Abstract
Pantothenate kinase-associated neurodegeneration (PKAN) is a rare, inborn error of metabolism characterized by iron accumulation in the basal ganglia and by the presence of dystonia, dysarthria, and retinal degeneration. Mutations in pantothenate kinase 2 (PANK2), the rate-limiting enzyme in mitochondrial coenzyme A biosynthesis, represent the most common genetic cause of this disorder. How mutations in this core metabolic enzyme give rise to such a broad clinical spectrum of pathology remains a mystery. To systematically explore its pathogenesis, we performed global metabolic profiling on plasma from a cohort of 14 genetically defined patients and 18 controls. Notably, lactate is elevated in PKAN patients, suggesting dysfunctional mitochondrial metabolism. As predicted, but never previously reported, pantothenate levels are higher in patients with premature stop mutations in PANK2. Global metabolic profiling and follow-up studies in patient-derived fibroblasts also reveal defects in bile acid conjugation and lipid metabolism, pathways that require coenzyme A. These findings raise a novel therapeutic hypothesis, namely, that dietary fats and bile acid supplements may hold potential as disease-modifying interventions. Our study illustrates the value of metabolic profiling as a tool for systematically exploring the biochemical basis of inherited metabolic diseases.
Keywords: PKAN, coenzyme A, mitochondria, mass spectrometry, cholesterol
1. Introduction
Neurodegeneration with Brain Iron Accumulation (NBIA) is a severe class of neurodegenerative disorders with clinical presentations ranging from early-onset neurodegeneration with premature fatality to adult-onset parkinsonism-dystonia [1]. These disorders follow an autosomal recessive pattern of inheritance, and distinct subclasses of disease are defined by mutations in specific genes: mutations in PANK2 (MIM*606157) are associated with Pantothenate Kinase-Associated Neurodegeneration (PKAN), PLA2G6 (MIM*256600) with PLA2G6-Associated Neurodegeneration (PLAN, also known as INAD), and FA2H (MIM*611026) with Fatty Acid Hydroxylase Neurodegeneration (FAHN).
PKAN is the most prevalent form of NBIA, and two distinct manifestations of this disease are observed. Classic PKAN patients present in the first decade of life with dystonia, dysarthria, rigidity, and pigmentary retinal degeneration. Eventually patients develop dysphagia, gastro-oesophageal reflux, and constipation. Periods of stability are interspersed with periods of rapid decline: patients typically do not survive past age 20 due to complications including malnutrition and pneumonia. Atypical PKAN does not present until the second or third decade of life and progresses more slowly than classic PKAN. Early signs include gait abnormality, speech defects, and some psychiatric manifestations. The main diagnostic criterion for both phenotypes is the presence of the “eye of the tiger” pattern in the medial globus pallidus on MRI. Many patients with NBIA show hypointensity in the globus pallidus, but PKAN patients exhibit additional hyperintensity in the antero medial region [2].
PKAN is an inborn error of Vitamin B5 metabolism [3]. Vitamin B5, otherwise known as pantothenate, is a micronutrient required for production of coenzyme A in cells. The vitamin is present in a variety of dietary sources including whole grains and meat as Coenzyme A and is processed to pantothenate in the intestine. Pantothenate is absorbed by endothelial cells via a sodium-dependent multivitamin transporter and then passes to the blood for delivery to the rest of the body [4]. At the cellular level, it is phosphorylated by pantothenate kinase, conjugated to cysteine, decarboxylated, conjugated to an adenosyl group and phosphorylated again to coenzyme A. All of these enzymatic activities have been detected in the cytosol and many in the mitochondrion; although the cysteine ligase and decarboxylation activities have not been detected explicitly in the mitochondrion, it has been proposed that an exclusively mitochondrial coenzyme A synthetic pathway exists [5,6,7]. The key rate-limiting step in this pathway is the phosphorylation of pantothenate by pantothenate kinase. Four isoforms of this enzyme are known: PANK1, PANK2, PANK3 and PANK4. It is unclear whether PANK4 is functional, but PANK1 and PANK3 are active in the cytosol, while PANK2 is localized to and active in the mitochondrion [6].
Coenzyme A is involved in a number of metabolic pathways, including the citric acid cycle, sterol and steroid biosynthesis, heme biosynthesis, amino acid synthesis, and β-oxidation. Mutations in PANK2, which encodes the key enzyme in this biosynthetic pathway, are expected to result in defective CoA biosynthesis, which could lead to a variety of metabolic defects. Recently it has been hypothesized that diminished CoA pools have a detrimental effect on histone and tubulin acetylation, contributing to the neurological phenotype of PKAN [8]. However, it is not known how mutations in PANK2 cause the spectrum of clinical symptoms exhibited by PKAN patients. Previous attempts to understand the mechanism of PKAN using animal models have met with limited success. A mouse model of PKAN exhibits retinal degeneration but lacks any neurological phenotype [9]. A Drosophilia model of PKAN does have a brain phenotype, but this involves the formation of vacuoles, not iron accumulation, and Drosophilia do not have as many pantothenate kinase isoforms as humans [10]. Given that the downstream product of PANK2, coenzyme A, is involved in a number of metabolic pathways, we considered global metabolic profiling as a useful approach to systematically characterize the biochemical abnormalities in this population.
2. Materials and Methods
2.1 Human Subjects
Patients were recruited based on clinical presentation, MRI findings and presence of PANK2 mutations, at the Unit of Child Neurology, IRCCS Foundation Neurological Institute “C.Besta” and at the Unit of Molecular Medicine, Bambino Gesù Pediatric Hospital; healthy controls were recruited at the “C. Besta” Institute. Subjects’ consent was obtained according to the Declaration of Helsinki: BMJ 1991; 302, 1194. In addition, we obtained institutional review board–approved informed consent from parents of all probands and controls before collecting blood for DNA extraction or performing skin biopsies. Our cohort includes 14 patients, one of whom (GHBE70) had blood drawn on three separate occasions, and two others (PAVA83 and PERO96) who had blood drawn on two separate occasions. Patient and control plasma samples were well-matched based on sex (22% male patients, 28% male controls), age (23 +/− 11 for patients, 27 +/− 4 for controls, P > 0.05) and BMI (20.9 +/− 6.9 for patients, 22.6 +/− 3.8 for controls, P > 0.05).
2.2 Cell culture
Fibroblasts derived from patients and controls were cultured at 37°C in a humidified 5% CO2 atmosphere, and were trypsinized once or twice a week. The culture medium, replaced twice per week, was Dulbecco's modified Eagle’s medium (DMEM) containing 4.5 g/L glucose, 10 % (V/V) foetal calf serum, 1 mM sodium pyruvate, 200 U/mL Penicillin G, 200 mg/mL streptomycin, and 4 mM glutamine. Control and patient fibroblasts were cultured for 6–8 passages, trypsinized and collected by centrifugation. The dry cell pellets were stored at −80°C until analysis.
2.3 Metabolic Profiling
Sample collection for metabolic profiling was performed on “non-fasted” patients and controls after 3 hours fasting, as previously described [11,12,13,14]. For each sample, 4 mL of blood was drawn and collected in K2 EDTA tubes, which were immediately centrifuged for 10 minutes at 2000g and 6°C. Plasma was divided into five 400 µL aliquots and stored at −80°C within 10 minutes. Plasma samples were coded and de-identified prior to metabolic profiling. Three separate LC-MS methods were used on each plasma sample: (1) a hydrophilic interaction liquid chromatography (HILIC) method for analysis of polar metabolites in the positive ion mode, (2) an ion pairing chromatography (IPC) method to measure polar metabolites in the negative ion mode, and (3) a reversed phase (RP) method to profile lipids. All LC-MS analyses were performed using a 4000 QTRAP triple quadrupole mass spectrometer (AB SCIEX; Foster City, CA) coupled to either a 1100 series pump or a 1200 Series pump (Agilent Technologies; Santa Clara, CA) and an HTS PAL autosampler (Leap Technologies; Carrboro, NC). A total of 36 plasma samples were analyzed in a blinded, random order.
For HILIC analyses, plasma samples (10 µL) were prepared via protein precipitation with the addition of nine volumes of 74.9:24.9:0.2 v/v/v acetonitrile/methanol/formic acid containing stable isotope-labeled internal standards (valine-d8, Isotec, Miamisburg, OH; and phenylalanine-d8, Cambridge Isotope Laboratories; Andover, MA). The samples were centrifuged (10 min, 10,000 rpm, 4°C), and the supernatants were injected directly onto a 150×2.1 mm Atlantis HILIC column (Waters; Milford, MA). The column was eluted isocratically at a flow rate of 250 µL/min with 5% mobile phase A (10 mM ammonium formate and 0.1% formic acid in water) for 1 minute followed by a linear gradient to 40% mobile phase B (acetonitrile with 0.1% formic acid) over 10 minutes. MS analyses were carried out using electrospray ionization and selective multiple reaction monitoring scans in the positive ion mode. De-clustering potentials and collision energies were optimized for each metabolite by infusion of reference standards before sample analyses. The ion spray voltage was 4.5 kV and the source temperature was 425°C.
