Abstract
Cyanobacteria are an important group of photoautotrophic organisms that can synthesize valuable bio-products by harnessing solar energy. They are endowed with high photosynthetic efficiencies and diverse metabolic capabilities that confer the ability to convert solar energy into a variety of biofuels and their precursors. However, less well studied are the similarities and differences in metabolism of different species of cyanobacteria as they pertain to their suitability as microbial production chassis. Here we assemble, update and compare genome-scale models (iCyt773 and iSyn731) for two phylogenetically related cyanobacterial species, namely Cyanothece sp. ATCC 51142 and Synechocystis sp. PCC 6803. All reactions are elementally and charge balanced and localized into four different intracellular compartments (i.e., periplasm, cytosol, carboxysome and thylakoid lumen) and biomass descriptions are derived based on experimental measurements. Newly added reactions absent in earlier models (266 and 322, respectively) span most metabolic pathways with an emphasis on lipid biosynthesis. All thermodynamically infeasible loops are identified and eliminated from both models. Comparisons of model predictions against gene essentiality data reveal a specificity of 0.94 (94/100) and a sensitivity of 1 (19/19) for the Synechocystis iSyn731 model. The diurnal rhythm of Cyanothece 51142 metabolism is modeled by constructing separate (light/dark) biomass equations and introducing regulatory restrictions over light and dark phases. Specific metabolic pathway differences between the two cyanobacteria alluding to different bio-production potentials are reflected in both models.
Introduction
Cyanobacteria represent a widespread group of photosynthetic prokaryotes [1]. By contributing oxygen to the atmosphere, they played an important role in the precambrian phase [2]. Cyanobacteria are primary producers in aquatic environments and contribute significantly to biological carbon sequestration, O2 production and the nitrogen cycle [3]–[5]. Their inherent photosynthetic capability and ease in genetic modifications are two significant advantages over other microbes in the industrial production of valuable bioproducts [6]. In contrast to other microbial production processes requiring regionally limited cellulosic feedstocks, cyanobacteria only need CO2, sunlight, water and a few mineral nutrients to grow [6]. Sunlight is the most abundant source of energy on earth. The incident solar flux onto the USA alone is approximately 23,000 terawatts which dwarfs the global energy usage of 3.16 terawatts [7]. Cyanobacteria perform photosynthesis more efficiently than terrestrial plants (3–9% vs. 2.4–3.7%) [8]. The short life cycle and transformability of cyanobacteria combined with a detailed understanding of their biochemical pathways are significant advantages of cyanobacteria as efficient platforms for harvesting solar energy and producing bio-products such as short chain alcohols, hydrogen and alkanes [6].
The genus Cyanothece includes unicellular cyanobacteria that can fix atmospheric nitrogen. Cyanothece sp. ATCC 51142 (hereafter Cyanothece 51142) is one of the most potent diazotrophs characterized and the first to be completely sequenced [9]. Studies show that it can fix atmospheric nitrogen at rates higher than many filamentous cyanobacteria and also accommodate the biochemically incompatible processes of photosynthesis and nitrogen fixation within the same cell by temporally separating them [10]. Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803), the first photosynthetic organism with a completely sequenced genome [11], is probably the most extensively studied model organism for photosynthetic processes [12]. It is also closely related to Cyanothece 51142 and shares many characteristics with all Cyanothece [9]. The genome of Cyanothece 51142 is about 35% larger than that of Synechocystis 6803 mostly due to the presence of nitrogen fixation and temporal regulation related genes in Cyanothece 51142 [9]. Synechocystis 6803 has been the subject of many targeted genetic manipulations (e.g., expression of heterologous gene products) as a photo-biological platform for the production of valuable chemicals such as poly-beta-hydroxybutyrate, isoprene, hydrogen and biofuels [12]–[20]. However, genetic tools for Cyanothece 51142 are still lacking thus hampering its wide use as a bio-production strain even though it has many attractive native pathways. For example, Cyanothece 51142 can produce (in small amounts) pentadecane and other hydrocarbons while containing a novel (though incomplete) non-fermentative pathway for producing butanol [21], [22].
