Abstract
Two yeast genes, FRE1 and FRE2 (encoding Fe(III) reductases) were placed under the control of the cauliflower mosaic virus 35S promoter and introduced into tobacco (Nicotiana tabacum L.) via Agrobacterium tumefaciens-mediated transformation. Homozygous lines containing FRE1, FRE2, or FRE1 plus FRE2 were generated. Northern-blot analyses revealed mRNA of two different sizes in FRE1 lines, whereas all FRE2 lines had mRNA only of the expected length. Fe(III) reduction, chlorophyll contents, and Fe levels were determined in transgenic and control plants under Fe-sufficient and Fe-deficient conditions. In a normal growth environment, the highest root Fe(III) reduction, 4-fold higher than in controls, occurred in the double transformant (FRE1 + FRE2). Elevated Fe(III) reduction was also observed in all FRE2 and some FRE1 lines. The increased Fe(III) reduction occurred along the entire length of the roots and on shoot sections. FRE2 and double transformants were more tolerant to Fe deficiency in hydroponic culture, as shown by higher chlorophyll and Fe concentrations in younger leaves, whereas FRE1 transformants did not differ from the controls. Overall, the beneficial effects of FRE2 were consistent, suggesting that FRE2 may be used to improve Fe efficiency in crop plants.
Fe uptake and storage are highly regulated processes. Because soils contain mainly insoluble Fe(III) oxides and hydroxides, plants have developed adaptive mechanisms to make Fe more available for uptake (for review, see Römheld, 1987; Guerinot and Yi, 1994). Two key mechanisms used by dicots and nongramineous monocots (Strategy I plants) are proton extrusion by activation of an ATPase-driven proton pump, thereby promoting solubility of Fe(III), and Fe(III) reduction by plasma membrane-bound Fe(III) reductases. Under Fe deficiency, elevated activity of Fe(III) reductases can be detected in specialized zones near the root tips.
The importance of Fe(III) reductases in Fe acquisition suggests that manipulation or addition of genes encoding such enzymes in plants may present an avenue for enhancing Fe uptake. Several Fe(III) reductases have been identified in plants using techniques that allow separation of PMs from other membrane fractions (for review, see Moog and Brüggemann, 1994). Recently, plant genes encoding putative Fe(III) reductases have been identified (Robinson et al., 1997a).
As in plants using Strategy I, reduction of Fe(III) is also essential for the utilization of Fe in yeast (Lesuisse et al., 1987). An Fe(III) reductase has been isolated from yeast PM fractions (Lesuisse et al., 1990) and the activity of the enzyme was increased upon Fe depletion. Two Fe(III) reductase genes, FRE1 and FRE2, have been isolated from the yeast Saccharomyces cerevisiae (Dancis et al., 1990, 1992; Georgatsou and Alexandraki, 1994) and a related gene, Frp1, has been isolated from Saccharomyces pombe (Roman et al., 1993). Dancis et al. (1992) identified FRE1 via complementation of a mutant yeast lacking externally directed PM reductase activity. The second Fe(III) reductase gene of S. cerevisiae, FRE2, was identified during sequencing of yeast chromosome XI (Georgatsou and Alexandraki, 1994). The combination of FRE1 and FRE2 was shown to account for nearly all membrane-associated Fe(III) reductase activity in this yeast (Georgatsou and Alexandraki, 1994). Even though FRE1 and FRE2 encode enzymes with similar functions, they do not show significant similarity at the nucleotide level and their deduced amino acid sequences have only 24.5% identity.
Because yeast genes can be successfully expressed in plants (Colau et al., 1987; Von Schaewen et al., 1990), incorporation of the FRE genes into the plant genome may lead to the formation of functional proteins and, consequently, enhanced Fe(III) reduction. Here we report on the genetic transformation of tobacco (Nicotiana tabacum L.) with FRE1 and FRE2 and the characterization of transformed plants with regard to Fe(III) reduction, chlorophyll content (SPAD reading), and Fe accumulation under Fe-sufficient and Fe-deficient conditions.
MATERIALS AND METHODS
Cloning of FRE1 and FRE2 into Plasmids
FRE1 was obtained from high-Mr Saccharomyces cerevisiae genomic DNA (provided by S. Johnston, The University of Texas Southwestern Medical School, Dallas) by PCR (92°C, 2 min; 52°C, 2 min; 72°C, 3 min; 40 cycles) using a 5′ primer with added BamHI and NcoI sites (CCGGGGATCCATGGTTAGAACCCGTGTATTATTCTGC) and a 3′ primer with an added EcoRI site (TTATGAATTCGGGGCCTTACCATGTAAAACTTTCTTC). The PCR product (2.1 kb) was cloned into the SmaI site of pUC18. Plasmid was digested with NcoI and HindIII to release the insert, which was then cloned into PTC:MT-1 (Carrington et al., 1987) to gain a 5′ Kozak sequence (Kozak, 1986). PTC:MT-1 plasmid with the FRE1 insert was digested with BamHI to release the FRE1-Kozak cassette, which was then cloned into pPEV containing the enhanced cauliflower mosaic virus 35S promoter and the NptII gene conferring kanamycin resistance (Lindbo and Dougherty, 1992).