For IPC analyses, plasma (130 µL) was extracted with 400 µL of 80:20 v/v methanol/water containing thymine-d4 and glutamate-13C5,15N (Cambridge Isotope Laboratories, Andover, MA) internal standards. After centrifugation, 430 µL of supernatant was transferred to a fresh tube and evaporated under nitrogen gas in a Turbovap LV (Caliper, Hopkinton, MA) at 37°C. Dried samples were resuspended in 80 µL of water containing glycocholate-d4 and inosine-15N4 internal standards (Sigma-Aldrich, St. Louis, MO) and injected onto a 100×3 mm Atlantis T3 column (Waters, Milford, MA). The column was eluted isocratically at a flow rate of 350 µL/min with 100% mobile-phase A (10mM tributylamine and 15mM acetic acid in water) for 2 minutes followed by a linear gradient to 98% mobile phase B (methanol) over 20 minutes. MS analyses were carried out as above, except in the negative ion mode, with ion spray voltage at −4.5 kV and a source temperature of 550°C.
Plasma samples (10 µL) were extracted for lipid analyses with 190 µL of isopropanol containing 1-dodecanoyl-2-tridecanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids; Alabaster, AL). After centrifugation, supernatants were injected directly onto a 150×3.0 mm Prosphere HP C4 column (Grace, Columbia, MD). The column was eluted isocratically with 80% mobile phase A (95:5:0.1 vol/vol/vol 10 mM ammonium acetate/methanol/acetic acid) for 2 minutes followed by a linear gradient to 80% mobile-phase B (99.9:0.1 vol/vol methanol/acetic acid) over 1 minute, a linear gradient to 100% mobile phase B over 12 minutes, then 10 minutes at 100% mobile-phase B. MS analyses were carried out using electrospray ionization and Q1 scans in the positive ion mode. Ion spray voltage was 5.0 kV and source temperature was 400°C. For each lipid analyte, the first number denotes the total number of carbons in the lipid acyl chain(s) and the second number (after the colon) denotes the total number of double bonds in the lipid acyl chain(s).
For each method, internal standard peak areas were monitored for quality control and individual samples with peak areas differing from the group by more than two standard deviations were re-analyzed. MultiQuant software (Version 1.1; AB SCIEX; Foster City, CA) was used for automated peak integration and metabolite peaks were manually reviewed for quality of integration and compared against a known standard to confirm identity.
2.4 Fasting lactate, pyruvate and alanine measurements
A second blood draw was performed on a subset of available patients and controls (n=10 individuals in each group), after an overnight fast and without using a tourniquet (a large rubber strap) secured above the vein. Lactic and pyruvic acids were assayed spectrophotometrically using lactate dehydrogenase [15]. Plasma alanine with corresponding stable-isotope standard was extracted by acetonitrile deproteinization [16]. Butylated derivatives were analyzed and quantified in MRM mode on an API 2000 LC/MS/MS system (PE SCIEX - Applied Biosystems) according to standard procedures.
2.5 Isotope dilution mass spectrometry analysis for sterols and oxysterols
Sterol and oxysterol measurements were performed on plasma collected from both fasting and non-fasting PKAN patients and controls. To a screw-capped vial sealed with a Teflon septum, 250 µl of plasma are added together with 200 ng D4-lathosterol, 500 ng D7-sitosterol and D7-campesterol, 100 ng D7-7α-hydroxycholesterol, D7-7β-hydroxycholesterol, D7-7ketocholesterol, D3-24S-hydroxycholesterol and D6-27-hydroxycholesterol as internal standards. To prevent auto oxidation, we added 50 µl butylated hydroxytoluene (5 g/L) and 50 µl EDTA (10 g/L) to each vial and flushed with nitrogen to remove air.
For sterol and oxysterol determination in cultured cells, the dry cell pellet was suspended in 1 mL of NaCl. 750 µL were used for sterol analysis and 250 µL for determination of protein with the Lowry method. Together with 50 µl butylated hydroxytoluene (5 g/L) and 50 µl EDTA (10 g/L) to a screw capped glass vials were added also 5 µg D6-cholesterol, 100ng of D4-lathosterol, 20ng of D7-24S-hydroxycholesterol.
Alkaline hydrolysis was allowed to proceed at room temperature (22°C) with magnetic stirring for 1 h in the presence of ethanolic 1M potassium hydroxide solution. After hydrolysis, the sterols were extracted twice with 5 mL cyclohexane. The organic solvents were evaporated under a gentle stream of argon and converted into trimethylsilyl ethers (pyridine:hexamethyldisilazane: trimethylchlorosilane 3:2:1 v/v/v).
Gas chromatography mass spectrometry (GC-MS) analysis was performed on a GC equipped with an Elite column (30m×0.32mm id×0.25 mm film; Perkin Elmer, USA) and injection was performed in splitless mode using helium (1 mL/min) as a carrier gas. The temperature program was as follows: initial temperature of 180°C was held for 1 min, followed by a linear ramp of 20°C/min to 270°C, and then a linear ramp of 5°C/min to of 290°C, which was held for 10 min.
The mass spectrometer operates in the selected ion-monitoring mode. Peak integration is performed manually, and sterols are quantified from selected-ion monitoring analyses against internal standards using standard curves for the listed sterols. Additional qualifier (characteristic fragment ions) ions were used for structural identification [17,18].
2.6 Fatty acids analysis in fibroblasts
For fatty acids determination in cultured cells, the dry cell pellet was suspended in 1 mL of NaCl. 750 µL were used for sterol analysis and 250 µL for determination of protein with the Lowry method. Together with 50 µl butylated hydroxytoluene (5 g/L) and 50 µl EDTA (10 g/L) to a screw capped glass vials was added 20 µg of heptadecanoic acid (C17:0). Alkaline hydrolysis was allowed to proceed at room temperature (22°C) with magnetic stirring for 1 h in the presence of ethanolic 1M potassium hydroxide solution. After hydrolysis, the sterols were extracted twice with 5 mL cyclohexane and twice with 5 ml of ethyl acetate. The organic solvents were evaporated under a gentle stream of argon and converted into trimethylsilyl ethers (pyridine:hexamethyldisilazane: trimethylchlorosilane 3:2:1 v/v/v).
Gas chromatography mass spectrometry (GC-MS) analysis was performed on a Perkin Elmer Clarus 600 GC-Mass Spectrometer system. The GC is equipped with an Elite column (30m×0.32mm id×0.25 mm film; Perkin Elmer, USA) and injection was performed in the split 1:10 mode and using helium (1 mL/min) as a carrier gas. For fatty acids determination the temperature program was as follows: initial temperature of 100°C was held for 1 min, followed by a linear ramp of 10°C/min to 300°C, and then was held for 10 min at 300°C. The mass spectrometer operates in scan (50–600 mz) mode. Peak integration is performed manually and fatty acids were quantified from total area of each compound against internal standard area using standard curves for the listed fatty acids. Each molecule was identified on the basis of the fragmentation pattern and retention time. The FA quantification was linear up to 100 µg and the mean within run variation was less than 5% (n = 10).
2.7 Univariate Analysis
For global metabolic profiling, the statistical significance of metabolic variation between patient and control plasma samples was calculated using the Wilcoxon Rank Sum test, ignoring NaN values for metabolites not robustly measured in a given sample. A significance threshold of 0.05 would be expected to yield approximately 11.5 false positives over the 230 metabolites (hypotheses) tested [19]. The significance of demographic differences between patients and controls was calculated using an unpaired, two-sided Student’s T Test assuming unequal variance. For single metabolite measurements, continuous data were inspected and tested to determine whether distributions were normal by Kolmogorov-Smirnov normality test, and compared using a two-sided Student’s T-Test assuming unequal variance or parametric Student’s T-Test, as appropriate. Values for statistical significance were set at P < 0.05. All analyses were performed with Matlab (The MathWorks Inc.) or Sigmastat 3.01 (SigmaAldrich, St Louis, MO, USA).
