Abstract
Coactivator-associated arginine methyltransferase 1 (CARM1) represents a valuable target for hormone-dependent tumors such as prostate and breast cancers. Here we report the enzyme and cellular characterization of the 1-benzyl-3,5-bis(3-bromo-4-hydroxybenzylidene) piperidin-4-one (7g) and its analogues 8a-l. Among them, 7g, 8e, and 8l displayed high and selective CARM1 inhibition, with lower or no activity against a panel of different PRMTs or HKMTs. In human LNCaP cells, 7g showed a significant dose-dependent reduction of the PSA promoter activity.
Arginine methylation of mainly nuclear proteins is a reversible post-translational modification process involved in structural remodeling of chromatin.1,2 Protein arginine methyltransferase (PRMT) enzymes remove the methyl group from the donor molecule S-adenosyl-L-methionine (AdoMet) generating the product S-adenosyl-L-homocystein (AdoHcy), and transfer this methyl residue to the terminal nitrogen atom(s) of the guanidinium side chain of an individual arginine residue in the target protein.3 PRMTs are ubiquitously expressed in most cell types and tissues of the human body with the unique exception of PRMT8, which appears to be restricted to neurons in the brain.4 Moreover, they differ in their substrate specificities, and are therefore probably involved in different physiological processes. Among PRMTs, PRMT4/CARM1 (coactivator-associated arginine methyltransferase 1) was the first to be identified as a transcriptional regulator.5 CARM1 methylates a number of proteins that are involved in transcription and RNA processing, including histone H3 (H3R17 and H3R26), amplified in breast cancer 1 (AIB1), p300/CBP (cAMP-responsive element binding protein [CREB] binding protein), poly(A)-binding protein 1 (PABP1), and co-activator of 150 kDa (CA150).1 CARM1 requires its enzymatic activity for all its in vivo functions.6 In cancer, CARM1 has been shown to regulate estrogen-stimulated MCF-7 breast cancer cell cycle progression through E2F1 upregulation.7 Moreover, CARM1 has been found upregulated in castration-resistant prostate cancer8 and in grade-3 breast tumors,9 and CARM1 knockdown by siRNA completely inhibited prostate cancer LNCaP cell proliferation by induction of apoptosis.10
All these findings prompted researchers to develop molecules able to inhibit CARM1 activity, as potential anticancer agents. Some pyrazole-containing compounds (1-4) as well as the benzo[d]imidazole (5) have been reported as inhibitors of CARM1,11-15 and the plant-derived ellagic acid (6)16 has been recently shown to selectively block methylation at Arg 17 of histone H3 (H3R17),16 the CARM1 histone site for methylation (Chart S1 in Supporting Information).
Despite the fact that all of these compounds showed submicromolar inhibitory activity against CARM1, no inhibitor has been demonstrated to exhibit cellular effects to date.
Pursuing our searches on design, synthesis, and biological validation of small molecule modulators of epigenetic targets,17 in 2008 we prepared and tested some bis(3-bromo-4-hydroxy- and 3,5-dibromo-4-hydroxyphenyl) compounds and their analogues against PRMT1,18 CARM1,18 SET7 (an histone lysine methyltransferase, HKMT),18 p300/CBP (an HAT enzyme),18,19 SIRT1, and SIRT2.18 Depending on the extent of bromination of the molecule (presence of four bromine atoms), and on the nature of the linker connecting the two dibromophenol moieties (penta-1,4-dien-3-one, 2,6-dimethylene(hetero)cycloalkanone, 1,1-(1,3-phenylene)diprop-2-en-1-one, and hepta-1,6-diene-3,5-dione), some of such compounds behaved as epigenetic multiple ligands (epi-MLs), they being active against all the tested enzymes.18 Differently, compounds carrying two or three bromine atoms in their structure or featuring a bis(3,5-dibromo-4-hydroxybenzamide) or bis(3,5-dibromo-4-hydroxyanilide) scaffold failed to be recognized as epi-MLs, and showed some degree of selectivity against a particular epigenetic target.
Thus, with the aim to identify CARM1-selective inhibitors among them, and taking in account the fluorograph data previously reported, we determined the IC50 values for selected bis(bromo- and dibromophenol) compounds 7a-m (see Figure S1 and Table S1 in Supporting Information) against PRMT1, CARM1, and the HKMT SET7.