FRE2 was obtained from the yeast genomic DNA by the same PCR procedures used to obtain FRE1. The 5′ primer CCAACGGGATCCATGCATTGGACGTCCATCTTGAGCGC and 3′ primer AAGTGGATCCTGATCACCAGCATTGATACTCTTCAAAG both contained BamHI sites. The PCR product was cut with BamHI and cloned into BamHI-cut pUC18. The 2.1-kb fragment was then cloned into dephosphorylated BamHI-cut pPEV. This plasmid was digested with XbaI and EcoRI and the insert, with the cauliflower mosaic virus 35S promoter, was cloned into the plant expression vector pGPTV-BAR, conferring bialaphos resistance (Becker et al., 1992).
Transformation of Tobacco
Agrobacterium tumefaciens EHA105 (Hood et al., 1993) was transformed with pPEV containing FRE1 and pGPTV-BAR containing FRE2 by the procedures of Hofgen and Willmitzer (1988). These EHA105/FRE1 and EHA105/FRE2 combinations were used for transformation of leaf discs of tobacco (Nicotiana tabacum L. cv Wisc. 38) by the methods described by Horsch et al. (1988). Shoots were selected on Murashige and Skoog medium (Murashige and Skoog, 1962) with 1 mg L−1 N6-BA, 0.1 mg L−1 α-naphthalene acetic acid, 400 mg L−1 Timentin (SmithKline-Beecham), and either 400 mg L−1 kanamycin for transformation with FRE1 or 5 mg L−1 bialaphos for transformation with FRE2. Resistant shoots were transferred to Murashige and Skoog medium with 400 mg L−1 Timentin plus 400 mg L−1 kanamycin (or 10 mg L−1 bialaphos) for rooting. Twenty-one independent putative FRE1 transformants were selected for production of T1 seed. As expected, T1 populations segregated for kanamycin resistance. Some fit a 3:1 (kanr:kans) segregation ratio; others had higher proportions of kanr plants likely due to two or more independent insertions. Plants from three independent families showing 3:1 segregation ratios (kanr:kans) (Table I) were selfed to generate T2 lines uniformly resistant to kanamycin. These were designated FRE1-A, FRE1-B, and FRE1-C. Eighteen independent FRE2 transformants were selected. Plants from three T1 families with 3:1 segregation ratios (bialaphosr:bialaphoss) (Table I) were selfed to give rise to T2 lines homozygous for bialaphos resistance, designated FRE2-A, FRE2-B, and FRE2-C. FRE1-C was used for retransformation with FRE2, and 16 T1 lines were generated. One of the T1 lines with a single FRE2 locus (Table I), FRE1+2-A, was selfed to obtain a transformed line with homozygous FRE1 and FRE2 loci. In addition, homozygous kanamycin-resistant (NI-1) and bialaphos-resistant (NI-2) control lines without FRE inserts were generated.
Table I.
Segregation (resistance:sensitivity) of FRE-transformed T1 populations and χ2 and P values based on 3:1 segregation ratios
| Line | Selective Agent | No. of Resistant Seedlings | No. of Sensitive Seedlings | χ2 (3:1) | P |
|---|---|---|---|---|---|
| FRE1-A | kana | 116 | 33 | 0.647 | 0.42 |
| FRE1-B | kan | 105 | 36 | 0.021 | 0.88 |
| FRE1-C | kan | 84 | 25 | 0.248 | 0.62 |
| FRE2-A | bial | 87 | 30 | 0.026 | 0.87 |
| FRE2-B | bial | 129 | 44 | 0.017 | 0.90 |
| FRE2-C | bial | 90 | 27 | 0.231 | 0.63 |
| FRE1+2-Ab | bial | 119 | 46 | 0.729 | 0.39 |
kan = 200 mg/L kanamycin; bial = 10 mg/L bialaphos.
FRE1+2-A was derived from a homozygous T2 of FRE1-C by transformation with FRE2.
PCR, Southern-Blot Analyses, and Northern-Blot Analyses of Transformed Plants
Genomic DNA was isolated from individual T2 seedlings using a modified hexadecyltrimethylammonium bromide procedure (Doyle and Doyle, 1990). Primers were designed to amplify a 1-kb region of DNA within the FRE1 gene (CAACTCGAGGCAAGGGTCTC and CAAACCAGCGGCTACACCTAC). PCR amplification was performed using 100 ng of DNA, 0.4 μm primers, 0.6 mm deoxyribonucleotide triphosphate, 2.5 mm MgCl2, and 2 to 3 units of Taq DNA polymerase in buffer supplied by the manufacturer (Promega). PCR conditions included one cycle of 5 min at 95°C, 1 min at 67°C, and 1.5 min at 72°C; and 30 cycles of 1 min at 94°C, 1.5 min at 67°C, and 1 min at 72°C. PCR products were analyzed on a 1.2% agarose gel.
For Southern-blot analyses, genomic DNA was extracted from 2 g of tobacco leaf tissue using the hexadecyltrimethylammonium bromide protocol (Doyle and Doyle, 1990). DNA (20 μg) was digested with a restriction enzyme, EcoRV for FRE1-containing lines and XbaI for FRE2-containing lines, separated on a 1.1% gel, and transferred by capillary transfer to a Zeta-Probe GT membrane following the manufacturer's instructions (Bio-Rad). (It should be noted that there is no EcoRV site in FRE1 and no XbaI site in FRE2.) The [α-32P]dCTP-labeled probe, covering the entire FRE1 or FRE2 gene, was synthesized using Ready-To-Go DNA-labeling beads (Pharmacia) following the manufacturer's protocol and purified on a Biospin 6 chromatography column (Bio-Rad). Membranes were incubated in 0.25 m Na2HPO4, pH 7.2, 7% SDS, and 1 mm EDTA at 65° for 30 min. Using fresh solution, membranes were hybridized with the labeled probe for 24 h. Membranes were washed for 45 min at 65°C in 0.04 m Na2HPO4, pH 7.2, 5% SDS, and 1 mm EDTA and then twice in 1% SDS, 40 mm NaHPO4, and 1 mm EDTA. Finally, they were exposed to x-ray film (Hyperfilm-MP, Amersham) at −80°C.