3. Results
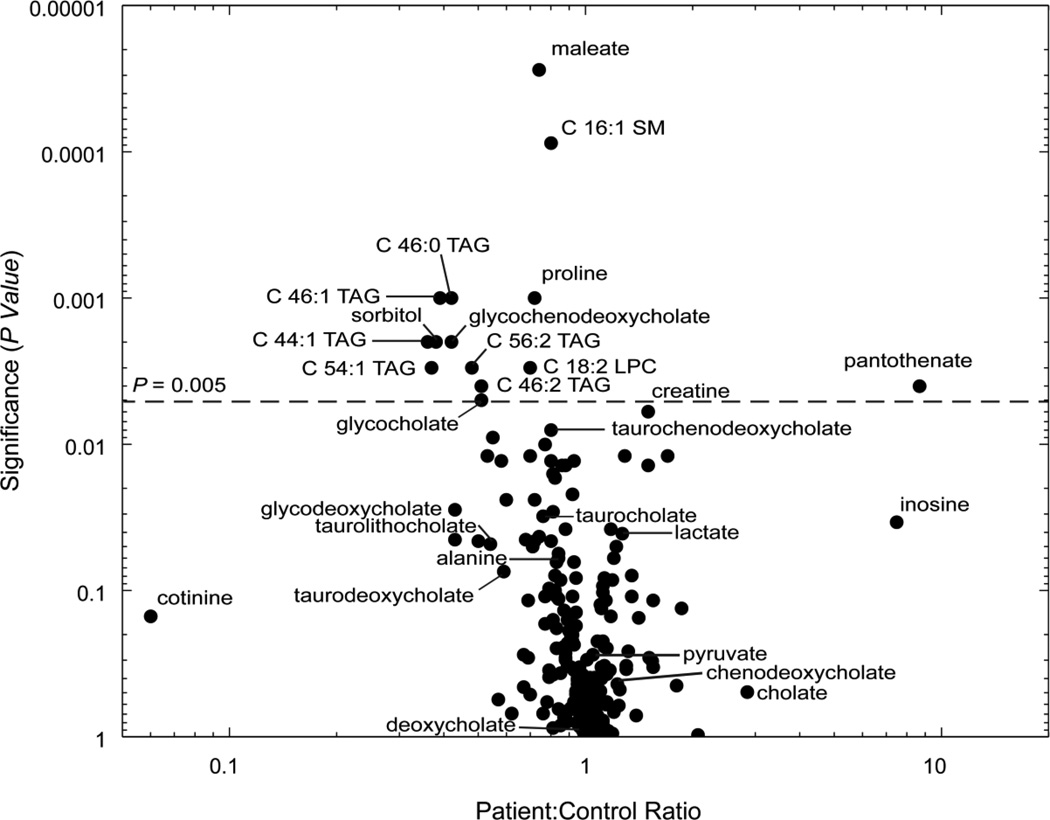
We performed global metabolic profiling on plasma samples obtained from a cohort of 14 genetically-verified PKAN patients and 18 healthy age- and gender-matched controls, taking advantage of plasma samples from multiple blood draws available for a subset of patients. We used a validated [11,12,13,14] analytical system that combines tandem mass spectrometry with high-performance liquid chromatography (HPLC-MS/MS) to measure the levels of 245 polar and nonpolar metabolites. Table 1 summarizes the clinical and genetic feautures of the 14 patients analyzed in this study, as well as a list of current medications at the time of sampling. We acknowledge the confounding effects of lifestyle factors, which we are unable to control in such small studies of patients with rare genetic disease. We determined the relative levels of 245 metabolites spanning the breadth of metabolism from amino acids and their derivatives to a variety of lipid species. Of the 245 metabolites targeted, 230 were detected and well measured in at least 80% of human plasma samples. The abundance of these 230 metabolites in patients versus controls is summarized in Figure 1 (full data in Supplementary Table 1).
Table 1.
Demographic, clinical, and genetic characteristics of patient cohort.
| Patient ID |
Gender | Age (yrs) |
Height (cm) |
Weight (kg) |
BMI | PKAN Presentation |
Mutation (cDNA) |
Mutation (Protein) |
Medications |
|---|---|---|---|---|---|---|---|---|---|
| ARFR80 | F | 28 | 133 | 32 | 18.1 | Classical | 569insA homozygous ex1 | Y190X | Trihexiphenidyl, Baclofen, Pimozide, Gabapentin, Valproic Acid, Diazepam, Omeprazole |
| ARRO78 | F | 31 | 154 | 51 | 21.5 | Classical | 569insA homozygous ex1 | Y190X | Trihexiphenidyl, Baclofen, Gabapentin |
| BOOLR2 | F | 27 | 150 | 48 | 21.3 | Atypical | IVS1+5 g>c ex1 + 635G>A ex.2 | ?+.E212G | Baclofen, CoEq |
| COVI02 | M | 5 | 107 | 17 | 14.8 | Classical | A1499T ex.5 et. + ivs2-1 G>A et. | N500I + ? | Trihexiphenidyl |
| DALA87 | F | 21 | 155 | 34 | 14.2 | Classical | 683T>C ex. 2 + 1648T>C ex.6 | F228S+F550L | Trihexiphenidyl, Pimozide, Baclofen IT, Ranitidine/Domperidone, Levetiracetam |
| DICA86 | F | 23 | 160 | 45 | 17.6 | Atypical | T332A homozygous ex.1 | L111Q | - |
| GHBE70 | F | 38 | 180 | 111 | 34.3 | Atypical | G1561A ete ex.6 + G775A et. ex.2 | G521R+G259R | - |
| MAAM02 | M | 7 | 115 | 20 | 15.1 | Classical | 1259 del G homozygous ex.4 | F419fsX472 | - |
| MAMO99 | M | 10 | 128 | 23 | 14.0 | Classical | 1259 del G homozygous ex.4 | F419fsX472 | - |
| PAVA83 | F | 26 | 162 | 62 | 23.6 | Classical | C1069T homozygous ex.3 | R357W | Diazepam, Paroxetine, Trihexiphenidyl, Gabapentin, coEq |
| PERO96 | F | 12 | 158 | 40 | 16.0 | Classical | 790C>T homozygous ex3 | R264W | Trihexiphenidyl, CoEq |
| ROST73 | M | 35 | 180 | 60 | 18.5 | Classical | 790C>T homozygous ex2 | R264W | Trihexiphenidyl, Baclofen, Tetrabenazine, Diazepam |
| SASA82 | F | 26 | 165 | 58 | 21.3 | Atypical | 790C>T ex2+ 856C>T ex.2 | R264W+R286C | Vitamin B5, Baclofen |
| SCFR94 | F | 12 | 120 | 25 | 17.4 | Classical | 821-822delCT / del ex 1-4 | L273fsX290/not translated | Trihexiphenidyl, Baclofen IT |
Abbreviations: M/F, Male/Female.
Figure 1.
Plasma metabolites in patients versus healthy controls. The figure depicts the 230 metabolites detected in > 80% of plasma samples. For each metabolite, the significance of fold change (Wilcoxon Rank Sum test) is plotted as a function of fold change in patients versus controls. For each lipid analyte (triacylgylcerols abbreviated as TAG, sphingomyelins as SM, and lysophosphatidylcholines as LPC), the number of carbons and double bonds in the acyl chain(s) are denoted.
3.1 Markers of mitochondrial dysfunction
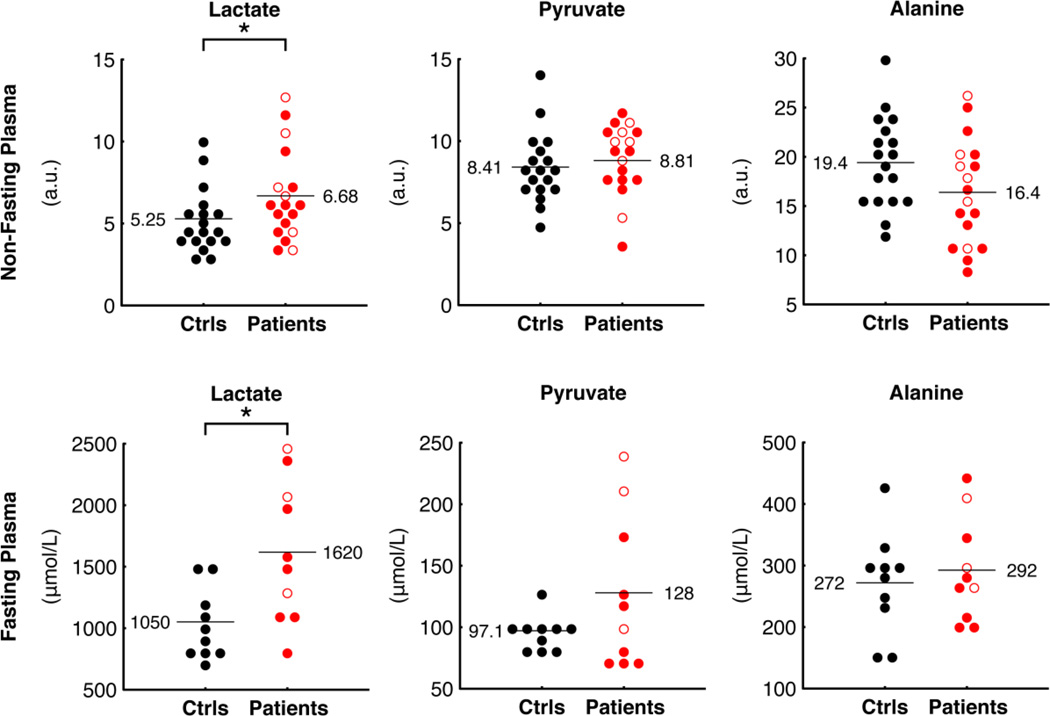
Given that PANK2 is a mitochondrial enzyme, we looked for biochemical evidence of mitochondrial dysfunction. Classic markers for mitochondrial dysfunction include elevations in lactate, alanine, and the lactate/pyruvate ratio [20]. Of these metabolites, we found that lactate was significantly elevated in patients compared to controls, with a patient:control ratio of 1.27 (P = 0.041) in our metabolic profiling analysis of non-fasted subjects (Figure 2). To confirm this observation, we performed a second blood draw on a subset of patients and controls after an overnight fast and analyzed lactate biochemically. We observed a patient:control ratio of 1.54 (P = 0.01) (Figure 2), confirming the metabolomic data. Levels of pyruvate and alanine were not significantly different between patients and controls in plasma from non-fasting subjects analyzed with metabolic profiling, or from fasting subjects analyzed biochemically (Figure 2).