A breakthrough in solar biofuel production will require following one of two strategies: 1) obtaining photosynthetic strains that naturally have high-throughput pathways analogous to those in known biofuel producers, or 2) creating cellular environments conducive for heterologous enzyme function. Despite its attractive capabilities including nitrogen fixation and H2 production [19], unfortunately genetic tools are not currently available to efficiently test engineering interventions directly for Cyanothece 51142. Therefore, a promising path forward may be to use Synechocystis 6803 as a “proxy” (for which a comprehensive genetic toolkit is available) and subsequently transfer knowledge gained during experimentation with Synechocystis 6803 to Cyanothece 51142. This requires high quality metabolic models for both organisms. Comprehensive genome-wide metabolic reconstructions include the complete inventory of metabolic transformations of a given cyanobacterial system. Comparison of the metabolic capabilities of Cyanothece 51142 and Synechocystis 6803 derived from their corresponding genome-scale models will provide valuable insights into their niche biological functions and also open up new avenues for economical biofuel production.
Genome-scale models (GSM) contain gene to protein to reaction associations (GPRs) along with a stoichiometric representation of all possible biotransformations known to occur in an organism combined with a set of appropriate regulatory constraints on each reaction flux [23], [24]. By defining the global metabolic space and flux distribution potential, GSMs can assess allowable cellular phenotypes under specific environmental conditions [23], [24]. The first genome-scale model for Cyanothece 51142 was recently published [25]. The authors addressed the complexity of the electron transport chain (ETC) and explored further the specific roles of photosystem I (PSI) and photosystem II (PSII). In contrast, Synechocystis 6803 has been the target for metabolic model reconstruction for quite some time [12], [26]–[32]. Most of these earlier efforts for Synechocystis 6803 focused on only central metabolism [26]–[28]. Knoop et al. [12] and Montagud et al. [30], [31] developed genome-scale models for Synechocystis 6803, analyzed growth under different conditions, identified gene knock-out candidates for enhanced succinate production and performed flux coupling analysis to detect potential bottlenecks in ethanol and hydrogen production. A more recent model describes in detail the photosynthetic apparatus, identifies alternate electron flow pathways and highlights the high photosynthetic robustness of Synechocystis 6803 during photoautotrophic metabolism [32]. All these efforts have brought about an improved understanding of the metabolic capabilities of Synechocystis 6803 and cyanobacterial systems in general.
This paper introduces high-quality genome-scale models for Cyanothece 51142 iCyt773 and Synechocystis 6803 iSyn731 (as shown in Table 1) that integrate all recent developments [25], [32], supplements them with additional literature evidence and highlights their similarities and differences (see Files S1, S2, S7 and S8 for detailed description of these models). As many as 322 unique reactions are introduced in the Synechocystis iSyn731 model and 266 in Cyanothece iCyt773. New pathways include, among many, a TCA bypass [33], heptadecane biosynthesis [21] and detailed fatty acid biosynthesis in iSyn731 and comprehensive lipid and pigment biosynthesis and pentadecane biosynthesis [21] in iCyt773. For the first time, not only extensive gene essentiality data [34] is used to assess the quality of the developed model (i.e., iSyn731) but also the allowable model metabolic phenotypes are contrasted against MFA flux data [35]. The diurnal rhythm of Cyanothece metabolism is modeled for the first time via developing separate (light/dark) biomass equations and regulating metabolic fluxes based on available protein expression data over light and dark phases [36].
Table 1. Synechocystis 6803 iSyn731 and Cyanothece 51142 iCyt773 model statistics.
| Synechocystis 6803 iSyn731 model | Cyanothece 51142 iCyt773 model | |
| Included genes | 731 | 773 |
| Proteins | 511 | 465 |
| Single functional proteins | 348 | 336 |
| Multifunctional proteins | 91 | 83 |
| Isozymes | 4 | 1 |
| Multimeric proteins | 32 | 22 |
| Othersa | 36 | 23 |
| Reactions | 1,156 | 946 |
| Metabolic reactions | 972 | 761 |
| Transport reactions | 127 | 128 |
| GPR associations | ||
| Gene associated (metabolic/transport) | 827 | 686 |
| Spontaneousb | 180 | 158 |
| Nongene associated (metabolic/transport) | 59 | 16 |
| No protein associated | 90 | 86 |
| Exchange reactions | 57 | 57 |
| Metabolites c | 996 | 811 |
| Cytosolic | 862 | 675 |
| Carboxisomic | 8 | 8 |
| Thylakoidic | 10 | 9 |
| Periplasmic | 59 | 62 |
| Extracellular | 57 | 57 |
Others include proteins involve in complex relationships, e.g. multiple proteins act as protein complex which is one of the isozymes for any specific reaction.