For northern-blot analyses, total RNA was isolated from 1 g of leaf tissue according to the TRIzol reagent protocol (GIBCO-BRL). RNA (30 μg) was separated on a formaldehyde gel (Davis et al., 1986). The suggested protocol (Bio-Rad) was used for capillary transfer to a Zeta-Probe membrane, and hybridization was performed as for the Southern blot.
Hydroponic Culture
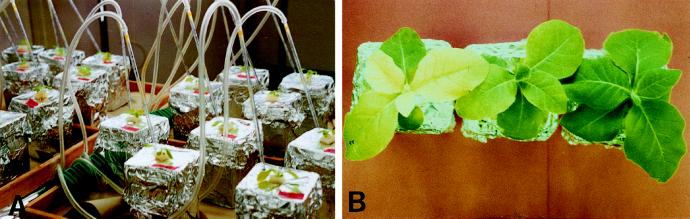
Seeds were surface sterilized with 70% ethanol (1 min) and 20% commercial bleach (15 min), and germinated on one-half-strength Murashige and Skoog mineral medium with 1.6 g L−1 Gelrite (Scott Laboratories, Carson, CA) in Magenta (Chicago, IL) boxes. After 3 weeks, seedlings were transferred to a hydroponic culture system, with one seedling per Magenta box filled with Hoagland nutrient solution containing 10 μm Fe-EDTA. During the first week in hydroponics, the seedlings were covered with glass jars to prevent desiccation. After 2 weeks in hydroponics, the Hoagland solution was replaced either with fresh Hoagland solution or with the nutrient solution devised by Chaney et al. (1992) containing 10 mm NaHCO3 and 2 μm Fe(III)-DTPA (Fe-deficient condition). Air enriched with 3% CO2 was used for aeration of the nutrient solution and stabilization of the pH (Chaney et al., 1992). The solution was replaced after 7 d; on the other days the level was adjusted with double-distilled water. Nutrient solution containing K2HPO4, NH4NO3, and H3BO3 was added daily to maintain proper nutrient levels as described by Chaney et al. (1992). Relative leaf chlorophyll concentrations were determined for the upper leaves on d 6 and 10 with a chlorophyll meter (model SPAD-502, Minolta, Ramsey, NJ). SPAD readings (on a scale of 0–50) determined by light reflection from two different light sources are linearly related to extractable chlorophyll (Marquard and Tipton, 1987). The upper, expanded leaf was designated leaf 1 at the beginning of the experiment, and the leaves formed next were designated leaves 2, 3, and 4, respectively. Fe(III) reduction and Fe content of leaves were determined after 10 d.
Fe(III) Reduction Assay
Fe(III) reduction was quantitatively determined using a liquid assay and qualitatively visualized on agarose plates (semisolid medium). The assay solution consisted of 0.3 mm BPDS, 0.1 mm Fe(III)-EDTA, 5 mm Mes buffer, and one-half-strength Murashige and Skoog mineral medium (Murashige and Skoog, 1962) in the dark. Because Co2+, Mn2+, Zn2+, and Cu2+ interfere with Fe(III) reduction (Römheld and Marschner, 1983), they were omitted from the assay medium. Experiments were performed on 8- to 10-week-old plants grown in flats with potting soil (Fe-sufficient conditions) in the greenhouse (22°C day, 18°C night) and on plants grown hydroponically. Roots were rinsed in double-distilled water and placed in assay solution. Shoot pieces (100 mg) from the greenhouse-grown plants were cut into eight slices and placed into assay medium. After 0.5 or 1 h, Fe reduction was determined spectrophotometrically from the A535 minus the blank (containing no tissue). Fe(II)-BPDS was quantified using a molar extinction coefficient of 22.14 mm−1 cm−1. Roots were blotted dry and weighed. Data were obtained from two replicate experiments each with four samples (greenhouse plants) or three samples (hydroponics).
For embedding in semisolid medium, roots or stem sections were placed on Whatman no. 1 filter paper in 90- × 90- × 15-mm Petri dishes. Assay medium (see above) containing 0.25% agarose (type I, Sigma) was cooled to 35°C and pipetted into the Petri plates (50 mL/plate).
To determine reduction by exudates, roots were incubated in assay solution without BPDS and Fe(III)-EDTA for 0.5 h, after which they were removed and the two compounds were added to the solution. Fe(III) reduction was determined as described above.
Fe Concentration of Leaves
Plants from two hydroponic experiments were pooled. The upper four leaves of all plants of a line were included. The samples were dried and mineral contents were determined with an Optima 3000 dual view inductively coupled plasma optical emission spectroscopy (Perkin-Elmer) at the Crop and Soil Science Analytical Laboratory at Oregon State University.