Figure 2.
Levels of plasma lactate, pyruvate, and alanine in patients and controls measured with global metabolic profiling of non-fasting subjects (top) and using clinical assays on fasting subjects (bottom). Open red circles represent patients with atypical PKAN, filled red circles represent patients with classic PKAN; black filled circles represent controls. Horizontal lines and accompanying values denote mean patient and control levels. For metabolites measured with metabolic profiling, levels are reported as chromatographic peak area (arbitrary units, a.u.). A star (*) denotes significant differences (P value < 0.05; Wilcoxon Rank Sum test for metabolic profiling measurements and two-tailed, unpaired Student’s T Test assuming unequal variance for clinical measurements).
3.2 Pantothenate levels are elevated in patients with PANK2 stop mutations
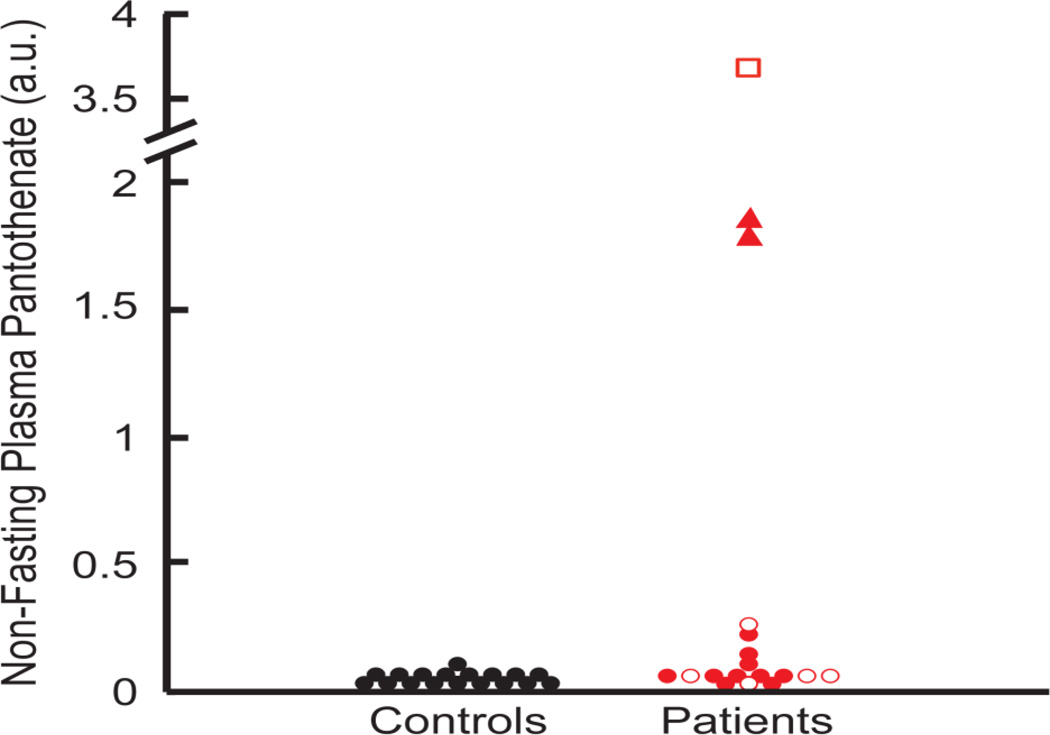
We found elevated plasma levels of pantothenate in patients relative to controls. Pantothenate is the direct substrate of PANK2, so an elevation in this metabolite might be expected, though it has not been reported previously. The highest level of pantothenate was found in the patient reported to be taking a Vitamin B5 supplement, indicating compliance with the supplement regimen, though failing to shed light on the efficacy of this treatment. Patients ARFR80 and ARRO78 also presented with high levels of pantothenate, although they have never taken and they were not taking pantothenate at the time of blood sampling. It is notable that these patients both harbor a premature Y190X stop mutation likely resulting in complete loss of PANK2 function. Although the majority of the difference in pantothenate levels was driven by these three individuals, pantothenate elevation was a general trend across all PKAN patients. In fact, by excluding the above three outliers, the patient:control ratio is 1.72, and remains statistically significant (P = 0.023) (Figure 3).
Figure 3.
(a) Plasma levels of pantothenate in 18 patient samples and 18 control samples detected with global metabolic profiling, reported as chromatographic peak area (arbitrary units, a.u.). Patients with the Y190X stop mutation are noted with a red triangle, and the one patient taking oral pantothenate supplements is noted with a red square. Open red shapes represent patients with atypical PKAN, filled red shapes represent patients with classic PKAN; black filled circles represent controls.
3.3 Global metabolic profiling suggests dysregulated bile acid synthesis and triacylglycerol (TAG) deficiency in PKAN patients
A number of TAGs were significantly reduced in patients compared to controls (Figure 1 and Supplementary Table 1). In addition, lysophosphatidylcholines (LPC), phosphatidylcholines (PC), and sphingomyelin (SM) were found to be significantly reduced. The metabolomic analysis revealed a general reduction of esterified fatty acids, especially esters with glycerol such as triglycerides and phospholipids (see Supplementary Table 1). This suggests that the synthesis of fatty acids, from acetyl-CoA and malonyl-CoA, is defective in the patients possibly because of a reduced CoA pool. We also highlighted a significant reduction of C 16:1 SM (Figure 1 and Supplementary Table 1), a sphingolipid species found in animal cell membranes.
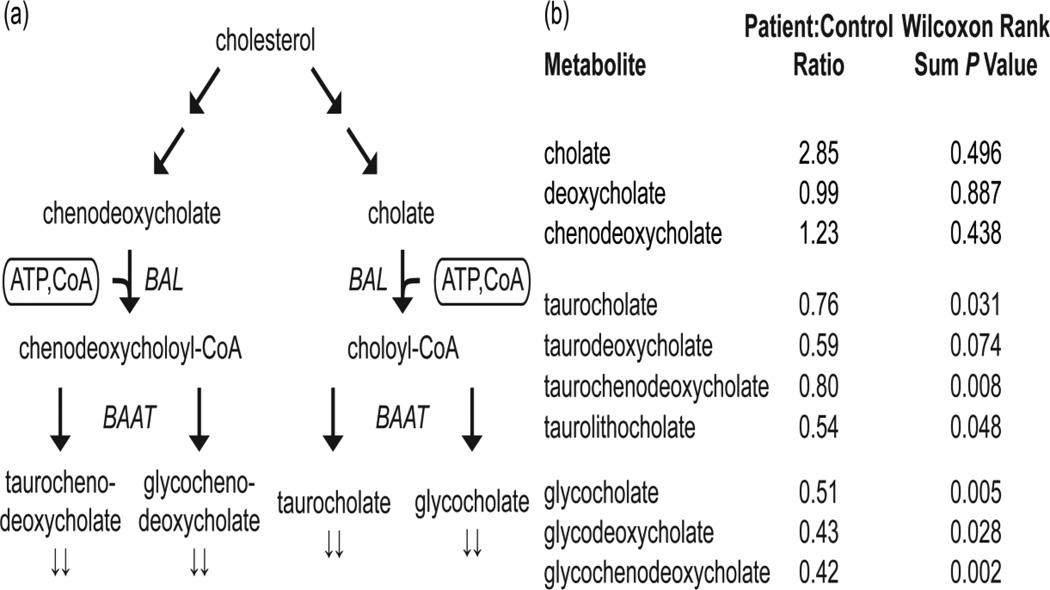
We detected significantly lower levels of tauro- and glyco-conjugated bile acids (Figure 4), which are only produced when coenzyme A is available as a cofactor. Of the seven conjugated bile acids measured here, six had patient:control ratios less than 0.9 with P < 0.05. This suggests dysregulated conjugation of bile acids in the patient population. Primary bile acids cholate and chenodeoxycholate were slightly but not significantly higher in PKAN patients compared to controls (Figure 4 and Supplementary Table 1).
Figure 4.
