyocytes during ischemia and hypertrophic disease are closely controlled by multiple miRNA which regulate cell survival, apoptosis, angiogenesis, fibrosis, and remodeling. Adapted from,[9] with permission. (B) Control of vascular function by microRNA. The widespread molecular pathologic events that occur in the diseased vasculature are closely controlled by multiple miRNA, which regulate vascular integrity and remodeling, some of which are highlighted here and reviewed by.[15,83]
PATHWAY-BASED DISCOVERY OF MIRNA-DEPENDENT ACTIONS IN UPSTREAM TRIGGERS OF PH
To speculate on their anticipated functions in PH, it is logical to predict the importance of specific miRNA if they carry already established functions related to the numerous upstream triggers of this disease. As discussed previously, etiologies of PH are varied and many have been challenging to study on the molecular level due to their biological complexities. Some of these include chronic thromboembolism, human immunodeficiency virus (HIV) or Schistosomiasis infections, and intracardiac shunting and increased pulmonary vascular flow, to name a few. Therefore, our insight into how miRNA influence these conditions remains limited. In contrast, triggers such as hypoxia, TGF/BMP signaling, and inflammation have been studied in more detail, and a number of miRNA have been found to play important and varied regulatory roles in those biological contexts.
Hypoxamirs in PH
Inadequate oxygen availability or hypoxia drives a complex set of cellular adaptations that influence cellular survival and function in the pulmonary vasculature, and is a common insult triggering PH. Many of these cellular adaptations are transcriptionally controlled by the master transcription factors, hypoxia-inducible factor 1-alpha (HIF-1α) and hypoxia-inducible factor 2-alpha (HIF-2α). In response to hypoxia, HIF levels increase and directly induce the transcription of >100 genes, influencing functions ranging from metabolism, survival, proliferation, migration, and angiogenesis, among others. Notably, HIF has been implicated as a central pathogenic factor across multiple clinical categories of PH (as reviewed in Reference 3). Numerous microRNA, now totaling over 100, have also been found to be modulated by hypoxia in a variety of biological and experimental contexts and consequently have been termed “hypoxamirs” (Tables 1 and 2).[24] A few hypoxamirs, such as miR-210, carry bona fide HIF-response elements in their promoters that, when bound by HIF, can robustly activate miRNA transcription and expression during hypoxia.[25] A great majority of hypoxamirs, however, are not directly driven by promoter binding to HIF but, rather, through indirect, hypoxia-associated stimuli,[24] such as inflammation (i.e., miR-146a/b[26] and miR-181b[27]). As expected for such a large number of unique and differentially regulated miRNA, hypoxamirs have been found to regulate a diverse set of targets and pathways known to be active in hypoxic adaptation, including proliferation, cell cycle response, apoptosis, metabolism, DNA repair, cellular hypertrophy, angiogenesis, hematopoiesis, fibrosis, and inflammatory response, among many others. Additionally, some hypoxamirs (e.g., miR-424[28]) carry important feedback actions in regulating HIF expression itself and, thus, affect global HIF-dependent activation of transcription.
Consequently, based on their known functions, hypoxamirs can be placed into molecular pathways that predict how they may regulate PH inception or progression. However, the more painstaking challenge remains of how to validate experimentally these actions in vivo. For example, such a roadmap could be construed to predict the direct actions of the prototypical HIF-dependent miRNA, miR-210, in PH, as it has been studied in multiple hypoxic or ischemic diseases in vivo, including ischemic heart disease,[29] wound healing,[30] and multiple forms of cancer.[31] As proven both in cell culture and in vivo, miR-210 plays critical roles in modulating cellular proliferation, survival, oxidative stress, and angiogenic potential.[24] Importantly, a number of these phenotypes in cell culture are directly linked to the ability of miR-210 to downregulate one of its primary target genes, the iron sulfur complex assembly 1/2 (ISCU1/2), in order to repress mitochondrial metabolism in favor of glycolysis (the so-called Pasteur effect).[32] Such a metabolic shift away from oxygen utilization for energy production is a critical adaptive step for cellular survival in acute hypoxia. However, prolonged repression of mitochondrial metabolism appears to have pathogenic consequences in chronic peripheral vascular disease, and such a pathogenic metabolic shift has been implicated in the induction of a hyper-proliferative and anti-apoptotic vascular state in PH.[33] Thus, it follows that upregulation of miR-210 in the hypoxic pulmonary vasculature may represent a sentinel pathogenic event that triggers down-regulation of ISCU1/2, repression of mitochondrial metabolism, and resultant pathogenic vascular dysregulation. Yet, whether this hypoxia/miR-210/ISCU/mitochondria axis or other predicted hypoxamir-dependent functions are even active in PH is unknown and awaits validation. With the advent of improving technology by which to manipulate miRNA pharmacologically in vivo as well as the increasing availability of genetic knockout mice to validate more definitively these hypotheses, we expect that the full breadth of the roles of hypoxamirs in controlling PH pathogenesis will become even more apparent in the upcoming years.
Inflammatory and BMP-related miRNA in PH
Beyond hypoxia, inflammatory insults and deficiencies of BMP signaling represent other well-established triggers of PH that are also tightly associated with miRNA functions. Furthermore, substantial overlap of shared miRNA exists among these functional categories and with hypoxia (Tables 1 and 2). While the direct actions of many of these miRNA await verification, there is growing appreciation that TGF-β and BMP superfamily signaling, in particular, are intricately related to miRNA biology at multiple mechanistic levels. Such a mechanistic connection is an especially appealing focus for research in PH, given the known genetic predisposition of patients for PH who are haploinsufficient for the BMP receptor type II (BMPR2). In that setting, unlike other cytokines which tend to regulate miRNA at the transcriptional level, BMPR2-specific signaling upregulates miR-21 expression through uniquely modulating post-transcriptional processing of the premature miRNA form to the mature, active form.[34,35] In turn, miR-21 directly represses BMPR2 expression in an autoregulatory feedback loop.[23] However, because of the substantial number of shared and overlapping miRNA that target BMPR2 and related BMP-signaling molecules, it is likely that various combinations of miRNA with miR-21 control the overall direction of TGF/BMP signaling in vivo.