Among the tested compounds, 7b showed high potency and selectivity in inhibiting PRMT1, whereas 7c,d,g,h,l,m preferably inhibited CARM1, 7g being the most potent (IC50 = 7.1 μM). With the exception of 7a,b, all the tested compounds displayed very low (if any) inhibition against SET7.
Accordingly, we chose 7g as our lead compound for selective CARM1 inhibition, and prepared some 3,5-bis(3-bromo-4-hydroxybenzylidene)-1-benzylpiperidin-4-one analogues 8a-l by insertion of a chlorine atom or a methyl or methoxy group at the ortho, meta, or para position of the N1-benzyl moiety, or by replacing such benzyl group with a 2-phenylethyl, 3-phenylpropyl, or 2-oxo-2-phenylethyl moiety at N1 (Figure 1). These new compounds were tested as CARM1-selective inhibitors, and two of them together with 7g were investigated in more detail in vitro and in vivo.
Figure 1.
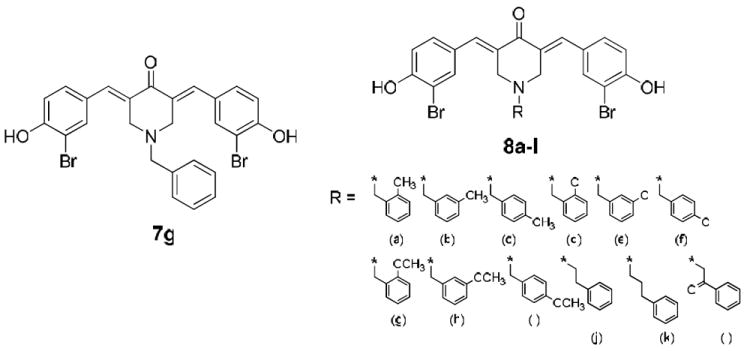
CARM1-selective inhibitors used in this study.
Chemistry
3,5-Bis(3-bromo-4-(methoxymethoxy)benzylidene)piperidin-4-one 9, the key intermediate of the title compounds, was prepared by condensation of 3-bromo-4-(methoxymethoxy)benzaldehyde18 with 4-piperidone in alkaline medium (barium hydrate). Alkylation reactions of 9, carried out at 60 °C with the opportune alkyl bromide in the presence of dry potassium carbonate in acetonitrile, furnished the N-arylalkyl-3,5-bis(3-bromo-4-(methoxymethoxy) benzylidene)piperidin-4-ones 10a-l that were subjected to acidic hydrolysis in methanolic 3 N HCl at 60 °C to afford the desired bis(3-bromo-4-hydroxybenzylidene) analogues 8a-l (Scheme S1 in Supporting Information).
Experimental procedures for compounds 9 and 10, and chemical and physical data (Tables S2-S4) for compounds 8-10 are reported as Supporting Information.
Results and Discussion
The new compounds were tested by fluorograph at 50 and 20 μM against CARM1 using PABP1 as a substrate,20 and at 50 μM against PRMT1 (substrate: the heterogeneous nuclear ribonucleoprotein NPL3)21 and SET7 (substrate: histone H3), to assess their potency and selectivity (Figure 2). At 20 μM, the 4-methyl- and the 2-, 3-, and 4-methoxybenzyl analogues of 7g (compounds 8c and 8g-i) as well as the 3-phenylpropyl-piperidone 8k showed no effect against the PABP1 methylation, thus the methoxy-containing compounds were excluded by IC50 calculation.
Figure 2.
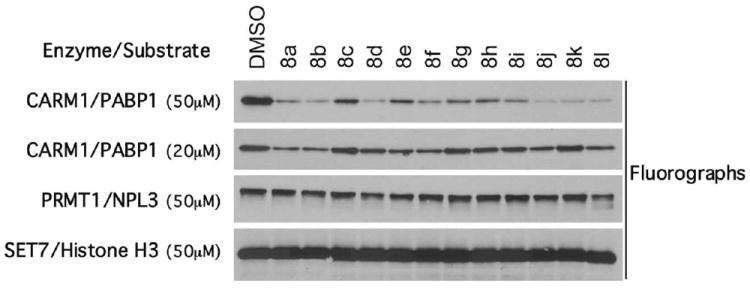
Inhibitory activities of compounds 8a-l against CARM1 using PABP1 as a substrate, PRMT1 using NPL3 as a substrate, and SET7 using histone H3 as a substrate. The concentration of the compounds used in each in vitro methylation assay is shown.