(a) A simplified schematic of the bile acid biosynthetic pathway. Cofactors are enclosed in rounded rectangles. Arrows (↓↓) denote metabolites detected with metabolic profiling to be significantly lower in patients relative to controls (Wilcoxon Rank Sum P < 0.05). Select enzymes in this pathway, bile acid CoA ligase (BAL) and bile acid-CoA:amino acid N-acyltransferase (BAAT), are shown in italics. (b) A summary of bile acids measured in this study.
3.4 Biochemical analysis of cholesterol and fatty acid biosynthesis
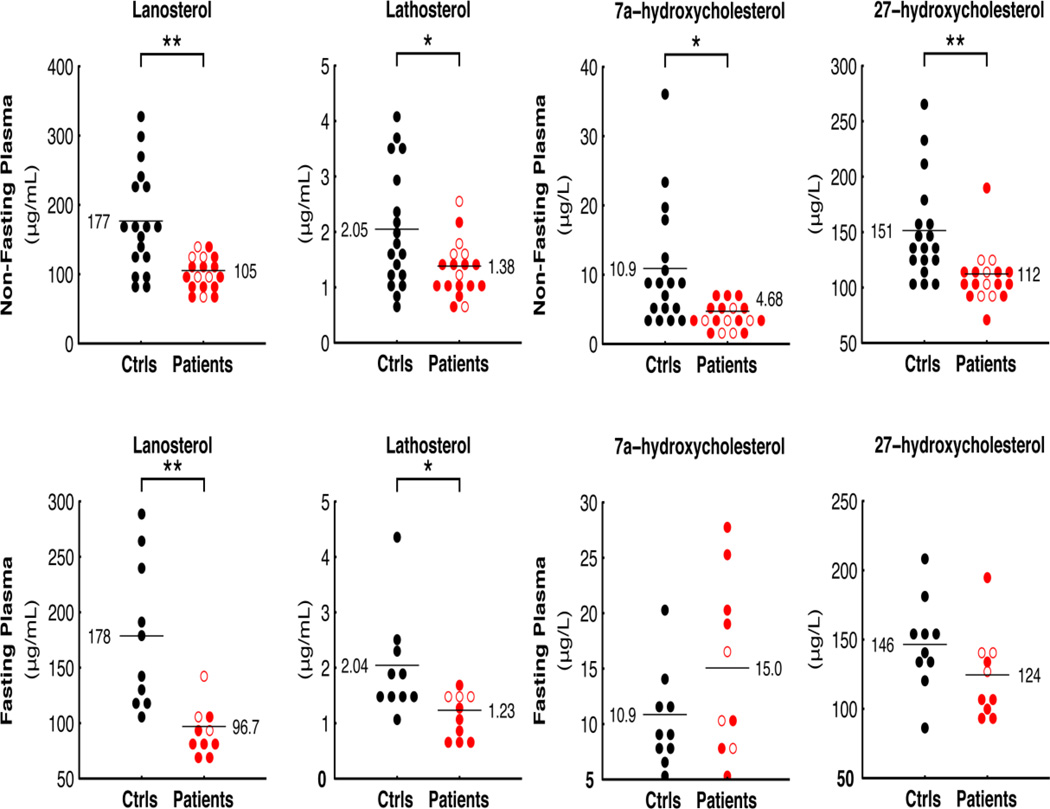
Coenzyme A is also required for cholesterol and fatty acid biosynthesis. Since the metabolic profiles suggested bile acid dysregulation, we used an alternative technology to measure markers of cholesterol biosynthesis both in non-fasted patients and controls, as well as in plasma from the fasting subset. We found that the cholesterol precursors lanosterol and lathosterol, considered markers of whole-body cholesterol biosynthesis [21] were significantly reduced in PKAN patients compared to controls, irrespective of fasting status (Figure 5).
Figure 5.
Plasma markers of sterol biosynthesis and absorption. Lathosterol and lanosterol (left) in patient and control plasma under non-fasting (top) and fasting conditions (bottom). 7a-hydroxycholesterol and 27-hydroxycholesterol (right) in patient and control plasma under non-fasting (top) and fasting conditions (bottom). Two stars (**) denote significant differences (P values < 0.005); other symbols as in Figure 2.
We also measured the plasma oxysterols 7α-hydroxycholesteorol (7a-OHC), formed in the first step of the neutral bile acid biosynthetic pathway, and 27-hydroxycholesterol (27OHC), formed mainly in the acidic pathway. In the non-fasted group, 27OHC was significantly lower in PKAN patients, while a mild reduction was found in the levels of 7a-OHC (Figure 5). No statistically significant differences were found in fasted subjects.
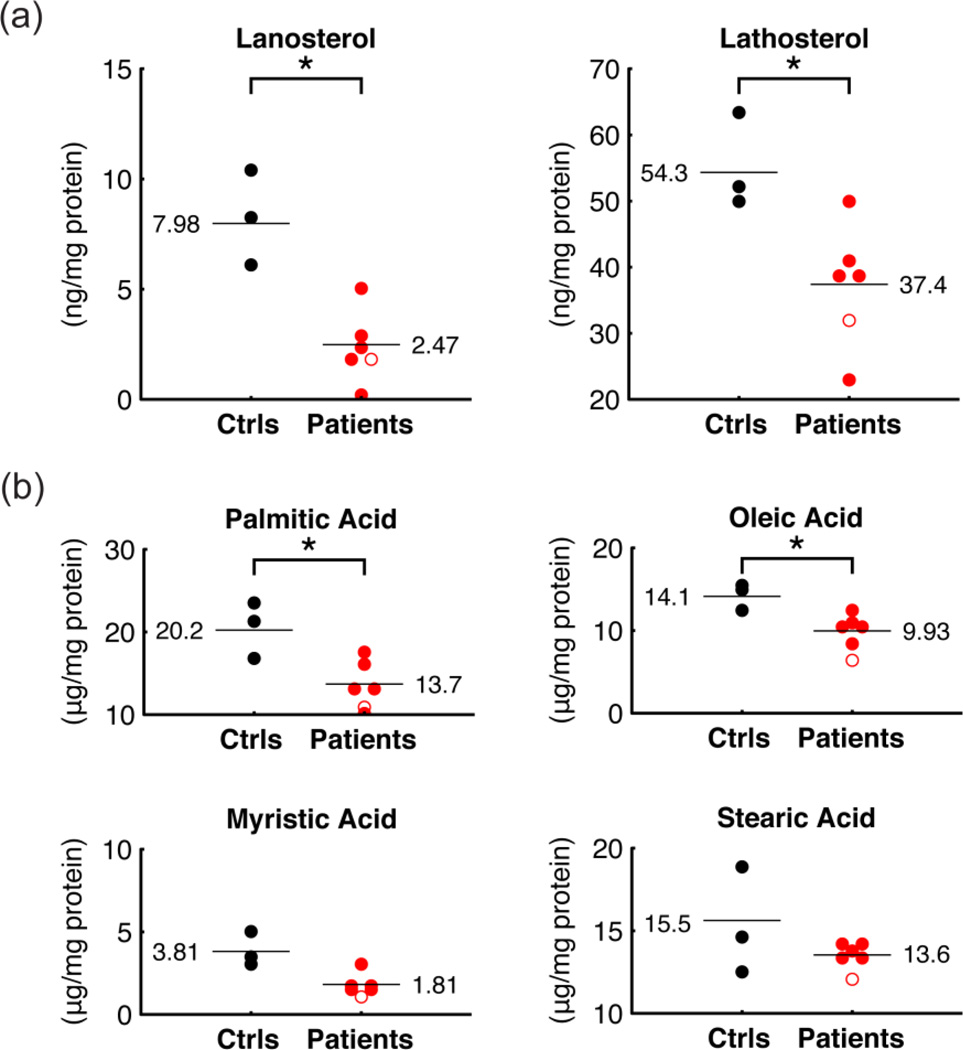
To complement the plasma analysis, we also measured markers of cholesterol synthesis and fatty acid levels in primary fibroblast cell lines from three controls and six PKAN patients, including four from our metabolomic analysis (ARFR80, MAAM02, ROST73, GHBE70. Table 1). The two additional cell lines derived from one patient with the compound heterozygous mutations R264W and S471N, and one patient homozygous for R286C. At the cellular level, we detected a reduction of lanosterol and lathosterol (Figure 6a), as well as a reduction of palmitic acid, the primary fatty acid synthesized by fatty acid synthase (FAS), and of myristic, oleic and stearic acids (Figure 6b). Taken together, these data support the hypothesis of reduced cholesterol and fatty acids synthesis as a result of mutations in PANK2.
Figure 6.
Cholesterol precursors and fatty acids in fibroblasts. (a) Lathosterol and lanosterol in fibroblasts derived from patients and healthy controls. (b) Palmitic, oleic, myristic and stearic acid levels in fibroblasts derived from patients and healthy controls. Symbols as in Figure 2.