Consequently, in addition to miR-21, other miRNA have been identified as intrinsically linked to regulation of the BMP pathway, notably the miR-17-92 cluster. As one of the most well-studied clusters of miRNA, the miR-17-92 cluster controls cell development, apoptosis, and proliferation in numerous biological contexts,[36] and directly regulates angiogenic potential in vascular endothelial cells in vivo.[37] For two such members of this cluster, miR-17 and miR-20a, the BMPR2 transcript is a predicted target. Correspondingly, the interleukin-6/STAT3 pathway upregulates this cluster, leading to downregulation of BMPR2 expression in cultured pulmonary vascular cell types.[38] Based on those clues, it was recently reported that an inhibitor of miR-17 attenuated the severity of PH in chronically hypoxic mice and in rats treated with monocrotaline. Similarly, an inhibitor of miR-20a reduced PH in hypoxic mice, potentially through a mechanism involving the rescue of BMPR2 expression. However, BMPR2 is likely not the only relevant target for these miRNA in this disease context, as miR-17 inhibition also suppressed expression of another target gene, the cyclin-dependent kinase inhibitor 1A (P21), thus promoting a hyperproliferative phenotype in cultured pulmonary artery smooth muscle cells (PASMCs). Taken together, these studies indicate an active role of the miR-17-92 cluster in the control of PH. Yet, the shared functionality of these and other miRNA converging upon the BMP pathway highlights the need for better tools to predict and validate the coordinated roles of miRNA in PH, rather than studying the actions of an individual miRNA in isolation.
“-OMICS”-BASED DISCOVERY THROUGH HIGH-THROUGHPUT MIRNA EXPRESSION SCREENING IN PH
A pathway-based discovery approach is by its very nature biased by the analysis of previously studied miRNA functions in other contexts. In contrast, many investigators have pursued high-throughput miRNA expression screening as a more objective and complete method of identifying miRNA that are dysregulated in PH in vivo. In the first and most comprehensive study of its type to date, Caruso and colleagues performed a high-throughput miRNA array analysis of whole lung tissue derived from chronically hypoxic rats and monocrotaline-treated rats at serial time points during the pathogenesis of PH.[39] Of 350 miRNA tested, five were similarly dysregulated across both models, suggesting the centrality of these miRNA in disease pathogenesis. Specifically, these included let-7f, miR-22, and miR-30c, which were downregulated as well as miR-322 and miR-451, which were upregulated. Conversely, perhaps reflecting separate pathways of disease progression unique to only certain models of disease, some notable discrepancies in miRNA expression profiles were observed comparing hypoxia-dependent and monocrotaline-dependent contexts. For example, miR-21 and let-7a expressions were significantly reduced only in monocrotaline-treated rats. Nonetheless, based on their robust level of dysregulation in one or both animal models, miR-322, miR-451, miR-21, miR-22, miR-30c, let-7f, and let-7a were selected for validation and further analysis. In cultured pulmonary vascular cells, stimulation with PH-associated mediators such as hypoxia, BMP4, or TGF-β, resulted in dysregulation of these miRNA in a similar pattern to those observed in vivo. Through RNA extraction and reverse transcription-polymerase chain reaction (RT-PCR) of the paraffin-embedded lungs of patients suffering from idiopathic pulmonary arterial hypertension (IPAH), miR-21 was shown to be downregulated while miR-451 was upregulated in the diseased lungs. Subsequently, Yang and colleagues have reported the results of a separate miRNA expression screen from lungs of mice chronically treated with hypoxia, which also revealed alterations in miR-21 and miR-451. Moreover, miR-210 and miR-144 were found to be upregulated by > 2-fold compared with the normoxic mouse lung in addition to others with expression levels changing by < 2-fold.[40] In total, these findings represent the first and most comprehensive screens of miRNA reported in this disease; however, none of these data sets were primarily based on human disease tissue.
Notably, since these results represent the net changes in a heterogeneous mixture of pulmonary cell types (e.g., whole lung), these profiles do not necessarily reflect the exact miRNA profiles in the specific diseased pulmonary vasculature in question. This may partially explain the fact that some of these same miRNA, such as miR-21, have since been found to manifest opposite expression changes in the diseased pulmonary vasculature in both rodents and humans.[23,35,40] This is especially emphasized in a recent study by Bockmeyer and colleagues comparing miRNA expression via in situ staining of plexiform lesions in human PH lung.[41] From the study of 12 PH patient samples as compared with eight healthy controls, an upregulation of miR-21 along with miR-126 was noted in plexiform lesions as compared with a downregulation of miR-204 and miR-143/145. The mechanistic significance of miR-21 and miR-204 have been further elucidated (as described below), but the importance of these other dysregulated miRNA in PH remains unclear.