IC50 values for compounds 7g and 8a-f,j-l were determined against CARM1 using PABP1 as a substrate, and against PRMT1 and SET7 using NPL3 and histone H3 as substrates, respectively (Table 1). The corresponding IC50 curves are reported in Supporting Information. All the tested compounds displayed low micromolar activity against CARM1, the insertion of methyl as well as chloro substituents at the N1-benzyl moiety having only modulator effects on enzyme inhibition. The preferred position to introduce a methyl group at the benzyl portion seems to be the ortho position (compound 8a), while for chlorine insertion the benzyl meta position afforded the highest inhibitory activity (compound 8e), similar to that of the lead compound 7g.
Table 1.
IC50 values of 7g and 8a-f,j-l against CARM1, PRMT1, and SET7.
| compd | IC50 (μM)
|
||
|---|---|---|---|
| CARM1/PABP1 | PRMT1/NPL3 | SET7/H3 | |
| 7g | 8.6 ± 0.8 | > 667 | > 667 |
| 8a | 10.3 ± 3.3 | > 667 | > 667 |
| 8b | 15.2 ± 0.9 | > 667 | > 667 |
| 8c | 11.9 ± 2.3 | > 667 | > 333 |
| 8d | 12.5 ± 6.1 | > 667 | > 667 |
| 8e | 8.1 ± 2.2 | > 667 | 174 ± 28 |
| 8f | 12.2 ± 3.0 | > 667 | > 600 |
| 8j | 14.8 ± 2.5 | > 667 | > 667 |
| 8k | 16.0 ± 3.8 | > 667 | > 667 |
| 8l | 14.4 ± 1.7 | > 667 | 149 ± 26 |
Values were determined from at least two separate experiments. The reaction contained 0.1 μM of GST-CARM1 and 0.5 μM of GST-PABP1, 0.15 μM of GST-PRMT1 and 0.5 μM GST-NPL3, or 0.15 μM of GST-SET7 and 1.1 μM of histone H3 with 0.22 μM [3H] AdoMet and different concentrations of each compound for IC50 determinations with different concentrations of each compound for IC50 determinations. The software that we used for fitting curves and determining IC50s is SigmPlot. The equation used for fitting is y=y0 + a/1+(x/x0)b.
All the tested compounds were selective towards CARM1, they showing very low (if any) activity against PRMT1 and SET7. Among them, we selected 7g, 8e, and 8l for further experiments: 7g and 8e were the most potent inhibitors of CARM1 with PABP1 as a substrate (see Table 1), while 8l was the only analogue carrying a structural diversity, the carbonyl group at the substituent at N1, that could influence someway its binding with the enzyme and its inhibitory behavior.
First, we repeated the CARM1 assay testing 7g, 8e, and 8l at 100 μM by fluorograph and using four different CARM1 substrates: PABP1, CA150,22 the spliceosome protein SmB,22 and histone H3 (Figure 3).
Figure 3.

Inhibitory activity of 7g, 8e, and 8l against CARM1 using PABP1, CA150, SMB, and histone H3 as substrates, and against a panel of PRMTs (PRMT1, PRMT3, PRMT5, and PRMT6) using indicated histone and/or non-histone substrates. The fluorographs are shown in the left panels, and the tritium count for each band is depicted in the right panels.
All the three tested compounds strongly inhibited the CARM1 activity on the various substrates; among these, CA150 was the most sensitive whereas the use of histone H3 yielded the lowest CARM1 inhibition. To check the real selectivity of 7g, 8e, and 8l against various PRMTs, we tested them at 100 μM against i) PRMT1 using NPL3 and histone H4 as a nonhistone and histone substrate, respectively, ii) PRMT3 using NPL3 and the ribosome protein rpS223 as substrates, iii) PRMT5 and histone H4 as a substrate, iv) PRMT6 using NPL3 and histone H3 as substrates (Figure 3).