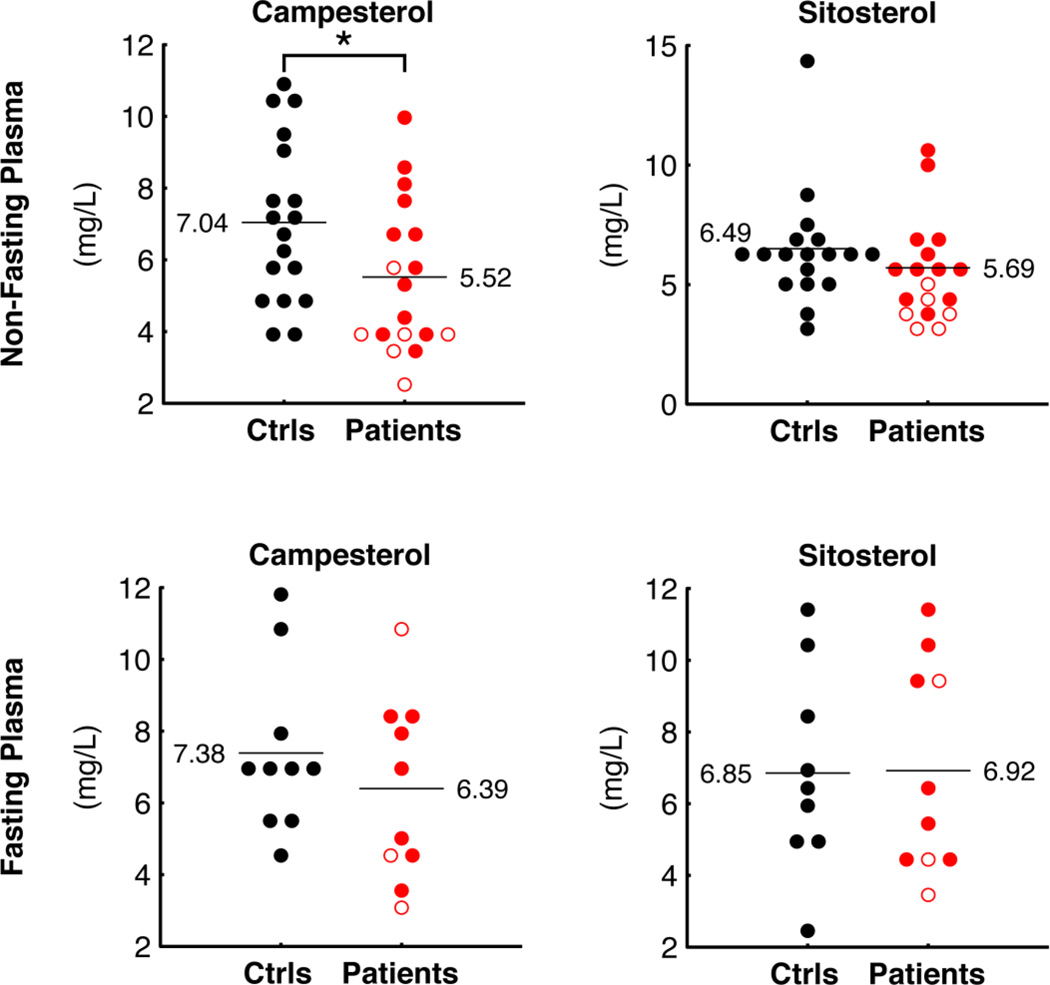
Finally, we measured the plant sterols campesterol and sitosterol (Figure 7), used as markers for nutrient absorption [22] and we found that campesterol was reduced in patient plasma compared to controls (P = 0.032). Sitosterol was also lower in patient plasma, though this did not reach statistical significance (P = 0.267. Under fasting conditions, these differences in sterol levels disappeared (Figure 7b).
Figure 7.
Markers of nutrient absorption. Plant sterols campesterol and sitosterol were measured in plasma derived from patients and controls under non-fasting (top) or fasting conditions (bottom). Symbols as in Figure 2.
4. Discussion
Here we present a preliminary metabolic analysis of plasma from PKAN patients. The severity and rarity of this disorder posed a challenge for patient enrollment and sample collection, but we consider our cohort of genetically and clinically defined patients large enough to support such an approach, and here report the results of our study and their implications for better understanding this devastating disease. Our cohort of patients includes classical and atypical cases but no obvious differences between these groups have emerged from our analyses. Although PKAN patients have not been previously reported to have mitochondrial respiratory chain deficiency, the elevation of lactate suggests possible mitochondrial dysfunction. We also report reduced lipid and cholesterol biosynthesis and impaired bile acid metabolism, two findings with potential therapeutic relevance. Although the patients were on heterogeneous drug regimens at the time of sampling, we are not aware of interactions between these medications and lipid and/or bile acid metabolism, though the precise mechanism of action of the anti-dystonic drug Baclofen remains unknown.
As highlighted in Figure 1, additional metabolites such as inosine, proline, sorbitol, maleate and cotinine show strong or significant differences between patients and controls. Cotinine, a tobacco metabolite [23], likely reflects a lifestyle difference. The relevance of the remaining metabolites is unclear at present; the implications of these metabolic differences may become apparent in future studies. Given the brain iron accumulation that characterizes PKAN, we had hoped to observe differences in iron-related metabolites. Our metabolic profiling method is not all-inclusive, which could explain the lack of iron-related metabolic differences observed. In addition, PKAN iron accumulation occurs in the brain, and our study was performed on plasma samples; sampling cerebrospinal fluid instead of plasma may have revealed differences in additional iron-related metabolites but was beyond the scope of this study.
Pantothenate kinase 2 catalyzes the key, rate-limiting step in coenzyme A biosynthesis, so mutations in the gene encoding this enzyme have the potential to disrupt a number of metabolic processes by disrupting cellular CoA pools. Elevations in lactate suggest that mutations in this mitochondrial enzyme could cause general mitochondrial dysfunction, in agreement with reports that respiratory chain dysfunction produces a similar elevation [11] (Figure 2). Our study suggests that the imbalance in coenzyme A pools created by PANK2 mutations may interfere with a number of metabolic pathways previously unstudied in the context of PKAN (Figure 1), including cholesterol, fatty acids, and bile acid biosynthesis.
Based on genetic analysis (Table 1), we would predict those patients with early stop codon mutations in PANK2 to have the most severe enzymatic defects, and perhaps to exhibit a metabolic blockade at this step in CoA biosynthesis. Indeed, patients ARFR80 and ARRO78 have the highest plasma pantothenate levels among patients not taking pantothenate supplements (Figure 3). Patients with non-stop codon loss-of-function mutations do not have such drastically elevated pantothenate levels, though their levels are still higher compared to controls. This data indicates a biochemical and metabolic heterogeneity among PKAN patients and suggests the presence of compensatory, but still unknown, mechanisms possibly exerted by other PANK proteins.
Cholesterol synthesis is a cytosolic pathway with a number of acetyl CoA-dependent steps. Acetyl-CoA enters the cytoplasm in the form of citrate via the tricarboxylate transport system. It is possible that reduced acetyl CoA synthesis in mitochondria might limit cholesterol biosynthesis by reducing the basic structural element for early cholesterol precursors, thereby limiting production of metabolites downstream of cholesterol, such as lanosterol and lathosterol. We detected significant reductions of these sterols, which are considered human plasma markers for whole body cholesterol synthesis [21,24,25], in both plasma and fibroblasts derived from PKAN patients (Figure 5 and 6).
Our global metabolic approach also highlighted a dysregulation of bile acid metabolism in PKAN patients. Bile acid synthesis from cholesterol proceeds via the intermediates 7a-OHC and 27OHC [26,27], which we found to be reduced in patient plasma (Figure 5). Once the primary bile acids cholate and chenodeoxycholate are synthesized, they are conjugated to either taurine or glycine [28,29]. This conjugation requires activation of the primary bile acids by bile acid CoA ligase (BAL) using ATP and CoA, prior to coupling with taurine or glycine (Figure 4a). Reduced levels of these tauro- and glyco-conjugated bile acids were observed in patient plasma (Figure 4). The reduced levels of tauro- and glyco-conjugated bile acids reported here could be due either to a reduction of available CoA or to reduced cholesterol synthesis. Along with eliminating cholesterol from the body, bile acids facilitate emulsion and uptake of lipids and fat-soluble vitamins in the intestine, as well as reduction of the bacterial flora found in the small intestine and biliary tract [30]. Moreover, bile acids have recently been identified as potential signaling molecules [31].
A low abundance of conjugated bile acids could be responsible for malabsorption of cholesterol and fat-soluble vitamins from the intestine, possibly explaining the lower levels of some lipids detected in patients compared to controls (Figure 1). We also observed low patients plasma levels of plantosterols (campesterol and sitosterol), considered markers of cholesterol absorption [22], suggesting that absorption of nutrients from the intestine is indeed impaired in these patients. This is further corroborated by the observation that plantosterol levels were not different between patients and controls under fasting conditions. It is formally possible that some of the clinical features of PKAN are due to nutritional and vitamin deficiencies related to this malabsorption. Based on the current results, we hypothesize that food supplementation with fat-soluble vitamins may ameliorate some of the pathology observed in these patients.