miR-204 controls PH
A similar high-throughput miRNA screening method was utilized by Courboulin and colleagues in studying cultured ex vivo PASMCs obtained from patients suffering from IPAH as compared with healthy individuals.[42] Of those found to be dynamically altered in IPAH-derived PASMCs, specific expression profiles were confirmed in lung biopsies derived from IPAH patients. In total, six miRNA were found to be consistently upregulated in disease, including miR-138, miR-367, miR-276, miR-302b, miR-145, and miR-450a, while miR-204 was the only miRNA exhibiting substantial downregulation. Depressed miR-204 expression has been previously observed in cancer cells and is associated with potassium homeostasis and membrane depolarization,[43,44] leading to a hyper-proliferative state. Correspondingly, in cultured nondiseased human PASMCs, miR-204 inhibition promoted a hyper-proliferative and anti-apoptotic cellular phenotype, consistent with primary IPAH-derived PASMCs in culture. Based on miRNA target algorithm predictions, miR-204 was found to target directly and repress the Src activating gene SHP2. Following inhibition of miR-204 in cultured PASMCs, the authors found that SHP2 is upregulated, thus driving Src activity, reflected by increased STAT3 phosphorylation and NFATc2 activity. Activity of this pathway was successfully suppressed in PAH-derived PASMCs in the presence of a miR-204 mimic oligonucleotide. In vivo, decreased miR-204 expression levels were reported in the lungs of chronically hypoxic mice and monocrotaline-treated rats. As final proof, hemodynamic and histological indices of monocrotaline-induced PH were pharmacologically reversed by intratracheal delivery of a synthetic miR-204 mimic, accompanied by downregulation of SHP2 and suppressed Src activity in the lungs of these miR-204-treated rats. Thus, through omics-based discovery and substantial mechanistic validation, this study was the first to reveal a miR-204/Src-STAT3/NFAT axis that controls the development of PAH and was the first to suggest a single miRNA as a viable therapeutic target in PH (Fig. 3).
Figure 3.
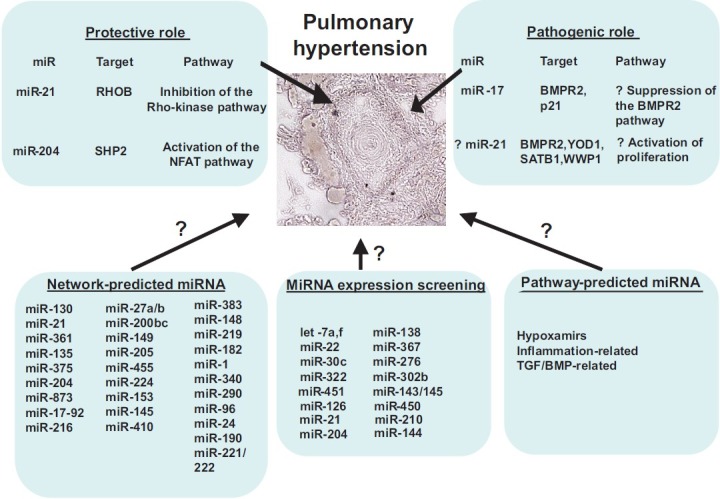
Predicted and established actions of miRNA in PH. MiRNA are listed that carry validated mechanisms of action in controlling PH in rodents and humans, as categorized by protective or pathogenic roles. As identified by network-based, “-omics”-based, or pathway-based approaches, additional miRNA are anticipated to hold critical regulatory roles in this disease
SYSTEMS-BASED DISCOVERY THROUGH NETWORK BIOLOGY IN PH
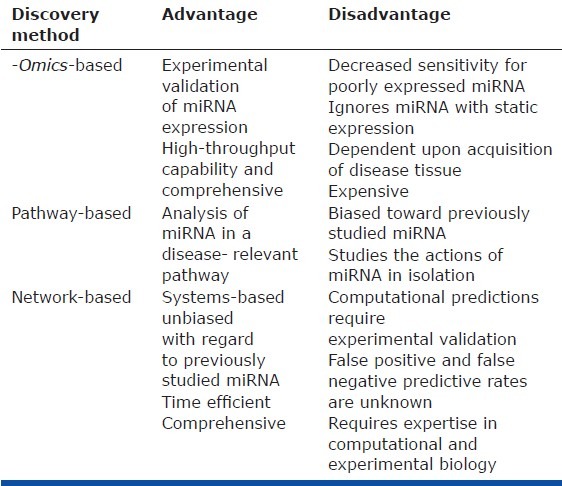
Although “-omics-based” approaches to miRNA discovery are useful, several caveats exist (Table 3). First, currently available technology for high-throughput miRNA screening is improving but still limited by sensitivity, thus preventing detection of poorly expressed miRNA that may nonetheless carry important biological functions. Second, because of the difficulty in direct sampling of PH-diseased tissue in small pulmonary arterioles, high-throughput screening from human samples either has entailed profiling of the whole lung or has necessitated substantial manipulation of lung tissue in order to obtain a purified population of diseased vascular cells. In the former case, analysis in whole lung biopsies obtained from PH patients is not adequately reflective of the pulmonary vascular environment, given the presence of heterogeneous cell populations. In the latter case, prolonged manipulation and/or culturing ex vivo risks the alteration of miRNA and mRNA expression patterns. Thus, in both scenarios, the resulting miRNA expression patterns may not accurately reflect the levels of specific miRNA in the relevant diseased tissue niches in vivo. Third, because miRNA-to-target stoichiometry is an important factor dictating the efficiency of target gene repression, high-throughput expression screens entirely miss changes in miRNA function (i.e., based on alterations of their target gene pool) in the absence of alterations in miRNA expression directly. Finally, a persistent problem of PH-relevant research guided by expression screening of any fashion stems from the paucity of available human tissue reflective of the inception and progression of disease. Instead, human lung samples are typically collected only during transplant or autopsy at severe/end-stage PH. Compounding this technical challenge, no small animal model of PH adequately recapitulates all aspects of human disease,[45] limiting the utility of expression screening during disease progression. Clearly, investigation of the molecular pathways active during the early stages would give novel insight into the largely unknown pathogenic events that coordinate the initial stages of human PH. Yet, given these drawbacks of miRNA expression screening in PH, alternative methods are needed to identify more comprehensively specific miRNA relevant to pathogenesis.