RESULTS
Genetic Characterization
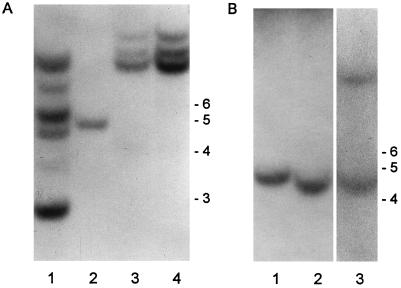
The presence of transgenes was confirmed by Southern blots of DNA digested with EcoRV and XbaI probed with FRE1 (Fig. 1A) and FRE2 (Fig. 1B), respectively. Five clear bands were visible in FRE1-A, one in FRE1-B, and three in FRE1-C when FRE1 was used as the probe (Fig. 1A). As expected, FRE1+2-A, a derivative of FRE1-C, also had three clear bands. When FRE2 was used as a probe, a single complementary fragment was detected in FRE2-A, FRE2-C (Fig. 1B), and FRE2-B (not shown). FRE1+2-A had two FRE2-positive bands. All 10 plants of each line analyzed had the same banding patterns, supporting the evidence from data on resistance to drugs that these progeny are homozygous. The control lines did not show any hybridizing bands with FRE1 or FRE2 as a probe (not shown).
Figure 1.
Southern blot of FRE-transformed lines of tobacco. A, Probed with FRE1. Lane 1, FRE1-A; lane 2, FRE1-B; lane 3, FRE1-C; and lane 4, FRE1+2-A. B, Probed with FRE2. Lane 1, FRE2-C; lane 2, FRE2-A; and lane 3, FRE1+2-A. DNA marker sizes (kilobases) are indicated at the right of each gel.
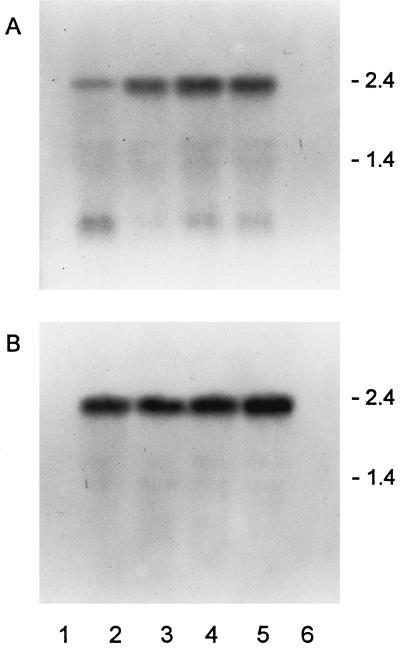
Northern blots of mRNA from transformed plants probed with FRE1 or FRE2 indicated that the corresponding messages were made in transformants (Fig. 2). In addition to the full-length mRNA, a lower band was clearly visible in the FRE1-containing lines. The relative abundance of this shorter message differed between lines and was highest in FRE1-A. No bands could be detected in wild-type and NI control lines after hybridization under stringent conditions.
Figure 2.
Northern blot of FRE-transformed lines of tobacco. A, Probed with FRE1. Lane 1, NI-1; lane 2, FRE1-A; lane 3, FRE1-B; lane 4, FRE1-C; lane 5, FRE1+2-A; and lane 6, wild type. B, Probed with FRE2. Lane 1, NI-2; lane 2, FRE2-A; lane 3, FRE2-B; lane 4, FRE2-C; lane 5, FRE1+2-A; and lane 6, wild type. RNA marker sizes (kilobases) are indicated at the right of each gel.
Fe(III) Reduction in Fe-Sufficient Conditions
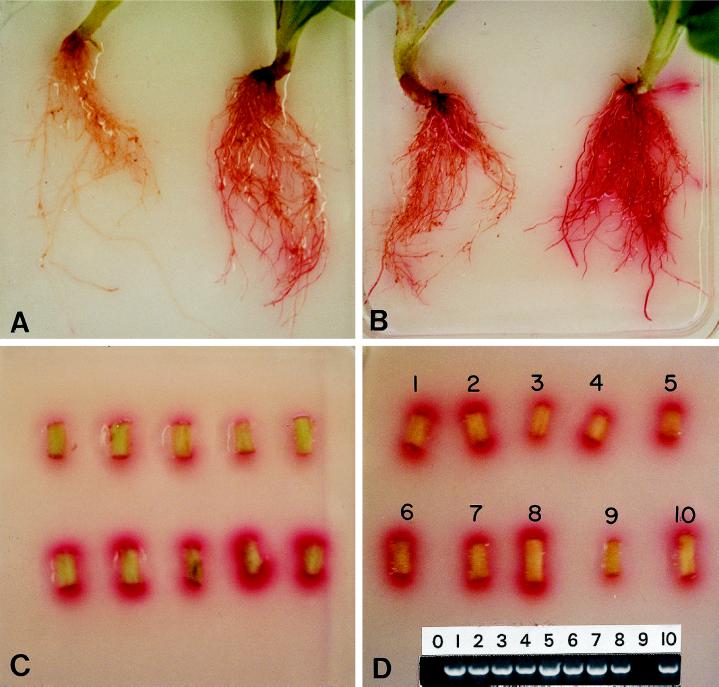
The presence of FRE1 and FRE2 resulted in increased Fe(III) reduction as determined both by visual inspection and quantitative measurement. When roots of greenhouse-grown plantlets were embedded in semisolid assay medium containing 25 μm Fe(III), the transformants started showing red color within 1 h and were bright red after 6 h (Fig. 3, A and B), at which time the controls were only slightly tinged. FRE1+2-A roots consistently had the most-intense red color development on and around the roots (Fig. 3B).