Spontaneous reactions are those without any enzyme as well as gene association.
Metabolites represent total number of metabolites with considering their compartmental specificity.
Materials and Methods
Measurement of Biomass Precursors
Growth conditions
Wild-type Synechocystis 6803 and Cyanothece 51142 were grown for several days from an initial OD730 of ∼0.05 to ∼0.4. Synechocystis 6803 was grown in BG-11 medium [92] and Cyanothece 51142 in ASP2 medium [93] with (+N) or without (−N) nitrate. All cultures were grown in shake flasks with continuous illumination of ∼100 µmol photons/m2/sec provided from cool white fluorescent tubes. Synechocystis was maintained at 30°C and Cyanothece at 25°C. For Synechocystis, the illumination was constant and doubling time was ∼24 hours. Cyanothece alternated between 12 hours of light and 12 hours of darkness, with a doubling time of ∼48 hours.
Pigments
1 mL of cells of both Synechocystis 6803 and Cyanothece 51142 (from light and dark phases) was pelleted and extracted twice with 5 mL 80% aqueous acetone and the extracts pooled. Spectra of this extract and of a sample of whole cells were taken on a DW2000 spectrophotometer (Olis, GA, USA) against 80% acetone or BG-11 media as a reference. Chlorophyll a contents were calculated as reported [94] from the acetone extract. Total carotenoid concentrations were also calculated from the acetone extract according to a published method [95]. The relative amounts of different carotenoids included in the biomass equation were estimated according to known ratios [96]. Concentrations of phycocyanin were estimated from the spectra of intact cells [97]. All measurements were taken in triplicate.
Amino acids
Total protein contents were measured using a Pierce BCA Assay kit. Amino acid proportions were determined according to published shotgun proteomics data for both Cyanothece 51142 and Synechocystis 6803 across a range of conditions [84] according to the following procedure: From peptide-level data, each mass spectral observation of a peptide was taken as an instance of a particular protein. The amino acid composition of each protein was taken from data in Cyanobase (http://genome.kazusa.or.jp/cyanobase) and thus the ‘proteome’ was taken to include all of the proteins whose peptides were observed in our data set, in proportion according to how often their peptides were observed. Amino acid frequencies were averaged across the proteome by a weighting factor of number of observations divided by the number of amino acids in the protein, similar to RPKM normalization for next-gen sequencing [98].
Other cellular components
The compositions of other cellular components of Synechocystis 6803 and Cyanothece 51142 were estimated based on values in the literature. DNA and RNA contents for Synechocystis 6803 were reported by Shastri and Morgan [27]. The remaining biomass components of Synechocystis 6803 (i.e., lipid, soluble pool and inorganic ions) were extracted from the measurements carried out by Nogales et al. [32]. For Cyanothece 51142, biochemical compositions of macromolecules such as lipids, RNA, DNA and soluble pool were extracted from the measurements reported by Vu et al. [25].
Model Simulations
Flux balance analysis (FBA) [99] was employed in both the model validation and model testing phases. Cyanothece iCyt773 and Synechocystis iSyn731 models were evaluated in terms of biomass production under several scenarios: light and dark phases, heterotrophic and mixotrophic conditions. Flux distributions for each one of these states were inferred using FBA:
Maximize 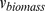
Subject to
| (1) |
| (2) |
Here, Sij is the stoichiometric coefficient of metabolite i in reaction j and vj is the flux value of reaction j. Parameters vj,min and vj,max denote the minimum and maximum allowable fluxes for reaction j, respectively. Light and dark phases in Cyanothece 51142 are represented via modifying the minimum or maximum allowable fluxes with the following constraints, respectively:
| (3) |
| (4) |
Here, vBiomass is the flux of biomass reaction and vGlytr, vGlyctr and vCO2tr are the fluxes of glycerol, glycogen and carbon dioxide transport reactions and vlight and vcf are the fluxes of light reactions and carbon fixation reactions. For light phase, constraint (3) was included in the linear model, whereas for dark phase constraint (4) was included.