Table 3.
Comparison of current miRNA discovery methods in PH
In that vein, the nascent paradigm of “network medicine” can offer insight into the pathogenic behavior of complex molecular interconnections that traditional methods fail to recognize, but whose utility in human disease is just emerging.[46,47] The study of miRNA and their overlapping networks of direct target genes may provide an ideal context to apply such network theory. Previously, we designed a bioinformatics approach to identify miRNA that robustly control the development and/or progression of PH, based on the evolving concept of shared functionality of multiple targets for a given miRNA (Fig. 4A). Target prediction algorithms such as TargetScan 5 (Conserved)[12,13] suggest that the repertoire of targets recognized by a single miRNA may rely upon a highly coordinated set of actions to orchestrate robust effects on a given biological phenotype. Likewise, in the cardiovascular system, examples of miRNA with functionally definable sets of direct targets are prevalent, such as the miR-29 family, which targets multiple collagen genes and matrix metalloproteinases in order to inhibit cardiac fibrosis.[48] Such capabilities suggest the need to consider the multiplicity, cooperativity, and redundancy of miRNA targets and how they work as an integrated network. Consequently, attempts have already commenced to identify comprehensively such cohesive networks of miRNA target genes in order to predict pervasive biological functions of particular miRNA.[49,50] Overall, there may be an advantage of such miRNA enrichment within a biological network in order to ensure the high fidelity of “buffering” of the cellular response to biologic or pathologic perturbations. Other more subtle reasons for such complexity may also emerge as we study these networks more fully.
Figure 4.
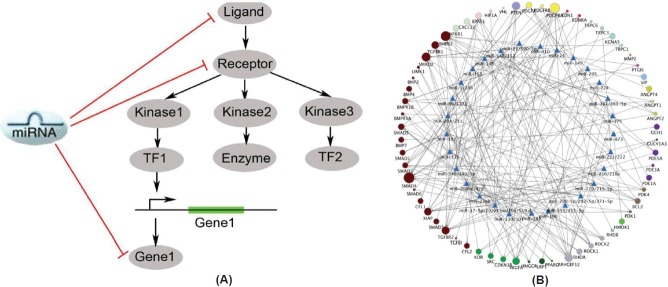
Network-based approaches to identify coordinate miRNA function in PH. (A) Shared functionality of multiple targets of a given miRNA. MiRNA are capable of sensitively controlling pathway function via the simultaneous regulation of multiple genes within its signaling cascade. (B) A network biology model to help elucidate the indirect involvement of microRNA in pulmonary hypertension. A bipartite map of the predicted associations by which multiple miRNA (blue triangles, inner circle) may regulate a network of gene targets (circles, outer circle colors represent distinct functional pathways) associated with pulmonary hypertension. Size of node is proportional to the number of miRNA groups (among those in the inner circle) predicted to target that particular gene
Based on this concept of shared functionality among miRNA and their targets, we utilized a network-based approach to identify systematically and rank those miRNA most likely to regulate PH by their predicted recognition of multiple targets in the same functional network of PH-associated genes. Specifically, a list of 131 genes was curated from the scientific literature of molecular factors that have been proven to influence PH. Using a set of consolidated databases of molecular interactions,[51–59] a network map, which we termed the “PH-network,” was created, representing the functional interconnectivity among the PH-relevant target genes. Importantly, both the total number of direct interconnections and the largest connected component (LCC) of the PH-network were substantially larger than those generated from random gene associations, thus indicating its propensity to act in a coordinated fashion and presenting a reasonable platform with which to rank PH-relevant miRNA preferentially regulating related gene targets. Using the highly sensitive and specific miRNA target prediction algorithm, TargetScan 5 (Conserved),[12] a hypergeometric analysis was then performed to rank miRNA according to the proportion of their predicted targets found within the PH-network. In doing so, 29 miRNA groups were found to have a less than 5% probability that the overlap of their predicted target list with the PH-network occurred by chance (Fig. 3) and, thus, are most likely to coordinate pathogenic effects within the PH-network.
Multiple pieces of evidence support the accuracy of these predictions. First, the PH-relevant targets of these 29 miRNA groups encompass pathways that are associated, by varying degrees, with known upstream triggers of PH,[3] including hypoxia, inflammation, and TGF/BMP signaling. Many of these miRNA had even been previously implicated in these signaling pathways. Furthermore, despite the nascent nature of this field, a number of miRNA that had been identified as direct mediators of PH independent of a network approach are also included in this list. These include miR-204[42] and the miR-17-92 family.[60] Additionally encompassed in this list are miR-145[61] and miR-21,[34,62,63] both of which were associated previously with cellular phenotypes relevant to PH pathology. Furthermore, of the 10 top-ranked miRNA, we found that five are expressed in the lung (miR-21, miR-20a, miR-27a, miR-130a, and miR-375) and are dynamically regulated in at least one rodent model of PH. Moreover, given the network predictions of both unique and shared miRNA targets in the PH network, these findings support the synergistic actions of miRNA to influence PH coordinately (Fig. 4B). Taken together, these data emphasize the utility of such bioinformatics analyses in expanding the predictive power regarding miRNA biology. The accuracy of this approach has deep roots in the connectedness of the PH network on which they were based.