The newly identified alterations in cholesterol and bile acid metabolism are interesting in the context of similar findngs in other neurological disorders. Over 25% of total body cholesterol is localized to the brain, where it is necessary for maturation of the central nervous system and participates in several key neurological processes including membrane trafficking, neurite overgrowth, synaptic transmission and synaptogenesis [32,33,34]. Reduced levels of the cholesterol precursors lathosterol and lanosterol have also been described in fibroblasts [35] and plasma [36,37] from humans with Huntington’s Disease (HD), as well as in the brains of rodent HD models [38,39]. Reduced levels of plasma lanosterol and lathosterol have also been reported in patients with cognitive impairment and Alzheimer’s disease [18]. Disturbances of cholesterol metabolism were found to affect the cleavage of Amyloid Precursor Protein in cellular models of Alzheimer’s disease, favoring the formation and deposition of the amyloidogenic fragment Aβ1–42 [40]. Finally, one of the typical clinical features of PKAN is the presence of axonal spheroids in the central nervous system. These spheroids represent swollen or distended axons, which could be generated by defects in axonal transport or membrane integrity, integrity that would be disrupted in the context of defective cholesterol biosynthesis [41].
Furthermore, several fatty acids were significantly or nearly significantly reduced in fibroblasts derived from PKAN patients (Figure 6b). These fatty acids, including myristic, palmitic, and oleic acids, constitute the majority of phospholipids in cellular membranes. Stearic acid, an important and abundant FA in cerebroside, was also slightly reduced in PKAN patient fibroblasts. Fatty acid biosynthesis is a cytosolic pathway and, similar to cholesterol synthesis, it requires both acetyl CoA and malonyl CoA to form stearic acid, the fatty acid that is further modified in the biogenesis of several classes of lipids. This acetyl CoA also comes from the mitochondrion in the form of citrate. In combination with the reduced plasma levels of several TAGs and phospholipids in PKAN patients, this data suggests an impairment of fatty acid synthesis that might cause deficiencies in several classes of lipids.
Finally, we found reduced levels of certain sphingomyelin species in plasma. Sphingomyelins are the principal component of the myelin sheath wrapping the axons of neuronal cells. It is now recognized that sphingomyelin and cholesterol have a high affinity for each other and are usually located together in membranes, in specific sub-domains called “lipid rafts.” In addition, recent evidence suggests that sphingomyelin and cholesterol metabolism are closely integrated [42]. Our results are in agreement with a 2005 report by Kotzbauer et al. [43] and suggest that alterations of mitochondrial and lipid metabolism may play a crucial role in the pathogenesis of PKAN, and possibly in other forms of NBIA. Consistent with the idea that low CoA disrupts lipid homeostasis, lipid dysregulation was also observed in Drosophila CoA mutants, including dPANK/fbl [44].
In conclusion, we have used global metabolic profiling to explore the metabolic consequences of mutations in pantothenate kinase 2 that are responsible for Pantothenate Kinase-Associated Neurodegeneration, a rare inborn error of metabolism with devastating consequences for affected individuals. This report highlights novel findings about PKAN patients with potential therapeutic relevance, and more generally illustrates the utility of global metabolic profiling as a tool for systematically exploring the biochemical basis of rare genetic syndromes.
Highlights.
We performed metabolic profiling of PKAN
We found lactate and pantothenate elevation
We found alterations in bile acid and lipid metabolism.
These findings raise a novel therapeutic hypothesis.
Metabolic profiling to explore biochemical basis of inherited metabolic diseases.
Supplementary Material
Acknowledgements
We would like to thank Mo Jain and Oded Shaham for helpful discussions.
Funding
The financial support of Mariani Foundation of Milan [grant n. R-10-84 to V.T.] is gratefully acknowledged. S.D. is supported by the Italian Foundation AISNAF (Associazione Italiana Sindromi Neurodegenerative Accumulo di Ferro). This work was supported by the Italian Minister of Health, Fondi per giovani Ricercatori 2008, to V. L. We thank the Cell line and DNA bank of paediatric movement disorders of the Telethon Genetic Biobank Network [project no. GTB07001] and the Bank for the Diagnosis and Research of Movement Disorders (MDB) of the EuroBiobank. L.S. was supported by a graduate student research fellowship from the National Science Foundation [Grant DGE – 0644491]. This work was supported by a grant from the Broad Institute Scientific Planning and Allocation of Resources Committee, by a gift from the Nestle Research Center to the Broad Institute, and by a grant [R01DK081457] from the National Institutes of Health to V.K.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kurian MA, McNeill A, Lin JP, Maher ER. Childhood disorders of neurodegeneration with brain iron accumulation (NBIA) Dev. Med. Child. Neurol. 2011;53:394–404. doi: 10.1111/j.1469-8749.2011.03955.x. [DOI] [PubMed] [Google Scholar]
- 2.McNeill A, Birchall D, Hayflick SJ, Gregory A, Schenk JF, Zimmerman EA, Shang H, Miyajima H, Chinnery PF. T2* and FSE MRI distinguishes four subtypes of neurodegeneration with brain iron accumulation. Neurology. 2008;70:1614–1619. doi: 10.1212/01.wnl.0000310985.40011.d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gregory A, Polster BJ, Hayflick SJ. Clinical and genetic delineation of neurodegeneration with brain iron accumulation. J. Med. Genet. 2009;46:73–80. doi: 10.1136/jmg.2008.061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spector R, Johanson CE. Vitamin transport and homeostasis in mammalian brain:focus on Vitamins B and E. J. Neurochem. 2007;103:425–438. doi: 10.1111/j.1471-4159.2007.04773.x. [DOI] [PubMed] [Google Scholar]
- 5.Daugherty M, Polanuyer B, Farrell M, Scholle M, Lykidis A, De Crécy-Lagard V, Osterman A. Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics. J. Biol. Chem. 2002;77:21431–21439. doi: 10.1074/jbc.M201708200. [DOI] [PubMed] [Google Scholar]
- 6.Hörtnagel K, Prokisch H, Meitinger T. An isoform of hPANK2, deficient in pantothenate kinase-associated neurodegeneration, localizes to mitochondria. Hum. Mol. Genet. 2003;12:321–327. doi: 10.1093/hmg/ddg026. [DOI] [PubMed] [Google Scholar]
- 7.Skrede S, Halvorsen O. Mitochondrial biosynthesis of coenzyme A. Biochem. Biophys. Res. Commun. 1979;91:1536–1542. doi: 10.1016/0006-291x(79)91239-7. [DOI] [PubMed] [Google Scholar]
- 8.Siudeja K, Srinivasan B, Xu L, Rana A, De Jong J, Nollen EAA, Jackowski S, Sanford L, Hayflick S, Sibon OCM. Impaired Coenzyme A metabolism affects histone and tubulin acetylation in Drosophilia and human cell models of pantothenate kinase associated neurodegeneration. EMBO Mol. Med. 2011;3:1–12. doi: 10.1002/emmm.201100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuo YM, Duncan JL, Westaway SK, Yang H, Nune G, Xu EY, Hayflick SJ, Gitschier J. Deficiency of pantothenate kinase 2 (Pank2) in mice leads to retinal degeneration and azoospermia. Hum. Mol. Genet. 2005;14:49–57. doi: 10.1093/hmg/ddi005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, Wu Z, Kuo YM, Zhou B. Dietary rescue of fumble--a Drosophila model for pantothenate-kinase-associated neurodegeneration. J. Inherit. Metab. Dis. 2005;28:1055–1064. doi: 10.1007/s10545-005-0200-0. [DOI] [PubMed] [Google Scholar]