In addition, 7g, 8e, and 8l were tested at 100 μM against a panel of HKMTs, namely SET7 (substrate: H3), DOTL1 (substrate: nucleosome), Suv39H1 (substrate: H3), and G9a (substrate: H3) (Figure 4). Against PRMTs, 7g and 8e were able to inhibit to some extent PRMT3, and 8e and 8l showed high inhibition of PRMT5 at 100 μM, nevertheless in all cases the observed inhibition values were weaker than those observed with CARM1 when used at the same concentration (see Figure 3). No significant activity at 100 μM was registered for 7g, 8e, and 8l against the tested HKMTs (see Figure 4).
Figure 4.
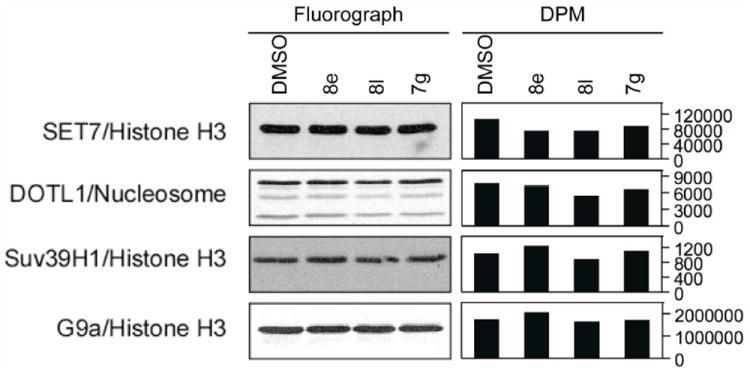
Inhibitory activities of 7g, 8e, and 8l against a panel of HKMTs (SET7, DOTL1, Suv39H1, and G9a) using the indicated histone and/or non-histone substrates.
Known CARM1 substrates such as PABP1 are hypermethylated in vivo and this methylation is very stable. To test the efficacy of potential PRMT inhibitors in cell may require days of treatment, while waiting for the methylated substrates to turn-over. Under these conditions, compounds with pleiotropic effects would be difficult to investigate in a cell-based assay. To reduce the exposure time of the compound to cells, and bypass this problem, we developed an Flag-tagged PABP1 inducible cell line obtained by engineering a tetracycline-controlled transrepressor protein (TetR) in human embryonic kidney HEK293 cells.24 The TetR protein binds to tet operator (tetO) sequences in absence but not in the presence of tetracycline, silencing the transcriptional activities at the promoter.
We can thus easily distinguish between the endogenous PABP1 and the induced Flag-tagged PABP1 due to its slower migration by SDS-PAGE. We tested 7g, 8e, and 8l in this reporter system. Upon addition of tetracycline, Flag-PABP1 is induced in HEK293 cells in the presence of the indicated compound, and its methylation status can be detected by the use of a methyl-specific PABP1 antibody generated in our lab.24 In this reporter system, only 7g was able to inhibit Flag-PABP1 methylation (Figure S4 in Supporting Information).
There is increasing evidence of the involvement of CARM1 in hormone responsive cancers such as prostate cancer. Thus, we determined the effect of 7g, 8e, and 8l on prostate-specific antigen (PSA) promoter in human prostate adenocarcinoma LNCaP cells by using PSA luciferase assay, relative to a CMV-Renilla control (Figure 5, top panel). In particular, we transfected PSA reporter into LNCaP cells, and then we treated the cells with increasing concentration of 7g, 8e, or 8l for two days.
Figure 5.