miR-21 controls PH
To further validate prospectively and independently one of these predictions, we chose to study mechanistically miR-21 due to its high ranking on the list (#2 ranking based on P-value) and the availability of reagents to test the hypothesis. Notably, miR-21 is an established molecular regulator in the pathogenesis of many human diseases,[64] including cancer and cardiovascular diseases, such as cardiac fibrosis and heart failure,[65] aortic aneurysm development,[66] and peripheral vascular remodeling.[62] It is upregulated in various cellular contexts by upstream triggers of PH, including hypoxia,[25,67] TGF-β and BMP signaling,[34] and proinflammatory cytokines such as interleukin-6 (IL-6).[68,69] With relevance to PH and the network-based analysis, miR-21 was predicted to regulate PH-network targets central to BMP and Rho/Rho kinase signaling, as well as functional pathways associated with hypoxia, inflammation, and genetic haploinsufficiency of BMPR2. Accordingly, we found that hypoxia and BMPR2 signaling independently induced miR-21 in cultured pulmonary arterial endothelial cells. Furthermore, miR-21 directly repressed RhoB expression and Rho kinase activity, leading to molecular reprogramming linked to decreased angiogenesis and vasodilation. BMPRII was also predicted as a key PH-relevant target, and miR-21 subtly repressed BMPRII expression, as well. However, unlike RhoB, BMPRII was predicted as a target of several PH-relevant miRNA, making miR-21's precise control of the BMP pathway more complex. Importantly, miR-21 was upregulated specifically in the diseased pulmonary vasculature from several rodent models of PH and in humans with PH. Upon induction of disease in miR-21-null mice, RhoB expression and Rho-kinase activity were increased, accompanied by exaggerated histological and hemodynamic manifestations of PH. Notably, since that report, the importance of RhoB as a pathogenic factor in PH was independently confirmed by Wojciak-Stothard and colleagues.[70] Thus, based on network-based bioinformatics, miR-21 was found to be regulated by hypoxia, inflammation, and BMP-dependent signaling, leading to direct repression of Rho kinase activation, and perhaps other pathways, as a protective brake against the development of PH in vivo.
In many respects, the integration of various PH-relevant stimuli by miR-21 correlates with the pleiotropic activities of this miRNA.[64] Importantly, however, miR-21-dependent protection against PH is somewhat surprising, given its actions as an anti-apoptotic, pro-proliferative “oncomiR,” directly targeting a number of tumor suppressor targets such as PDCD4 to facilitate cellular transformation in cancer.[64] Yet, in vivo, PDCD4 in the pulmonary vasculature is not substantially altered in miR-21-null mice with PH,[23] consistent with independent data of cardiac-specific PDCD4 in a separate miR-21-null mouse.[71] Thus, control of each direct target by miR-21 clearly differs depending on the biological context, thus giving credence to the idea that miR-21 can hold dramatically different functions, depending on the specific cell type or milieu. Notably, a protective effect of miR-21 against PH in the pulmonary vasculature is further supported by independent mechanistic studies demonstrating that miR-21 can drive a nonproliferative and nonmigratory “contractile” phenotype in PASMCs[34,72] and PAECs.[35] Correspondingly, mutations in the BMP signaling pathway in both humans[35] and mice[23] can decrease miR-21 expression and subsequently predispose to PH. Thus, these data support a possible mechanism explaining how genetic deficiencies in BMP signaling predispose to PAH: via decreased miR-21 expression in response to injury and loss of protection against disease progression (Fig. 3).
Although the actions of miR-21 in controlling PH are substantial, reports of its patterns of expression and functions have not been entirely consistent. For example, Caruso and colleagues have reported a downregulation, rather than upregulation, of miR-21 in lung homogenate in rodent and human PH.[39] Differences in the clinical context of disease, especially in regard to BMPR2 genotype as mentioned above, may partially explain these results. Furthermore, some studies have indicated that miR-21 can drive a pro-proliferative state in hypoxic PASMCs in culture.[63] A pro-proliferative function has also been proposed in relation to the use of antisense inhibitors of miR-21 in vivo to decrease pulmonary vascular remodeling in hypoxic rodents.[40] The in vivo use of miRNA inhibitors may partially explain these discrepancies, as the particular chemical makeup of these oligonucleotides may have dramatic differences in efficiency and perhaps specificity of action.[73] In vivo inhibitors for miR-21, in particular, have recently come under substantial scrutiny, in light of discrepancies in the study of cardiac fibrosis using different inhibitor technologies as opposed to the “gold-standard” of genetic knockout lines.[74] Alternatively, given the pleiotropic nature of this molecule, it would not be entirely surprising that slight alterations in disease context may ultimately allow for drastically different miR-21-dependent actions. Thus, although the reasons for these discrepancies are not fully explained, it is clear that miR-21 carries a central, albeit complex, function in controlling the progression of PH.
CONCLUSIONS AND FUTURE DIRECTIONS
Based on their ubiquitous and critical actions in other biological systems and diseases, miRNA are predicted to regulate numerous essential molecular pathways important in the genesis, progression, and perhaps attenuation or prevention of PH. Yet, the actions of only a few miRNA have been experimentally validated for this disease (Fig. 3). While a substantial opportunity for scientific discovery certainly exists over the next years, challenges still remain to identify rigorously the networks of miRNA and their targets that carry the most robust actions in the control of PH in vivo.