Once the Synechocystis iSyn731 model was validated, it was further tested for in silico gene essentiality. The following constraint(s) was included individually in the linear model to represent any mutant:
| (5) |
Here, vmutant represents flux of reaction(s) associated with any genetic mutation.
Flux variability analysis [100] for the reactions (for which photoautotrophic 13C MFA measurements [35] were available) was performed based on the following formulation:
Maximize/Minimize 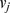
Subject to
| (6) |
| (7) |
| (8) |
Here, 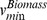 is the minimum level of biomass production. In this case we fixed it to be the optimal value obtained under light condition for the Synechocystis iSyn731 model.
is the minimum level of biomass production. In this case we fixed it to be the optimal value obtained under light condition for the Synechocystis iSyn731 model.
CPLEX solver (version 12.1, IBM ILOG) was used in the GAMS (version 23.3.3, GAMS Development Corporation) environment for implementing GapFind and GapFill [39] and solving the aforementioned optimization models. All computations were carried out on Intel Xeon E5450 Quad-Core 3.0 GH and Intel Xeon E5472 Quad-Core 3.0 GH processors that are the part of the lionxj cluster (Intel Xeon E type processors and 96 GB memory) of High Performance Computing Group of The Pennsylvania State University.
Results and Discussion
Model Components
Biomass composition and diurnal cycle
The biomass equation approximates the dry biomass composition by draining all building blocks or precursor molecules in their physiologically relevant ratios. Most of the earlier genome-scale modeling efforts [12], [29], [30] of Synechocystis 6803 contain approximate biomass equations completely or partially adopted from other species without direct measurements. This can adversely affect the accuracy of maximum biomass yield calculations, gene essentiality predictions and knockouts for overproduction.
Biomass composition for Synechocystis iSyn731 and Cyanothece iCyt773 models were generated by defining all essential cellular biomass content values by experimental measurement or collection from existing literature (see ‘Materials and Methods’ for detail). Macromolecules present in both cyanobacteria such as protein, carbohydrates, lipids, DNA, RNA, pigments, soluble pool and inorganic ions were assigned to their corresponding metabolic precursors (e.g., L-glycine, glucose, 16C-lipid, ATP, dGTP, beta-carotene, coenzyme A and potassium respectively) (see File S3 for the complete list of biomass components). Based on the experimental measurements of precursor molecules needed to form a gram of the biomass, stoichiometric coefficients were assigned. For Synechocystis 6803 we measured compositions of proteins and pigments and extracted compositions of the remaining biomass macromolecules from the model by Nogales et al. [32]. Thereby we developed biomass equations for three different conditions: photoautotrophic, mixotrophic and heterotrophic (see File S3). Experimental measurements (described in the Materials and Methods section and also in File S3) showed that biomass composition (i.e., mainly pigments) varies for Cyanothece 51142 between light and dark conditions and nitrogen supplementation. Since pigments such as chlorophyll, carotenoids and phycocyanobilin play important roles in photosynthetic processes their quantities are consequently higher under light conditions. In the presence of light Cyanothece 51142 uses photosynthesis to store solar energy in the form of carbohydrates (i.e., glycogen), while in dark it expends that energy to fix nitrogen. Surprisingly, no significant change was measured in the carbohydrate pool between light and dark phases due to infinitesimal contribution of photosynthetically stored carbohydrates to total carbohydrate content in the biomass of Cyanothece 51142. Aggregate quantities of the remaining biomass macromolecules for Cyanothece 51142 such as lipids, RNA, DNA and soluble pool were extracted from the most recent Cyanothece 51142 model by Vu et al. [25] to develop biomass equations for light and dark phases (File S3).
An earlier characterization study for Cyanothece 51142 revealed that 113 proteins are expressed in higher abundance in the light phase while 137 are expressed in higher abundance in dark conditions [36]. The constructed model spans 26 light-specific proteins, associated with 36 reactions mainly involved in fatty acid, pigment, and amino acid metabolism and 11 dark-specific proteins accounting for 16 reactions from glycolysis, purine, pyrimidine, pyruvate, and amino acid metabolism (Files S4). Separate biomass equations as well as two regulatory structures for the model were derived in order to represent diurnal metabolic differences for Cyanothece 51142 (File S4). In contrast, diurnal differences observed in Synechocystis 6803 [37] are less pronounced (i.e., observed for only 54 genes) and less well functionally annotated (i.e., 32 genes with ‘unassigned’ functions). When compared to existing biomass equations of Synechocystis 6803 [12], [30] we found significantly lower values for the percent weight contribution of proteins towards the biomass pool (i.e., 52% for Synechocystis 6803 and 53% for Cyanothece 51142 vs. 84% [12] and 66% [30], respectively). The new protein biomass contribution is in better agreement with the previously reported value of 55% for Cyanothece 51142 [38].