- 11.Shaham O, Slate NG, Goldberger O, Xu Q, Ramanathan A, Souza AL, Clish CB, Sims KB, Mootha VK. A plasma signature of human mitochondrial disease revealed through metabolic profiling of spent media from cultured muscle cells. Proc. Natl. Acad. Sci. U S A. 2010;107:1571–1575. doi: 10.1073/pnas.0906039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis GD, Farrell L, Wood MJ, Martinovic M, Arany Z, Rowe GC, Souza A, Cheng S, McCabe EL, Yang E, Shi X, Deo R, Roth FP, Asnani A, Rhee EP, Systrom DM, Semigran MJ, Vasan RS, Carr SA, Wang TJ, Sabatine MS, Clish CB, Gerszten RE. Metabolic signatures of exercise in human plasma. Sci. Transl. Med. 2010;2:33–37. doi: 10.1126/scitranslmed.3001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O'Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, Yang E, Farrell L, Fox CS, O'Donnell CJ, Carr SA, Vasan RS, Florez JC, Clish CB, Wang TJ, Gerszten RE. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J. Clin. Invest. 2011;121:1402–1411. doi: 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noll F. In: Methods of enzymatic analysis. Bergmeyer HU, editor. vol. III. New York: Verlag Chemie, Weinheim/Academic Press, Inc; 1974. pp. 1475–1479. [Google Scholar]
- 16.Mineri R, Rimoldi M, Burlina AB, Koskull S, Perletti C, Heese B, Von Döbeln U, Mereghetti P, Di Meo I, Invernizzi F, Zeviani M, Uziel G, Tiranti V. Identification of new mutations in the ETHE1 gene in a cohort of 14 patients presenting with ethylmalonic encephalopathy. J. Med. Genet. 2008;45:473–478. doi: 10.1136/jmg.2008.058271. [DOI] [PubMed] [Google Scholar]
- 17.Leoni V, Mariotti C, Tabrizi SJ, Valenza M, Wild EJ, Henley SM, Hobbs NZ, Mandelli ML, Grisoli M, Björkhem I, Cattaneo E, Di Donato S. Plasma 24S-hydroxycholesterol and caudate MRI in pre-manifest and early Huntington's disease. Brain. 2008;131:2851–2859. doi: 10.1093/brain/awn212. [DOI] [PubMed] [Google Scholar]
- 18.Solomon A, Leoni V, Kivipelto M, Besga A, Oksengård AR, Julin P, Svensson L, Wahlund LO, Andreasen N, Winblad B, Soininen H, Björkhem I. Plasma levels of 24S-hydroxycholesterol reflect brain volumes in patients without objective cognitive impairment but not in those with Alzheimer's disease. Neurosci. Lett. 2009;462:89–93. doi: 10.1016/j.neulet.2009.06.073. [DOI] [PubMed] [Google Scholar]
- 19.Benjamini Y, Yekutieli D. Quantitative trait Loci analysis using the false discovery rate. Genetics. 2005;171:783–790. doi: 10.1534/genetics.104.036699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morava E, Van Den Heuvel L, Hol F, De Vries MC, Hogeveen M, Rodenburg RJ, Smeitink JA. Mitochondrial disease criteria: diagnostic applications in children. Neurology. 2006;67:1823–1826. doi: 10.1212/01.wnl.0000244435.27645.54. [DOI] [PubMed] [Google Scholar]
- 21.Kempen HJ, Glatz JF, Gevers Leuven JA, Van Der Voort HA, Katan MB. Serum lathosterol concentration is an indicator of whole-body cholesterol synthesis in humans. J. Lipid. Res. 1988;29:1149–1155. [PubMed] [Google Scholar]
- 22.Baumgartner S, Mensink RP, Plat J. Plant sterols and stanols in the treatment of dyslipidemia: new insights into targets and mechanisms related to cardiovascular risk. Curr. Pharm. Des. 2011;17:922–932. doi: 10.2174/138161211795428795. [DOI] [PubMed] [Google Scholar]
- 23.Yildiz D. Nicotine, its metabolism and an overview of its biological effects. Toxicon. 2004;43:619–632. doi: 10.1016/j.toxicon.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 24.Bloch K, Clayton RB, Schneider PB. Synthesis of lanosterol in vivo. J. Biol. Chem. 1957;224:175–183. [PubMed] [Google Scholar]
- 25.Matthan NR, Raeini-Sarjaz M, Lichtenstein AH, Ausman LM, Jones PJ. Deuterium uptake and plasma cholesterol precursor levels correspond as methods for measurement of endogenous cholesterol synthesis in hypercholesterolemic women. Lipids. 2000;35:1037–1044. doi: 10.1007/s11745-000-0616-9. [DOI] [PubMed] [Google Scholar]
- 26.Javitt NB. Bile acid synthesis from cholesterol: regulatory and auxiliary pathways. FASEB J. 1994;8:1308–1311. doi: 10.1096/fasebj.8.15.8001744. [DOI] [PubMed] [Google Scholar]
- 27.Javitt NB. Cholesterol, hydroxycholesterols, and bile acids. Biochem. Biophys. Res. Commun. 2002;292:1147–1153. doi: 10.1006/bbrc.2001.2013. [DOI] [PubMed] [Google Scholar]
- 28.Monte MJ, Marin JJ, Antelo A, Vazquez-Tato J. Bile acids: chemistry, physiology, and pathophysiology. World. J. Gastroenterol. 2009;15:804–816. doi: 10.3748/wjg.15.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiang JY. Bile acids: regulation of synthesis. J. Lipid. Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philipp B. Bacterial degradation of bile salts. Appl. Microbiol. Biotechnol. 2011;89:903–915. doi: 10.1007/s00253-010-2998-0. [DOI] [PubMed] [Google Scholar]
- 31.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug. Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 32.Dietschy JM, Turley SD. Thematic review series: brain Lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. J. Lipid. Res. 2004;45:1375–1397. doi: 10.1194/jlr.R400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Björkhem I, Meaney S. Brain cholesterol: long secret life behind a barrier. Arterioscl. Thromb. Vasc. Biol. 2004;24:806–815. doi: 10.1161/01.ATV.0000120374.59826.1b. [DOI] [PubMed] [Google Scholar]
- 34.Liu JP, Tang Y, Zhou S, Toh BH, McLean C, Li H. Cholesterol involvement in the pathogenesis of neurodegenerative diseases. Mol. Cell. Neurosci. 2010;43:33–42. doi: 10.1016/j.mcn.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Valenza M, Rigamonti D, Goffredo D, Zuccato C, Fenu S, Jamot L, Strand A, Tarditi A, Woodman B, Racchi M, Mariotti C, Di Donato S, Corsini A, Bates G, Pruss R, Olson JM, Sipione S, Tartari M, Cattaneo E. Dysfunction of the cholesterol biosynthetic pathway in Huntington's disease. J. Neurosci. 2005;25:9932–9939. doi: 10.1523/JNEUROSCI.3355-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leoni V, Mariotti C, Nanetti L, Salvatore E, Squitieri F, Bentivoglio AR, Bandettini Del Poggio M, Piacentini S, Monza D, Valenza M, Cattaneo E, Di Donato S. Whole body cholesterol metabolism is impaired in Huntington's disease. Neurosci. Lett. 2011;494:245–249. doi: 10.1016/j.neulet.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 37.Leoni V, Caccia C. Oxysterols as biomarkers in neurodegenerative diseases. Chem. Phys. Lipids. 2011;164:515–524. doi: 10.1016/j.chemphyslip.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 38.Valenza M, Leoni V, Tarditi A, Mariotti C, Bjökhem I, Di Donato S, Cattaneo E. Progressive dysfunction of the cholesterol biosynthesis pathway in the R6/2 mouse model of Huntington's disease. Neurobiol. Dis. 2007;28:133–142. doi: 10.1016/j.nbd.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 39.Valenza M, Leoni V, Karasinska JM, Petricca L, Fan J, Carroll J, Pouladi MA, Fossale E, Nguyen HP, Riess O, MacDonald M, Wellington C, Di Donato S, Hayden M, Cattaneo E. Cholesterol defect is marked across multiple rodent models of Huntington's disease and is manifest in astrocytes. J. Neurosci. 2010;30:10844–10850. doi: 10.1523/JNEUROSCI.0917-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prasanthi JR, Huls A, Thomasson S, Thompson A, Schommer E, Ghribi O. Differential effects of 24-hydroxycholesterol and 27-hydroxycholesterol on beta-amyloid precursor protein levels and processing in human neuroblastoma SH-SY5Y cells. Mol. Neurodegener. 2009;6 doi: 10.1186/1750-1326-4-1. 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweitzer JK, Krivda JP, D'Souza-Schorey C. Neurodegeneration in Niemann-Pick Type C disease and Huntington's disease: impact of defects in membrane trafficking. Curr. Drug. Targets. 2009;10:653–665. doi: 10.2174/138945009788680437. [DOI] [PubMed] [Google Scholar]
- 42.Milhas D, Clarke CJ, Hannun YA. Sphingomyelin metabolism at the plasma membrane: Implications for bioactive sphingolipids. FEBS Letts. 2010;584:1887–1894. doi: 10.1016/j.febslet.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kotzbauer PT, Truax AC, Trojanowski JQ, Lee VM. Altered neuronal mitochondrial coenzyme A synthesis in neurodegeneration with brain iron accumulation caused by abnormal processing, stability, and catalytic activity of mutant pantothenate kinase 2. J. Neurosci. 2005;25:689–698. doi: 10.1523/JNEUROSCI.4265-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bosveld F, Rana A, Van Der Wouden PE, Lemstra W, Ritsema M, Kampinga HH, Sibon OC. De novo CoA biosynthesis is required to maintain DNA integrity during development of the Drosophila nervous system. Hum. Mol. Genet. 2008;17:2058–2069. doi: 10.1093/hmg/ddn105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.