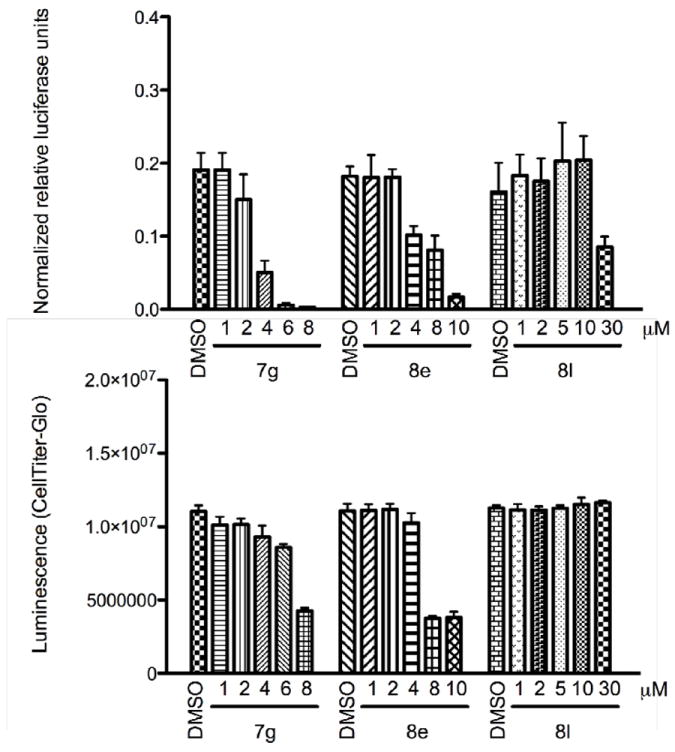
Effects of increasing concentrations of 7g, 8e, and 8l on PSA promoter activity by luciferase assay in LNCaP cells, relative to a CMV-Renilla control (top panel), and on cell viability based on quantitation of the ATP present, which is an indicator of metabolically active cells, and is used to determine the viability of cells in culture (bottom panel). The results are presented as mean ± SD that were calculated from triplicate luciferase assays.
As seen in Figure 5, a dose-dependent decrease of the reporter activity was observed with 7g and 8e up to 8-10 μM, while 8l was effective only at 30 μM. In parallel, we measured the cell viability through Cell Titer-Glo (CTG), based on quantitation of the ATP present (Figure 5, bottom panel). This was done to confirm that the observed PSA effects were the results of CARM1 inhibition, and to rule out involvement of other targets and/or cell death. 7g and 8l displayed no or little effects on cell viability, at concentrations that impacted the luciferase assay.
In conclusion, we have reported on the ability of the 1-substituted-3,5-bis(3-bromo-4- hydroxybenzylidene)piperidin-4-ones 7g and 8a-l to selectively inhibit CARM1 activity. Compounds 7g, 8e, and 8l were able to inhibit CARM1-mediated methylation of different substrates (PABP1, CA150, SmB, and H3) up to single-digit micromolar level, displaying low inhibitor activity (if any) against a panel of different PRMTs or HKMTs. In human prostate cancer LNCaP cells, 7g showed a significant dose-dependent reduction of the PSA promoter activity, at concentration that did not affect cell viability.
Experimental Section
Chemistry
Melting points were determined on a Buchi 530 melting point apparatus and are uncorrected. 1H NMR and 13C NMR spectra were recorded at 400 MHz on a Bruker AC 400 spectrometer; chemical shifts are reported in δ (ppm) units relative to the internal reference tetramethylsilane (Me4Si).
EIMS spectra were recorded with a Fisons Trio 1000 spectrometer; only molecular ions (M+) and base peaks are given. All solvents were reagent grade and, when necessary, were purified and dried by standard methods. Organic solutions were dried over anhydrous sodium sulfate. Elemental analysis has been used to determine purity of the described compounds, that is >95%. Analytical results are within ± 0.40% of the theoretical values (See Table S3 in Supporting Information). All chemicals were purchased from Aldrich Chimica, Milan (Italy), or from Alfa Aesar, Milan (Italy), and were of the highest purity.
General Procedure for the Synthesis of N-substituted-3,5-bis(3-bromo-4-hydroxybenzylidene)piperidin-4-ones (8a-l). Example: 3,5-Bis(3-bromo-4-(hydroxybenzylidene)-1-(3-chlorobenzyl)piperidin-4-one (8e)
A solution of 10e (0.42 mmol, 0.3 g) in methanol (5 mL) and 3 N hydrochloric acid (5 mL) was stirred at 60 °C for 3 h, then the suspension was neutralized with 1 N sodium hydrogen carbonate, the precipitated solid was filtered and washed with water (3 × 10 mL) and diethyl ether (3 × 10 mL) to give the pure compound 8e as a yellow powder. 1H NMR (DMSO-d6, 400 MHz, δ ppm) δ 4.43-4.48 (s, 6H, PhCH2 and CH2 piperidone), 7.05-7.74 (m, 12H, PhCH and benzene protons), 11.13 (bs, 2H, OH) ppm; 13C NMR (DMSO-d6, 400 MHz, δ; ppm) δ 53.4 (2C), 63.9, 113.7 (2C), 118.0 (2C), 126.0, 126.9, 127.3, 128.7 (2C), 129.6 (2C), 131.3 (2C), 132.2, 134.0, 136.9, 140.6 (2C), 145.9 (2C), 155.8 (2C), 186.0 ppm; MS (EI): m/z: 588.95 (M)+.