Figure 3.
Fe(III) reductase assays of tobacco roots and shoot segments of 8-week-old plants grown in the greenhouse. A, Roots of control NI-1 (left) and transformant FRE1-C (right) after 6 h in semisolid medium with 25 μm Fe(III)-EDTA and 0.3 mm BPDS. B, Roots of FRE2-B (left) and FRE1+2-A (right) after 6 h in semisolid medium with 25 μm Fe(III)-EDTA and 0.3 mm BPDS. C, Shoot segments of control NI-1 (top) and transformant FRE1+2-A (bottom) after 4 h in semisolid medium with 0.1 mm Fe(III)-EDTA and 0.3 mm BPDS. D, Shoot segments of 10 seedlings of a family segregating for FRE1 in semisolid medium after 4 h in semisolid medium with 0.1 mm Fe(III)-EDTA and 0.3 mm BPDS; PCR analyses of the same seedlings and water control (0) with FRE1 primers are shown at bottom.
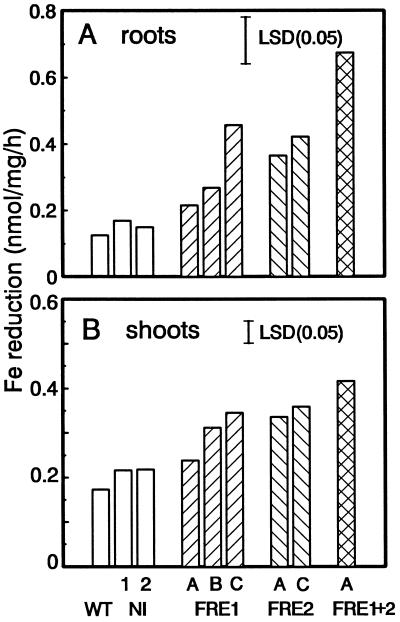
In liquid Fe(III) reduction assays performed with roots excised from plants grown in greenhouse potting soil, FRE-containing lines had generally increased reduction, with up to 4-fold higher activity than control lines in a 1-h assay with 100 μm Fe(III)-EDTA (Fig. 4A). The FRE1+2 line had the highest Fe-reduction activity. The two FRE-transformed lines that did not show enhanced Fe(III) reduction compared with the controls were FRE1-A and FRE1-B. FRE1-A also showed the highest level of truncated mRNA in the northern blots (Fig. 2A, lane 2). Fe(III) reduction of exudates did not differ between lines; it contributed about 25% to the total Fe(III) reduction of the wild type and relatively less to the transformants. Therefore, differences between transformed and control lines reflect Fe(III) reduction by the roots. Essentially the same results were obtained with roots of plants grown in Hoagland solution hydroponically. FRE1 lines varied in their Fe(III) reduction levels, whereas FRE2 lines had consistently higher Fe(III) reduction levels than the controls (data not shown).
Figure 4.
Fe(III) reduction in 0.1 mm Fe(III)-EDTA and 0.3 mm BPDS for 1 h of control (wild type [WT], NI-1, and NI-2) and FRE-transformed plants. A, Fe(III) reduction by roots. B, Fe(III) reduction by shoot sections. The lsd (P = 0.05) is indicated by a bar.
Assays with stem segments also confirmed the higher Fe(III) reduction in transformants than in the controls. Intense color formation was seen at the cut ends (Fig. 3C). Fe(III) reduction in liquid assays (Fig. 4B) was also significantly higher in transformants than in controls. Generally, shoot Fe(III) reduction correlated with that obtained for roots, indicating consistent expression of FRE1 and FRE2 in roots and shoots. Again, the double transformant had the highest reduction and the FRE1-A line was not significantly different from the control.
The correlation between the presence of FRE and increased Fe reduction was further confirmed by examining 10 seedlings from a T2 family segregating for FRE1. The one shoot segment (no. 9) that produced weak color intensity typical of nontransformed shoots in the plate assay was also the only one negative in the PCR test (Fig. 3D).
Chlorophyll Formation and Fe(III) Reduction at Fe-Deficient Conditions
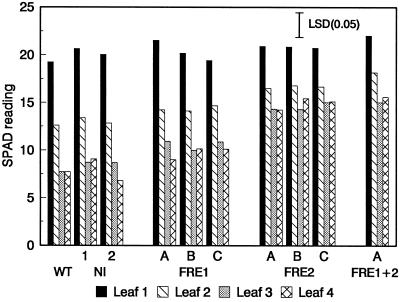
Plants were grown in hydroponic culture (Fig. 5A) to manipulate the Fe availability. Tiny seedlings obtained from seeds germinated under aseptic conditions were first acclimated in Hoagland nutrient solution for 2 weeks and then transferred to Fe-deficient culture solution. Fe deficiency was created by the presence of 10 mm NaHCO3, which increased the pH to 7.8. Leaf chlorosis appeared on control plants after 4 to 5 d. Younger leaves became progressively more yellow and by d 6, significant differences in chlorophyll concentrations (SPAD readings) of the upper two leaves were detected between FRE2 and control plants (data not shown). By d 10, the upper leaves of the controls were highly chlorotic, whereas only slight yellowing occurred among the FRE2 transformants (Fig. 5B) and the double transformants (not shown). The visual difference was reflected by the SPAD readings (Fig. 6). In contrast, the phenotype and chlorophyll concentrations of FRE1 transformants were not significantly different from those of controls. Older leaves, such as leaf 1, which were about fully expanded before Fe stress was applied, were unaffected and remained green on all plants.