Identification and correction of network gaps
Upon ensuring biomass formation, GapFind [39] was applied to assess network connectivity and blocked metabolites. By applying Gapfill [39] putative reconnection hypotheses were identified for blocked metabolites. Only the suggested modifications that were independently corroborated using literature sources and also did not lead to the introduction of thermodynamically infeasible cycle were included in the model. For Synechocystis iSyn731 model, GapFind [39] identified 207 blocked metabolites. Note that there exist 125 blocked metabolites in the iJN678 model [32]. GapFill [39] identified unblocking hypotheses for 138 blocked metabolites. However, 88 of them led to the generation of infeasible thermodynamic cycles and thus were excluded. For only 5 blocked metabolites corroborating evidence for reconnection was obtained by adding 10 reactions (i.e., 2 metabolic, 4 transport and 4 exchange reactions). The added metabolic reactions have unknown gene associations (see File S1 for detailed information) while all 4 added transport reactions involve passive diffusion and thus are not associated with any specific gene(s) or protein(s). Ultimately, the 45 remaining blocked metabolites with GapFill suggested (but unconfirmed) reconnection mechanisms along with 69 blocked metabolites with no reconnection hypotheses were retained in the model iSyn731, while metabolites such as ubiquinone, which was proposed as an alternate substrate for succinate dehydrogenase [32] was excluded from iSyn731.
For the Cyanothece iCyt773 model, 74 blocked metabolites were found after applying GapFind [39]. Note that there are 66 blocked metabolites in iCce806 [25]. Two exchange reactions were added to allow the uptake of glucose and thyaminose ensuring biomass production under heterotrophic or mixotrophic conditions. Four blocked metabolites directly adopted from iCce806 (during the draft model creation phase) were linked to five reactions with spurious gene associations and thus both metabolites and reactions were removed from iCyt773. GapFill [39] suggested re-connection mechanisms for 52 blocked metaboloites (out of a total of 70). However, for 12 blocked metabolites the re-connection model modifications led to the creation of thermodynamically infeasible cycles and thus were discarded. Corroborating evidence for the reconnection of 30 blocked metabolites was identified through the addition of 19 GapFill suggested reactions (i.e., 8 metabolic, 7 transport and 4 exchange reactions). Of the eight added metabolic reactions we found direct literature evidence for five, homology-based evidence for one while two reactions are spontaneous (see File S2 for detailed information). All seven added transport reactions are through passive diffusion and thus are not connected with any specific gene(s) or protein(s). Ten remaining blocked metabolites with GapFill suggested reconnection hypotheses (along with 22 with no reconnection hypotheses) were left blocked in iCyt773 as no information to corroborate the GapFill suggested changes was found in the published literature and databases. For example, biotin is produced in Cyanothece 51142; however, there is no literature evidence to support the presence of the initial step of the primary production pathway (i.e., conversion of pimeloyl-CoA from pimelate) and the intermediate step (i.e., biotransformation of 7,8-diamino-nonanoate from 8-amino-7-oxononanoate). This indicates that Cyanothece 51142 may utilize a currently unknown pathway for producing biotin. The six other blocked metabolites are involved in the nonfermentative alcohol production pathway (as explained in model comparison section) known to be incomplete in Cyanothece 51142. Table 2 summarizes the results related to connectivity restoration of Synechocystis iSyn731 and Cyanothece iCyt773 models.