Importantly, although already invaluable to comprehensively assessing the systems-wide effects of miRNA in other human diseases (i.e., cancer), pathway-based or high-throughput “-omics” technology in PH is currently limited by the inaccessibility of diseased vasculature compounded by the lack of a robust and genetically tractable animal model of PH that recapitulates human disease (Table 3). As a result, further applications of network biology may prove an efficient scientific approach in identifying novel miRNA and target genes/pathways. For example, the so-called “linkage” method in network biology enables the identification of unique factors important in human disease via their direct connection to known disease genes.[46] Thus, a simple extension of current PH-gene network[75] to include all first degree (i.e., direct) interacting genes may uncover novel disease genes as well as unique miRNA which target them that are centrally involved in disease pathogenesis. Consequently, further in silico pursuit of network biology that can guide traditional experimental methods should facilitate more rapid characterization of critical points of coordinate miRNA regulation in PH, many of which would be missed by reductionistic methods alone.
In addition to characterizing the direct functions of candidate miRNA in PH, another challenge includes the determination of the exact cellular location of these actions in vivo. Given the important but distinct roles of endothelial, smooth muscle, fibroblast, inflammatory cells, platelets, and even progenitor cells in the diseased pulmonary vasculature, the unique miRNA profile and consequent target engagement may differ in each cellular niche and likely factor heavily in pathogenesis. The growing population of tissue-specific (Cre-lox) genetic knockout lines for specific miRNA[76] represent the most rigorous reagents to study these vascular niches.[74] However, these studies are time consuming and expensive, and, thus, have yet to be pursued extensively in the vasculature. Furthermore, despite the growing use in vivo of either oligonucleotide inhibitors or mimics of miRNA, current delivery methods cannot dictate tissue or cellular specificity.
The function of “circulating” miRNA in PH also awaits definitive characterization. Recent studies suggest that extracellular forms of miRNA are released into the circulating bloodstream and could be transported into closely neighboring recipient cells in culture[77–80] and perhaps in vivo.[81] Changes in miR-21 have been reported in blood samples from human PH.[39] However, it remains unknown whether dynamic changes in plasma-based miRNA may be used as PH-specific biomarkers. Perhaps, more importantly, it is unclear whether these extracellular forms may act as paracrine messengers communicating among diseased vascular cell types or even endocrine messengers shuttling among the pulmonary vasculature and distant anatomic sites.
Finally, if the importance of miRNA in the pathobiology of PH is realized, therapeutic targeting of miRNA in the pulmonary vasculature should also rapidly evolve. Presently, the most promising results have stemmed from the use of chemically modified oligonucleotide antisense inhibitors (e.g., antagomirs or anti-miRs). Yet, current drawbacks of such inhibitors include the inability to target-specific vascular tissue and the need to utilize high concentrations for vascular delivery with substantial risk for off-target effects. The rapid pace of development in miRNA therapeutics should overcome these deficits. However, it remains to be seen if such tissue and concentration specificity can allow not only for fine control of a single miRNA but also for such control over a network of miRNA that converges upon the same pulmonary vascular phenotype.
In summary, over the next years, we anticipate a substantial increase in our understanding of the molecular actions of various miRNA in PH. Given their pleiotropic actions in modulating multiple gene targets, we suspect that many of these may even serve as central molecular “lynchpins” connecting diverse upstream disease triggers to a common pathologic pulmonary vascular result. By better integrating network and systems biology with evolving technologies in cell culture and in vivo experimentation with miRNA, this discovery process can be further accelerated in order to identify more comprehensively all relevant miRNA important in this highly morbid condition.
ACKNOWLEDGMENTS
We thank Ms. Stephanie Tribuna for expert administrative assistance and Dr. Sabrina Rabello for aid in constructing the bipartite map in Figure 4B.
Footnotes
Source of Support: This work was supported in part by the NIH, the Pulmonary Hypertension Association, Gilead Sciences, and the Lerner, Harris, and Watkins funds (Stephen Y Chan); NIH grants HL061795, HL48743, HL107192, HL70819, and HL108630 (Joseph Loscalzo).
Conflict of Interest: None declared.
REFERENCES
- 1.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C.elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–46. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 3.Chan SY, Loscalzo J. Pathogenic mechanisms of pulmonary arterial hypertension. J Mol Cell Cardiol. 2008;44:14–30. doi: 10.1016/j.yjmcc.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrell NW, Adnot S, Archer SL, Dupuis J, Jones PL, MacLean MR, et al. Cellular and molecular basis of pulmonary arterial hypertension. J Am Coll Cardiol. 2009;54:S20–31. doi: 10.1016/j.jacc.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuder RM. Pathology of pulmonary arterial hypertension. Semin Respir Crit Care Med. 2009;30:376–85. doi: 10.1055/s-0029-1233307. [DOI] [PubMed] [Google Scholar]
- 6.Wu X, Chang M, Mitsialis S, Kourembanas S. Hypoxia regulates bone morphogenetic protein signaling through C-terminal-binding protein 1. Circ Res. 2006;99:240–7. doi: 10.1161/01.RES.0000237021.65103.24. [DOI] [PubMed] [Google Scholar]