Experimental procedures for compounds 9 and 10, and chemical and physical data (Tables S2-S4) for compounds 8-10 are reported as Supporting Information.
Plasmids and antibodies
In Vitro Methylation Assay and IC50 determination
The assays have been described in detail previously.25 Briefly, all methylation reactions were performed in a final volume of 30-μL of PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.4) and in the presence of S-adenosyl-l-[methyl-3H]methionine ([3H]AdoMet, 85 Ci/mmol from a 0.5 mCi/mL in dilute HCl/ethanol 9:1, pH 2.0–2.5, PerkinElmer Life Sciences). The reaction contained 0.5–1.5 μM of substrate and 0.1-0.2 μM of recombinant enzyme with 100 μM of each indicated compound for fluorograph (Figures 3 and 4) or different doses of each compound for IC50 determination (Table 1). The reaction was incubated at 30 °C for 90 min and then separated by SDS/PAGE, transferred to a PVDF membrane, sprayed with Enhance (PerkinElmer Life Sciences), and exposed to film overnight for fluorograph. After fluorograph, the same PVDF membrane stained by Ponceau S, and cut the visualized bands of substrate to count dpm by using liquid scintillation analyzer (Tri-Carb; Packard) for graphic depiction or IC50 value determination.
Cell Lines and Cultures
Luciferase assay
LNCaP cells were cultured in phenol-red-free RPMI1640 supplemented with 10% charcoal-stripped fetal calf serum. Approximately 20 h before transfection, cells were seeded into each well of 24-well culture dishes. The cells in each well were transfected with Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer’s protocol. For each transfection, 300 ng of PSA(ARE)-LUC and 2 ng humanized CMV-Renilla internal control were used. After 12 h of transfection, cells were treated with 20 nM DHT to induce PSA-firefly and indicated amount of compound. After 42–44 h, the cells were washed twice with PBS and harvested. 5/6 Cells were used to perform luciferase assay using the Dual Luciferase Assay System (Promega) (Figure 5 top panel), and 1/6 cells were used to determine cell viability using CellTiter-Glo luminescent reagent (Promega) according to the manufacturer’s protocol (Figure 5 bottom panel).
Supplementary Material
Acknowledgments
This work was partially supported by grants from Fondazione Roma (AM), COST Action TD09/05 Epigenetics (AM), and by an institutional NIEHS center grant ES007784 and ES015188 (MTB).