Table 2. Summary of connectivity restoration in Synechocystis 6803 iSyn731 and Cyanothece 51142 iCyt773 models.
| Synechocystis 6803 iSyn731 | Cyanothece 51142 iCyt773 | |
| Number of blocked metabolites | 207 | 74 |
| Number of metabolites with GapFill [39] suggested reconnection strategies | 138 | 52 |
| Number of metabolites whose reconnection forms a cycle | 88 | 12 |
| Number of metabolites with validated reconnection mechanisms | 5 | 30 |
| Number of added reactions to the model | 10 | 19 |
GPR associations and elemental and charge balancing
GPR associations connect genotype to phenotype by linking gene(s) that code for the protein(s) that catalyze a particular reaction. They are important to trace correctly as they provide the means to target at the gene level any change in the network desired at the reaction level. This is critical because genes may catalyze multiple reactions in multiple pathways. Many earlier models for Synechocystis 6803 do not provide in detail complex GPR associations, rather list only gene(s) and enzyme(s) involved in a specific reaction [12], [30], [31]. For both iCyt773 and iSyn731 models, we included comprehensive GPR associations (see Table 1 for detail information). All four intracellular compartments (i.e., periplasm, cytosol, thylakoid lumen and carboxysome) were assumed to have the same pH (7.2) and subsequently, metabolites were assigned appropriate protonation states corresponding to this pH and each reaction was elementally and charge balanced.
Under high light intensity in photoautotrophic conditions, Cyanothece iCyt773 model produces 0.026 mole biomass/mole carbon fixed whereas Synechocystis iSyn731 yields 0.021 mole biomass/mole carbon fixed. These yields are almost identical to the ones calculated using the most recent models of Cyanothece 51142 [25] and Synechocystis 6803 [32]. Experimental measurements of biomass yields are in the same order of magnitude with model predictions for the two organisms (i.e., 0.072 [25], [40] and 0.082 mole biomass/mole carbon fixed [41]), respectively.
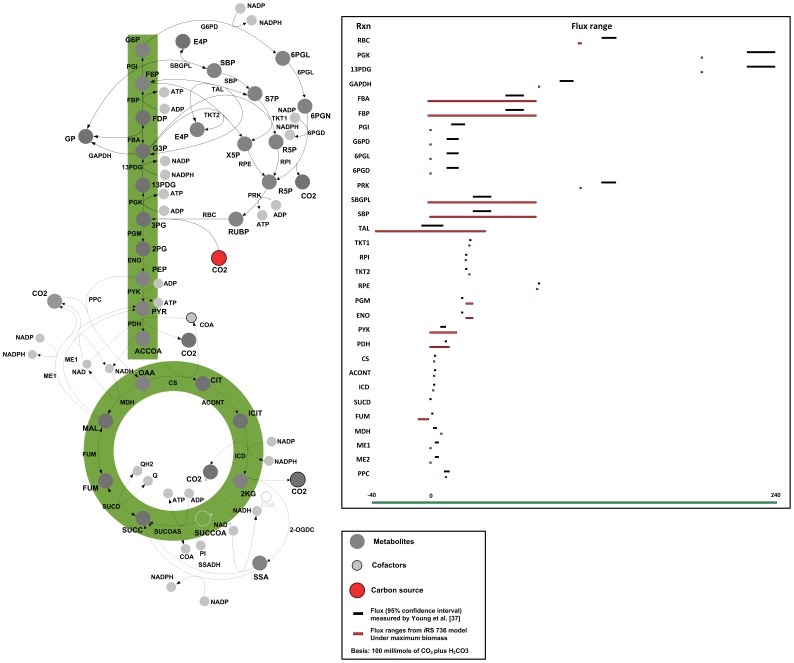
Comparison of iSyn731 Model Predicted Flux Ranges Against Experimental Measurements
We superimposed photoautotrophic flux measurements [35] for Synechocystis 6803 onto iSyn731 model to assess if the measurements are consistent with the model and whether the biomass maximization assumption correctly apportions fluxes to the metabolic network. For each reaction that was assigned a flux we calculated the flux-range under the maximum biomass assumption. Table 3 and Figure 1 summarize the obtained results for a basis of 100 millimole of CO2 plus H2CO3 uptake [35]. In seven (out of thirty one) cases the measured flux is fully contained within the model predicted ranges obtained upon maximizing biomass formation implying model consistency with MFA measurements. In contrast, under the maximum biomass assumption for thirteen fluxes the ranges underestimate and for four fluxes the ranges overestimate the experimentally deduced flux ranges while for seven fluxes the model derived flux ranges partially overlap with the experimental ones.