- 7.Hagen M, Fagan K, Steudel W, Carr M, Lane K, Rodman DM, et al. Interaction of interleukin-6 and the BMP pathway in pulmonary smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2007;292:L1473–9. doi: 10.1152/ajplung.00197.2006. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–42. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 11.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimson A, Farh K, Johnston W, Garrett-Engele P, Lim L, Bartel D. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, et al. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–78. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdellatif M. Differential expression of microRNAs in different disease states. Circ Res. 2012;110:638–50. doi: 10.1161/CIRCRESAHA.111.247437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrell NW, Yang X, Upton PD, Jourdan KB, Morgan N, Sheares KK, et al. Altered growth responses of pulmonary artery smooth muscle cells from patients with primary pulmonary hypertension to transforming growth factor-beta(1) and bone morphogenetic proteins. Circulation. 2001;104:790–5. doi: 10.1161/hc3201.094152. [DOI] [PubMed] [Google Scholar]
- 17.Takemoto M, Sun J, Hiroki J, Shimokawa H, Liao J. Rho-kinase mediates hypoxia-induced downregulation of endothelial nitric oxide synthase. Circulation. 2002;106:57–62. doi: 10.1161/01.cir.0000020682.73694.ab. [DOI] [PubMed] [Google Scholar]
- 18.Eddahibi S, Humbert M, Fadel E, Raffestin B, Darmon M, Capron F, et al. Serotonin transporter overexpression is responsible for pulmonary artery smooth muscle hyperplasia in primary pulmonary hypertension. J Clin Invest. 2001;108:1141–50. doi: 10.1172/JCI12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Zhang X, Leathers R, Makino A, Huang C, Parsa P, et al. Notch3 signaling promotes the development of pulmonary arterial hypertension. Nat Med. 2009;15:1289–97. doi: 10.1038/nm.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen AH, Hanson K, Morris K, Fouty B, McMurty IF, Clarke W, et al. Inhibition of cyclic 3′-5′-guanosine monophosphate-specific phosphodiesterase selectively vasodilates the pulmonary circulation in chronically hypoxic rats. J Clin Invest. 1996;97:172–9. doi: 10.1172/JCI118386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giaid A, Yanagisawa M, Langleben D, Michel RP, Levy R, Shennib H, et al. Expression of endothelin-1 in the lungs of patients with pulmonary hypertension. N Engl J Med. 1993;328:1732–9. doi: 10.1056/NEJM199306173282402. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan CC, Du L, Chu D, Cho AJ, Kido M, Wolf PL, et al. Induction of pulmonary hypertension by an angiopoietin 1/TIE2/serotonin pathway. Proc Natl Acad Sci, USA. 2003;100:12331–6. doi: 10.1073/pnas.1933740100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parikh VN, Chan SY. Inflammatory Mechanisms in Pulmonary hypertension. In: Wang YX, editor. Recent Advances in Pulmonary Vascular Biology. Kerala, India: Research Signpost; 2012. [Google Scholar]
- 24.Chan SY, Loscalzo J. MicroRNA-210: A unique and pleiotropic hypoxamir. Cell Cycle. 2010;9:107–83. doi: 10.4161/cc.9.6.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–67. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taganov K, Boldin M, Chang K, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun X, Icli B, Wara AK, Belkin N, He S, Kobzik L, et al. MicroRNA-181b regulates NF-kappaB-mediated vascular inflammation. J Clin Invest. 2012;122:1973–90. doi: 10.1172/JCI61495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D, et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. J Clin Invest. 2010;120:4141–54. doi: 10.1172/JCI42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu S, Huang M, Li Z, Jia F, Ghosh Z, Lijkwan MA, et al. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation. 2010;122:S124–31. doi: 10.1161/CIRCULATIONAHA.109.928424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biswas S, Roy S, Banerjee J, Hussain SR, Khanna S, Meenakshisundaram G, et al. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc Natl Acad Sci U S A. 2010;107:6976–81. doi: 10.1073/pnas.1001653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35:856–67. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10:273–84. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuder RM, Davis LA, Graham BB. Targeting energetic metabolism: a new frontier in the pathogenesis and treatment of pulmonary hypertension. Am J Respir Crit Care Med. 2011;185:260–6. doi: 10.1164/rccm.201108-1536PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drake KM, Zygmunt D, Mavrakis L, Harbor P, Wang L, Comhair SA, et al. Altered microRNA Processing in Heritable Pulmonary Arterial Hypertension: an Important Role for Smad-8. Am J Respir Crit Care Med. 2011;184:1400–8. doi: 10.1164/rccm.201106-1130OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–22. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–3. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 38.Brock M, Trenkmann M, Gay RE, Michel BA, Gay S, Fischler M, et al. Interleukin-6 modulates the expression of the bone morphogenic protein receptor type II through a novel STAT3-microRNA cluster 17/92 pathway. Circ Res. 2009;104:1184–91. doi: 10.1161/CIRCRESAHA.109.197491. [DOI] [PubMed] [Google Scholar]
- 39.Caruso P, MacLean MR, Khanin R, McClure J, Soon E, Southgate M, et al. Dynamic changes in lung microRNA profiles during the development of pulmonary hypertension due to chronic hypoxia and monocrotaline. Arterioscler Thromb Vasc Biol. 2010;30:716–23. doi: 10.1161/ATVBAHA.109.202028. [DOI] [PubMed] [Google Scholar]
- 40.Yang S, Banerjee S, Freitas Ad, Cui H, Xie N, Abraham E, et al. miR-21 regulates chronic hypoxia-induced pulmonary vascular remodeling. Am J Physiol Lung Cell Mol Physiol. 2012;302:L521–2s9. doi: 10.1152/ajplung.00316.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bockmeyer CL, Maegel L, Janciauskiene S, Rische J, Lehmann U, Maus UA, et al. Plexiform vasculopathy of severe pulmonary arterial hypertension and microRNA expression. J Heart Lung Transplant. 2012;31:764–72. doi: 10.1016/j.healun.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 42.Courboulin A, Paulin R, Giguère NJ, Saksouk N, Perreault T, Meloche J, et al. Role for miR-204 in human pulmonary arterial hypertension. J Exp Med. 2011;208:535–48. doi: 10.1084/jem.20101812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee Y, Yang X, Huang Y, Fan H, Zhang Q, Wu Y, et al. Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLoS Comput Biol. 2010;6:e1000730. doi: 10.1371/journal.pcbi.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang FE, Zhang C, Maminishkis A, Dong L, Zhi C, Li R, et al. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24:1552–71. doi: 10.1096/fj.08-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryan J, Bloch K, Archer SL. Rodent models of pulmonary hypertension: Harmonisation with the world health organisation's categorisation of human PH. Int J Clin Pract Suppl. 2011;172:15–34. doi: 10.1111/j.1742-1241.2011.02710.x. [DOI] [PubMed] [Google Scholar]