Figure 5.
Plants grown in hydroponics. A, Hydroponic system 5 d before the start of the experiment. B, Plants of NI-2 (left), FRE2-A (middle), and FRE2-C (right) after 10 d in nutrient solution with 10 mm NaHCO3 and 2 μm Fe-DTPA.
Figure 6.
SPAD leaf readings of control and transformed plants after 10 d in Fe-deficient nutrient solution with 10 mm NaHCO3 and 2 μm Fe-DTPA. Leaf 1 was the lowest leaf included in the measurements and the only fully expanded leaf at the beginning of the experiment; leaf 4 was the uppermost leaf at the completion of the experiment. The lsd (P = 0.05) is indicated by a bar.
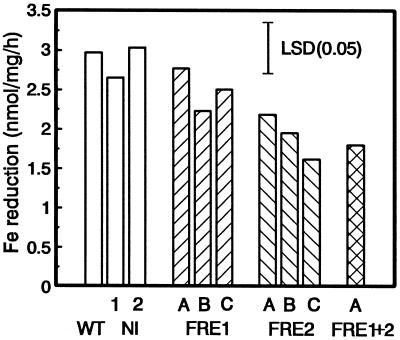
Fe(III) reduction was measured after 10 d (Fig. 7). It is interesting that Fe(III) reduction was higher in the controls than in the FRE2 transformants. Compared with plants grown in Fe-sufficient conditions, the controls showed a 10- to 15-fold increase in reduction, whereas the increase in the FRE1+2 transformant was only 2-fold. Root Fe reduction was inversely related to leaf chlorosis and seemed to reflect the severity of Fe stress sensed by the plant.
Figure 7.
Root Fe(III) reduction in 0.1 mm Fe(III)-EDTA and 0.3 mm BPDS of control (wild type [WT], NI-1, and NI-2) and FRE-transformed plants after 10 d in Fe-deficient nutrient solution with 10 mm NaHCO3 and 2 μm Fe-DTPA. The lsd (P = 0.05) is indicated by a bar.
Leaf Fe Concentrations
The Fe concentrations in the upper three leaves of plants grown under Fe-sufficient and Fe-deficient conditions were measured (Table II). The same sets of Fe-stressed plants were used as were used for SPAD readings and Fe-reduction determinations. For a comparison, plants of the same lines were grown in Hoagland solution for the same duration. Clearly, the Fe concentrations were significantly lower under Fe deficiency. However, under both conditions, FRE2 plants (including the double transformant) had about 50% higher Fe than the controls. FRE1 lines had Fe concentrations closer to those of the controls than FRE2 lines when grown under Fe deficiency and were more variable when Fe supply was sufficient.
Table II.
Fe concentrations in the upper three leaves of FREtransformed and control tobacco plants grown in Fe-sufficient (Hoagland) solution or Fe-deficient (Chaney) solution for 10 d
| Line | Fe Concentration
|
|
|---|---|---|
| Hoagland solution | Chaney solutiona | |
| μg/g dry wt | ||
| NI-1 | 134 | 28 |
| NI-2 | 123 | 24 |
| FRE1-A | 149 | 30 |
| FRE1-B | 190 | 33 |
| FRE1-C | 123 | 29 |
| FRE2-A | 190 | 45 |
| FRE2-B | 213 | 40 |
| FRE2-C | 193 | 44 |
| FRE1+2-A | 223 | 41 |
Overall, FRE2 lines were consistent in all parameters measured. These included higher Fe concentrations under both Fe-deficient and Fe-sufficient conditions, higher leaf chlorophyll and lower Fe(III) reduction under Fe stress, but higher Fe(III) reduction under Fe sufficiency.
DISCUSSION
Comparisons between transgenic and control tobacco plants in Fe-sufficient conditions indicated that the yeast FRE genes are functional in plants. The increase in Fe(III) reduction at Fe sufficiency (up to 4-fold) was constitutive, occurring in both transformed roots and shoots. Fe(III) reduction varied between FRE1-containing lines, whereas FRE2 plants uniformly showed higher activity. Although variability in transgene expression is a common phenomenon and the presence of multiple gene copies can lead to gene silencing (Meyer, 1995), the variability may be caused by other factors. One of the possibilities may be instability or premature truncation of FRE1 mRNA, as illustrated by the occurrence of a second, shorter-than-expected message. The relatively large amount of this shorter mRNA in FRE1-A, the line without any increased Fe(III) reductase activity, seems to lend support to this hypothesis. The highest reduction activity was found in the line containing both FRE1 and FRE2. This may be due to additive effects of the FRE1 and FRE2 genes, as in yeast, in which the two genes together account for the total Fe(III) reduction activity (Georgatsou and Alexandrakis, 1994).