Table 3. Comparison of 13C MFA flux measurements [35] vs. model-predicted flux ranges.
| Reaction | Flux measurements by Young et al., 2011 [35] | Flux ranges predicted by iJN678 model (With max biomass) | Flux ranges predicted by iSyn731 model (With max biomass) | |||
| 95% LB | 95% UB | LB | UB | LB | UB | |
| RBC | 123.00 | 132.00 | 109.02 | 109.10 | 102.49 | 106.33 |
| PGK | 219.00 | 237.00 | 187.11 | 187.25 | 182.70 | 182.92 |
| 13PDG | 219.00 | 237.00 | 187.11 | 196.36 | 182.70 | 201.96 |
| GAPDH | 90.00 | 99.00 | 74.98 | 75.07 | 73.40 | 73.50 |
| FBA | 53.00 | 66.00 | −0.17 | 74.85 | −0.08 | 73.17 |
| FBP | 53.00 | 66.00 | 0.00 | 74.85 | 0.00 | 73.17 |
| PGI | 15.00 | 24.00 | 0.68 | 0.73 | 0.82 | 0.84 |
| G6PD | 12.00 | 21.00 | 0.00 | 0.05 | 0.00 | 0.03 |
| 6PGL | 12.00 | 21.00 | 0.00 | 0.05 | 0.00 | 0.03 |
| 6PGD | 12.00 | 21.00 | 0.00 | 0.05 | 0.00 | 0.03 |
| PRK | 123.00 | 132.00 | 109.02 | 109.10 | 106.24 | 106.32 |
| SBGPL | 29.00 | 43.00 | −0.17 | 74.85 | −0.08 | 73.17 |
| SBP | 29.00 | 43.00 | 0.00 | 74.85 | 0.00 | 73.17 |
| TAL | −6.00 | 9.00 | −36.74 | 38.28 | −35.93 | 37.32 |
| TKT1 | 37.20 | 37.50 | 36.57 | 36.60 | 36.66 | 36.79 |
| RPI | 35.40 | 35.70 | 35.18 | 35.21 | 35.82 | 35.86 |
| TKT2 | 35.40 | 35.70 | 37.25 | 37.28 | 36.18 | 36.23 |
| RPE | 75.50 | 76.20 | 73.83 | 73.88 | 72.01 | 72.10 |
| PGM | 22.90 | 23.60 | 26.83 | 26.95 | 25.92 | 29.79 |
| ENO | 23.40 | 23.80 | 26.84 | 26.95 | 25.92 | 29.79 |
| PYK | 7.90 | 11.10 | 0.00 | 13.88 | 0.00 | 16.72 |
| PDH | 11.50 | 12.00 | 0.00 | 8.97 | 0.00 | 13.46 |
| CS | 3.00 | 3.40 | 2.15 | 2.21 | 1.35 | 1.37 |
| ACONT | 3.00 | 3.40 | 2.15 | 2.21 | 1.35 | 1.37 |
| ICD | 3.00 | 3.00 | 2.15 | 2.21 | 1.32 | 1.37 |
| SUCD | 0.00 | 0.40 | 0.00 | 0.00 | 0.00 | 0.00 |
| FUM | 1.70 | 2.00 | −5.44 | 1.55 | −7.26 | 1.49 |
| MDH | 1.90 | 5.20 | 5.35 | 5.61 | 7.15 | 7.32 |
| ME1 | 3.70 | 6.90 | 0.00 | 0.17 | 0.00 | 0.08 |
| ME2 | 3.70 | 6.90 | − | − | 0.00 | 0.08 |
| PPC | 9.90 | 13.20 | 11.74 | 11.98 | 12.25 | 12.37 |
Figure 1. Comparison of model derived and experimentally measured [35] flux ranges for Synechocystis 6803 under the maximum biomass condition.
Basis is 100 millimole of CO2 plus H2CO3.