ABBREVIATIONS
- AdoHcy
S-adenosyl-L-homocystein
- AdoMet
S-adenosyl-L-methionine
- AIB1
amplified in breast cancer 1
- CA150
co-activator of 150 kDa
- CREB
cAMP-responsive element binding protein
- NPL3
heterogeneous nuclear ribonucleoprotein
- PABP1
poly(A)-binding protein 1
- rpS2
ribosome protein
- PSA
prostate-specific antigen
- SmB
spliceosome protein
- tetO
tet operator
- TetR
tetracycline-controlled transrepressor protein
Footnotes
Supporting Information. Chemistry, Experimental Section. IC50 curves for 7g and 8a-l against CARM1/PABP1, PRMT1/NPL3, and SET7/H3. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Smith BC, Denu JM. Chemical mechanisms of histone lysine and arginine modifications. Biochim Biophys Acta. 2009;1789:45–57. doi: 10.1016/j.bbagrm.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taneda T, Miyata S, Kousaka A, Inoue K, Koyama Y, Mori Y, Tohyama M. Specific regional distribution of protein arginine methyltransferase 8 (PRMT8) in the mouse brain. Brain Res. 2007;1155:1–9. doi: 10.1016/j.brainres.2007.03.086. [DOI] [PubMed] [Google Scholar]
- 5.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 6.Kim D, Lee J, Cheng D, Li J, Carter C, Richie E, Bedford MT. Enzymatic activity is required for the in vivo functions of CARM1. J Biol Chem. 2010;285:1147–1152. doi: 10.1074/jbc.M109.035865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frietze S, Lupien M, Silver PA, Brown M. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 2008;68:301–306. doi: 10.1158/0008-5472.CAN-07-1983. [DOI] [PubMed] [Google Scholar]
- 8.Hong H, Kao C, Jeng MH, Eble JN, Koch MO, Gardner TA, Zhang S, Li L, Pan CX, Hu Z, MacLennan GT, Cheng L. Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status. Cancer. 2004;101:83–89. doi: 10.1002/cncr.20327. [DOI] [PubMed] [Google Scholar]
- 9.El Messaoudi S, Fabbrizio E, Rodriguez C, Chuchana P, Fauquier L, Cheng D, Theillet C, Vandel L, Bedford MT, Sardet C. Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the Cyclin E1 gene. Proc Natl Acad Sci U S A. 2006;103:13351–13356. doi: 10.1073/pnas.0605692103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majumder S, Liu Y, Ford OH, 3rd, Mohler JL, Whang YE. Involvement of arginine methyltransferase CARM1 in androgen receptor function and prostate cancer cell viability. Prostate. 2006;66:1292–1301. doi: 10.1002/pros.20438. [DOI] [PubMed] [Google Scholar]
- 11.Purandare AV, Chen Z, Huynh T, Pang S, Geng J, Vaccaro W, Poss MA, Oconnell J, Nowak K, Jayaraman L. Pyrazole inhibitors of coactivator associated arginine methyltransferase 1 (CARM1) Bioorg Med Chem Lett. 2008;18:4438–4441. doi: 10.1016/j.bmcl.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 12.Huynh T, Chen Z, Pang S, Geng J, Bandiera T, Bindi S, Vianello P, Roletto F, Thieffine S, Galvani A, Vaccaro W, Poss MA, Trainor GL, Lorenzi MV, Gottardis M, Jayaraman L, Purandare AV. Optimization of pyrazole inhibitors of Coactivator Associated Arginine Methyltransferase 1 (CARM1) Bioorg Med Chem Lett. 2009;19:2924–2927. doi: 10.1016/j.bmcl.2009.04.075. [DOI] [PubMed] [Google Scholar]
- 13.Allan M, Manku S, Therrien E, Nguyen N, Styhler S, Robert MF, Goulet AC, Petschner AJ, Rahil G, Robert Macleod A, Deziel R, Besterman JM, Nguyen H, Wahhab A. N-Benzyl-1-heteroaryl-3-(trifluoromethyl)-1H-pyrazole-5-carboxamides as inhibitors of co-activator associated arginine methyltransferase 1 (CARM1) Bioorg Med Chem Lett. 2009;19:1218–1223. doi: 10.1016/j.bmcl.2008.12.075. [DOI] [PubMed] [Google Scholar]
- 14.Therrien E, Larouche G, Manku S, Allan M, Nguyen N, Styhler S, Robert MF, Goulet AC, Besterman JM, Nguyen H, Wahhab A. 1,2-Diamines as inhibitors of co-activator associated arginine methyltransferase 1 (CARM1) Bioorg Med Chem Lett. 2009;19:6725–6732. doi: 10.1016/j.bmcl.2009.09.110. [DOI] [PubMed] [Google Scholar]