Transgenic FRE2 plants were more tolerant of Fe-deficient conditions. The chlorophyll content was higher in new leaves of FRE2 and FRE1+FRE2 transformants than in comparable leaves of control plants. However, no increased resistance to Fe-deficient conditions was found for FRE1 lines, including FRE1-C, which showed significantly increased Fe(III) reduction. The reason for this is uncertain, but it is possible that FRE1 protein has only low affinity for the Fe(III) substrate, resulting in enzyme activity at the high substrate concentration (100 μm) of the Fe(III) reductase assay but not at low substrate availability (Fe-deficient conditions in hydroponics). Also intriguing is the inverse relationship between tolerance of Fe deficiency and root Fe reduction under low Fe availability. It is possible that, since their Fe levels are higher, FRE2-containing plants sense less stress than controls, and, therefore, the native inducible Fe(III) reductase system is less active. This interpretation would be compatible with the hypothesis that signals from the shoots stimulate Fe reduction in roots (Grusak and Pezeshgi, 1996). In addition, constitutive expression of FRE2 in shoots and leaves may increase the level of available Fe for chlorophyll synthesis in these tissues. FRE2 plants may show Fe(III) reduction levels as high as or higher than those of controls if left under stress conditions for a longer period, but this could not be tested in our present system because of the small size of the containers relative to the size of the plants. Finally, the higher chlorophyll level in FRE2-containing plants may be related to other, as-yet-undetermined functions of the FRE genes. For yeast, Lesuisse et al. (1996) have provided evidence that the FRE proteins may not be Fe(III) reductases themselves but rather a part of a multicomponent electron-transport chain. However, the observation that expression of FRE1 from a high-copy plasmid leads to increased Fe reductase activity (Shatwell et al., 1996) indicates clearly that FRE1 is an important and limiting component in Fe reduction, regardless of its exact function.
Posttranslational modifications and targeting may also affect the activity of FRE proteins in plants. The FRE1 sequence contains several potential glycosylation sites (Dancis et al., 1992), and glycosylation of the protein was demonstrated by in vitro transcription/translation products formed in the presence and absence of canine microsomes (Samuelsen, 1996). Fully active FRE1 may require a flavin or heme group, as suggested by data reported by Shatwell et al. (1996). It is not known at present if such modifications occur in transgenic plants. In theory, effective Fe reductase should be located in the PM of epidermal cells. The fact that most transgenic lines exhibited higher Fe reductase activity under nonstress conditions than controls would suggest that some of the FRE protein is correctly targeted. However, ectopic expression of high levels of protein may lead to altered localization and function. To answer some of these questions, monoclonal antibodies to selected portions of the FRE proteins are being generated.
Yamaguchi et al. (1995) reported no detectable effect of a similar FRE1 construct in tobacco. Although in our tests the FRE1 gene also had no effect on tolerance to Fe-deficient conditions, it did enhance Fe(III) reduction under Fe sufficiency in some of the transformed lines. It is not clear how many lines were examined by Yamaguchi et al. (1995), but the lack of transgene effect may be attributable to random variation among lines. Moreover, a slightly different gene construct could have led to relatively high mRNA truncation, as in line FRE1-A, and significant reduction of functional gene product.
Although the exact mechanism of the FRE proteins in mediating tolerance of Fe deficiency remains to be determined, from a practical point of view, FRE2 and other Fe-related genes are promising candidates for improving Fe utilization in plants. These may include the Arabidopsis froh genes (Robinson et al., 1997a, 1997b) and genomic clones with some homology to the FRE1 gene (Yi et al., 1994). Because frohC transcript accumulated in roots and leaves under Fe-deficient conditions, it was suggested that the protein may be a membrane-bound Fe reductase involved in Fe transport. It will be interesting to determine if controlled expression of such genes in specific tissues will alter Fe reduction and uptake.
ACKNOWLEDGMENTS
We thank Dr. Stephen Johnston (University of Texas Southwestern Medical Center, Dallas) for his gift of yeast genomic DNA and Dr. Detlef Becker (Hygienie Institut der Freien und Hansestadt Hamburg, Germany) for providing the vector pGPTV-BAR. We thank Dr. Walter Ream and Dr. Joyce Loper for their helpful suggestions on the manuscript.
Footnotes
This research was supported by the U.S. Department of Agriculture (USDA)-National Research Initiative Competitive Grants Program (grant no. 95-37100-1566), the Oregon Agricultural Experiment Station (paper no. 11,169), and USDA Special Grant: Oregon/Massachusetts Biotechnology Partnership.
Abbeviations: BPDS, bathophenanthrolinedisulfonic acid; DTPA, diethylenetriaminepentaacetic acid; PM, plasma membrane; SPAD, Specialty Products Agricultural Division.