- 46.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: A network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan SY, White K, Loscalzo J. Deciphering the molecular basis of human cardiovascular disease through network biology. Curr Opin Cardiol. 2012;27:202–9. doi: 10.1097/HCO.0b013e3283515b31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–32. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shirdel EA, Xie W, Mak TW, Jurisica I. NAViGaTing the micronome--using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS One. 2011;6:e17429. doi: 10.1371/journal.pone.0017429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gennarino VA, D’Angelo G, Dharmalingam G, Fernandez S, Russolillo G, Sanges R, et al. Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res. 2012;22:1163–72. doi: 10.1101/gr.130435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–8. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 52.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–68. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 53.Cusick ME, Yu H, Smolyar A, Venkatesan K, Carvunis AR, Simonis N, et al. Literature-curated protein interaction datasets. Nat Methods. 2009;6:39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–31. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, et al. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2009;38:D532–9. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wingender E, Chen X, Fricke E, Geffers R, Hehl R, Liebich I, et al. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001;29:281–3. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diella F, Gould CM, Chica C, Via A, Gibson TJ. Phospho. ELM: A database of phosphorylation sites--update 2008. Nucleic Acids Res. 2008;36:D240–4. doi: 10.1093/nar/gkm772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–61. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 59.Hong KH, Lee YJ, Lee E, Park SO, Han C, Beppu H, et al. Genetic ablation of the BMPR2 gene in pulmonary endothelium is sufficient to predispose to pulmonary arterial hypertension. Circulation. 2008;118:722–30. doi: 10.1161/CIRCULATIONAHA.107.736801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pullamsetti SS, Doebele C, Fischer A, Savai R, Kojonazarov B, Dahal BK, et al. Inhibition of microRNA-17 improves lung and heart function in experimental pulmonary hypertension. Am J Respir Crit Care Med. 2011;185:409–19. doi: 10.1164/rccm.201106-1093OC. [DOI] [PubMed] [Google Scholar]
- 61.Joshi SR, Abe K, Oka M, McMurtry IF, Gerthoffer WT. Mir-145 in vascular smooth muscle phenotype in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2011;183:A3448. [Google Scholar]
- 62.Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–88. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 63.Sarkar J, Gou D, Turaka P, Viktorova E, Ramchandran R, Raj JU. MicroRNA-21 plays a role in hypoxia-mediated pulmonary artery smooth muscle cell proliferation and migration. Am J Physiol Lung Cell Mol Physiol. 2010;299:L861–71. doi: 10.1152/ajplung.00201.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–4. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 66.Maegdefessel L, Azuma J, Toh R, Deng A, Merk DR, Raiesdana A, et al. MicroRNA-21 blocks abdominal aortic aneurysm development and nicotine-augmented expansion. Sci Transl Med. 2012;4:122ra22. doi: 10.1126/scitranslmed.3003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gorospe M, Tominaga K, Wu X, Fahling M, Ivan M. Post-transcriptional control of the hypoxic response by RNA-binding proteins and microRNAs. Front Mol Neurosci. 2011;4:7. doi: 10.3389/fnmol.2011.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Löffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermüller J, Kretzschmar AK, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–3. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 69.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wojciak-Stothard B, Zhao L, Oliver E, Dubois O, Wu Y, Kardassis D, et al. Role of RhoB in the regulation of pulmonary endothelial and smooth muscle cell responses to hypoxia. Circ Res. 2012;110:1423–34. doi: 10.1161/CIRCRESAHA.112.264473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, et al. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest. 2010;120:3912–6. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kang H, Davis-Dusenbery BN, Nguyen PH, Lal A, Lieberman J, Van Aelst L, et al. Bone morphogenetic protein 4 promotes vascular smooth muscle contractility by activating microRNA-21 (miR-21), which down-regulates expression of family of dedicator of cytokinesis (DOCK) proteins. J Biol Chem. 2012;287:3976–86. doi: 10.1074/jbc.M111.303156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thum T, Chau N, Bhat B, Gupta SK, Linsley PS, Bauersachs J, et al. Comparison of different miR-21 inhibitor chemistries in a cardiac disease model. J Clin Invest. 2011;121:461–2. doi: 10.1172/JCI45938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morrisey EE. The magic and mystery of miR-21. J Clin Invest. 2010;120:3817–9. doi: 10.1172/JCI44596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parikh VN, Jin RC, Rabello S, Gulbahce N, White K, Hale A, et al. MicroRNA-21 Integrates pathogenic signaling to control pulmonary hypertension: Results of a network bioinformatics approach. Circulation. 2012;125:1520–32. doi: 10.1161/CIRCULATIONAHA.111.060269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Prosser HM, Koike-Yusa H, Cooper JD, Law FC, Bradley A. A resource of vectors and ES cells for targeted deletion of microRNAs in mice. Nat Biotechnol. 2011;29:840–5. doi: 10.1038/nbt.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133–44. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 78.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A. 2010;107:6328–33. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hergenreider E, Heydt S, Tréguer K, Boettger T, Horrevoets AJ, Zeiher AM, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14:249–56. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 82.van Rooij E, Olson E. MicroRNAs: Powerful new regulators of heart disease and provocative therapeutic targets. J Clin Investig. 2007;117:2369–76. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–8. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]