- 15.Wan H, Huynh T, Pang S, Geng J, Vaccaro W, Poss MA, Trainor GL, Lorenzi MV, Gottardis M, Jayaraman L, Purandare AV. Benzo[d]imidazole inhibitors of Coactivator Associated Arginine Methyltransferase 1 (CARM1)-Hit to Lead studies. Bioorg Med Chem Lett. 2009;19:5063–5066. doi: 10.1016/j.bmcl.2009.07.040. [DOI] [PubMed] [Google Scholar]
- 16.Selvi BR, Batta K, Kishore AH, Mantelingu K, Varier RA, Balasubramanyam K, Pradhan SK, Dasgupta D, Sriram S, Agrawal S, Kundu TK. Identification of a novel inhibitor of coactivator-associated arginine methyltransferase 1 (CARM1)-mediated methylation of histone H3 Arg-17. J Biol Chem. 2010;285:7143–7152. doi: 10.1074/jbc.M109.063933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Mai A, Massa S, Rotili D, Simeoni S, Ragno R, Botta G, Nebbioso A, Miceli M, Altucci L, Brosch G. Synthesis and biological properties of novel, uracil-containing histone deacetylase inhibitors. J Med Chem. 2006;49:6046–6056. doi: 10.1021/jm0605536. [DOI] [PubMed] [Google Scholar]; b) Mai A, Rotili D, Tarantino D, Ornaghi P, Tosi F, Vicidomini C, Sbardella G, Nebbioso A, Miceli M, Altucci L, Filetici P. Small-molecule inhibitors of histone acetyltransferase activity: identification and biological properties. J Med Chem. 2006;49:6897–6907. doi: 10.1021/jm060601m. [DOI] [PubMed] [Google Scholar]; c) Pasco MY, Rotili D, Altucci L, Farina F, Rouleau GA, Mai A, Neri C. Characterization of sirtuin inhibitors in nematodes expressing a muscular dystrophy protein reveals muscle cell and behavioral protection by specific sirtinol analogues. J Med Chem. 2010;53:1407–1411. doi: 10.1021/jm9013345. [DOI] [PubMed] [Google Scholar]; d) Binda C, Valente S, Romanenghi M, Pilotto S, Cirilli R, Karytinos A, Ciossani G, Botrugno OA, Forneris F, Tardugno M, Edmondson DE, Minucci S, Mattevi A, Mai A. Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2. J Am Chem Soc. 2010;132:6827–6833. doi: 10.1021/ja101557k. [DOI] [PubMed] [Google Scholar]
- 18.Mai A, Cheng D, Bedford MT, Valente S, Nebbioso A, Perrone A, Brosch G, Sbardella G, De Bellis F, Miceli M, Altucci L. epigenetic multiple ligands: mixed histone/protein methyltransferase, acetyltransferase, and class III deacetylase (sirtuin) inhibitors. J Med Chem. 2008;51:2279–2290. doi: 10.1021/jm701595q. [DOI] [PubMed] [Google Scholar]
- 19.Costi R, Di Santo R, Artico M, Miele G, Valentini P, Novellino E, Cereseto A. Cinnamoyl compounds as simple molecules that inhibit p300 histone acetyltransferase. J Med Chem. 2007;50:1973–1977. doi: 10.1021/jm060943s. [DOI] [PubMed] [Google Scholar]
- 20.Lee J, Bedford MT. PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 2002;3:268–273. doi: 10.1093/embo-reports/kvf052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McBride AE, Cook JT, Stemmler EA, Rutledge KL, McGrath KA, Rubens JA. Arginine methylation of yeast mRNA-binding protein Npl3 directly affects its function, nuclear export, and intranuclear protein interactions. J Biol Chem. 2005;280:30888–30898. doi: 10.1074/jbc.M505831200. [DOI] [PubMed] [Google Scholar]
- 22.Cheng D, Cote J, Shaaban S, Bedford MT. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 23.Swiercz R, Cheng D, Kim D, Bedford MT. Ribosomal protein rpS2 is hypomethylated in PRMT3-deficient mice. J Biol Chem. 2007;282:16917–16923. doi: 10.1074/jbc.M609778200. [DOI] [PubMed] [Google Scholar]
- 24.Deuschle U, Meyer WK, Thiesen HJ. Tetracycline-reversible silencing of eukaryotic promoters. Mol Cell Biol. 1995;15:1907–1914. doi: 10.1128/mcb.15.4.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mai A, Valente S, Cheng D, Perrone A, Ragno R, Simeoni S, Sbardella G, Brosch G, Nebbioso A, Conte M, Altucci L, Bedford MT. Synthesis and biological validation of novel synthetic histone/protein methyltransferase inhibitors. ChemMedChem. 2007;2:987–991. doi: 10.1002/cmdc.200700023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