LITERATURE CITED
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Parks TD, Cary SM, Dougherty WG. Vectors for cell-free expression and mutagenesis of protein-coding sequences. Nucleic Acids Res. 1987;15:10066. doi: 10.1093/nar/15.23.10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaney RL, Coulombe BA, Bell PF, Angle SJ. Detailed method to screen dicot cultivars for resistance to Fe-chlorosis using FeDTPA and bicarbonate in nutrient solutions. J Plant Nutr. 1992;15:2063–2083. [Google Scholar]
- Colau D, Negrutiu I, Van Montagu M, Hernalsteens J-P. Complementation of a threonine dehydratase-deficient Nicotiana plumbaginifolia mutant after Agrobacterium tumefaciens-mediated transfer of the Saccharomyces cerevisiae ILV1 gene. Mol Cell Biol. 1987;7:2552–2557. doi: 10.1128/mcb.7.7.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancis A, Klausner AD, Hinnebusch AG, Barriocanal JG. Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2294–2301. doi: 10.1128/mcb.10.5.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancis A, Roman DG, Anderson GJ, Hinnebusch AG, Klausner RD. Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proc Natl Acad Sci USA. 1992;89:3869–3873. doi: 10.1073/pnas.89.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LG, Dibner MD, Battey JF (1986) Basic Methods in Molecular Biology. Elsevier, New York
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Georgatsou E, Alexandraki D. Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3065–3073. doi: 10.1128/mcb.14.5.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusak MA, Pezeshgi S. Shoot-to-root signal transmission regulates root Fe(III) reductase activity in the dgl mutant of pea. Plant Physiol. 1996;110:329–334. doi: 10.1104/pp.110.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y. Iron: nutritious, noxious, and not readily available. Plant Physiol. 1994;104:815–820. doi: 10.1104/pp.104.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofgen R, Willmitzer L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988;16:9877. doi: 10.1093/nar/16.20.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood EE, Gelvin SB, Melchers LS, Hoekema A. New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 1993;2:208–218. [Google Scholar]
- Horsch RB, Fry J, Hoffmann N, Neidermeyer J, Rogers SG, Fraley RT. Leaf disc transformation. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1988. pp. 1–9. [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Lesuisse E, Casteras-Simon M, Labbe P. Evidence for the Saccharomyces cerevisiae ferrireductase system being a multicomponent transport chain. J Biol Chem. 1996;271:13578–13583. doi: 10.1074/jbc.271.23.13578. [DOI] [PubMed] [Google Scholar]
- Lesuisse E, Crichton RR, Labbe P. Iron-reductases in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 1990;1038:253–259. doi: 10.1016/0167-4838(90)90213-y. [DOI] [PubMed] [Google Scholar]
- Lesuisse E, Raguzzi F, Crichton RR. Iron uptake by the yeast Saccharomyces cerevisiae: involvement of a reduction step. J Gen Microbiol. 1987;133:3229–3236. doi: 10.1099/00221287-133-11-3229. [DOI] [PubMed] [Google Scholar]
- Lindbo J, Dougherty WG. Pathogen-derived resistance to a potyvirus: immune and resistant phenotypes in transgenic tobacco expressing altered forms of a potyvirus coat protein nucleotide sequence. Mol Plant Microbe Interact. 1992;5:144–153. doi: 10.1094/mpmi-5-144. [DOI] [PubMed] [Google Scholar]
- Marquard RD, Tipton JL. Relationship between extractable chlorophyll and an in situ method to estimate leaf greenness. HortScience. 1987;22:1327. [Google Scholar]
- Meyer P. Variation of transgene expression in plants. Euphytica. 1995;85:359–366. [Google Scholar]
- Moog PR, Brüggemann W. Iron reductase systems on the plant plasma membrane: a review. Plant Soil. 1994;165:241–260. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Robinson NJ, Sadjuga MR, Groom QJ. The froh gene family from Arabidopsis thaliana: putative iron-chelate reductases. Plant Soil. 1997a;196:245–248. [Google Scholar]
- Robinson NJ, Wilson JR, Turner JS, Fordham-Skelton AP, Groom QJ. Metal-gene-interactions in roots: metallothionein-like genes and iron reductases. In: Anderson HM, Barlow PW, Clarkson DT, Jackson MB, Shewry PR, editors. Plant Roots: From Cells to Systems. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997b. pp. 117–130. [Google Scholar]
- Roman DG, Dancis A, Anderson GJ, Klausner RD. The fission yeast ferric reductase gene Frp1+ is required for ferric iron uptake and is homologous to the gp91-phox subunit of the human NADPH phagocyte oxidoreductase. Mol Cell Biol. 1993;13:4342–4350. doi: 10.1128/mcb.13.7.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römheld V. Existence of two different strategies for the acquisition of iron in higher plants. In: Winkelmann G, van der Helm D, Neilands JB, editors. Iron Transport in Microbes, Plants and Animals. Weinheim, Germany: VCH; 1987. pp. 353–374. [Google Scholar]
- Römheld V, Marschner H. Mechanism of iron uptake by peanut plants. I. Fe III reduction, chelate splitting, and release of phenolics. Plant Physiol. 1983;71:949–954. doi: 10.1104/pp.71.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsen AI. Transformation of tobacco with the yeast FRE1 and FRE2 genes: characterization of transformants and discovery of a temperature-dependent morphological mutant. PhD thesis. Corvallis: Oregon State University; 1996. [Google Scholar]
- Shatwell KP, Dancis A, Cross AR, Klausner RD, Segal AW. The FRE1 ferric reductase of Saccharomyces cerevisiae is a cytochrome b similar to that of NADPH oxidase. J Biol Chem. 1996;271:14240–14244. doi: 10.1074/jbc.271.24.14240. [DOI] [PubMed] [Google Scholar]
- Von Schaewen A, Stitt M, Schmidt R, Sonnewald U, Willmitzer L. Expression of a yeast-derived invertase in the cell wall of tobacco and Arabidopsis plants leads to accumulation of carbohydrate and inhibition of photosynthesis and strongly influences growth and phenotype of transgenic tobacco plants. EMBO J. 1990;9:3033–3044. doi: 10.1002/j.1460-2075.1990.tb07499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Fujiwara T, Mori S (1995) Genetic introduction of the gene coding yeast ferric reductase into tobacco plants. In International Conference on BioIron, Asheville, NC (Abstracts), p 84
- Yi Y, Saleeba JA, Guerinot ML (1994) Iron uptake in Arabidopsis thaliana. In JA Manthey, DE Crowley, DG Luster, eds, Biochemistry of Metal Micronutrients in the Rhizosphere. Lewis Publishers, Boca Raton, FL, pp 295–307