Perhaps the most informative discrepancy is for the CO2 fixing RuBisCO (RBC) reaction, which has a measured flux range of (123.00 to 132.00) vs. the model-calculated range of (102.49 to 106.33). In both cases the increased RBC flux (in comparison to the basis of 100 millimole of CO2 plus H2CO3 uptake) is needed to counteract the carbon loss due to the CO2 releasing reactions such as isocitrate dehydrogenase (ICD) and pyruvate dehydrogenase (PDH). We find that flux ranges, under the maximum biomass production assumption, of reactions such as glucose 6-phosphate dehydrogenase (G6PD), 6-phosphogluconolactonase (6PGL) and phosphogluconate dehydrogenase (6PGD) in oxidative pentose phosphate (OPP) pathway are negligible (0.00 to 0.03). In contrast, the experimentally derived range for OPP is (12 to 21). This is approximately equal to the difference between the model-predicted vs. experimentally deduced RBC reaction range implying the persistence of OPP flux even under the photoautotrophic condition [35] despite the presence of a more efficient NADPH production route through photosynthesis as predicted by the model (under max biomass). The high values Young et al. [35] obtained for the OPP fluxes were surprising as OPP is not a very efficient route for cyanobacteria to generate reducing power. This may reflect some inherent biological constraint that is not captured by the optimality assumption.
Model predicted lower flux ranges for RBC are propagated to seven other reactions in the Calvin cycle (i.e., phosphoglycerate kinase (PGK), glyceraldehyde-3-phosphate dehydrogenase (13PDG), triose-phosphate isomerase (TPI), transketolase (TKT1), ribose-5-phosphate isomerase (RPI), ribulose 5-phosphate 3-epimerase (RPE) and phosphoribulokinase (PRK). The remaining six reaction fluxes with lower model predicted fluxes compared to measurements [35] are all in the TCA cycle (i.e., citrate synthase (CS), aconitase (ACONT), isocitrate dehydrogenase (ICD), succinate dehydrogenase (SUCD) and malic enzyme (ME1 and ME2) reactions). Even under the max biomass assumption, SUCD is not required to carry any flux due to the presence of other succinate dehydrogenases (as part of respiratory chain) in the iSyn731 model. Furthermore, in contrast with experimental observations, under the maximum biomass assumption, the model predicts no flux through the malic enzyme (ME) reactions presumably because it is a less energy-efficient route (i.e., phosphoenolpyruvate → oxaloacetate → malate → pyruvate) for pyruvate generation than the pyruvate kinase (PYK) reaction [35].
There are nine reactions with experimentally derived ranges completely subsumed within the ones derived under the maximum biomass assumption. Five of them are in the Calvin cycle (i.e., fructose-bisphosphate aldolase (FBA), fructose-bisphosphatase (FBP), Sedoheptulose 1,7-bisphosphate D-glyceraldehyde-3-phosphate-lyase (SBGPL), sedoheptulose-bisphosphatase (SBP) and bidirectional transaldolase (TAL)). The first four reactions are essential with experimentally deduced flux ranges of (53.00 to 66.00) for FBA and FBP and (29.00 to 43.00) for SBGPL and SBP. In contrast, the calculated flux ranges (−0.08 to 73.17) for FBA and SBGPL and (0.00 to 73.17) for FBP and SBP imply that they are in silico non-essential. As depicted in Figure 1, these reactions are involved in the production of sedoheptulose 7-phosphate (S7P) from fructose 1,6-bisphosphate (FDP). An alternative production route for S7P is afforded in the model through the bidirectional transaldolase (TAL) reaction from fructose 6-phosphate (F6P) alluding to an explanation for the wider flux ranges derived using the model. Experimental and model predicted flux ranges for TAL are (−6.00 to 9.00) and (−35.93 and 37.32), respectively. Upon restricting the TAL flux ranges in the calculations to the ones found experimentally, the flux variability analysis shrinks the flux ranges for FBA and FBP to (28.22 to 43.27) and (28.22 to 43.33) and for SBGPL and SBP to (29.82 to 44.87) and (29.82 to 44.87), respectively which are very close to the experimentally measured ranges. This is indicative that in addition to the maximization of biomass formation, additional restrictions (e.g., photosynthetic efficiency and relative selectivity of RuBISCO for carboxylation over oxidation) limit the range of fluxes that the aforementioned glycolytic fluxes may span in vivo. Note that the presence of experimentally measured fluxes is important to test the model and the adopted maximization principle. We were fortunate in this case to have access to such data as for most organisms they are absent.
Phosphoglycerate mutase (PGM) and enolase (ENO) reactions have very similar model derived and experimentally obtained flux ranges. Model-predicted flux values of the remaining two reactions, pyruvate kinase (PYK) and pyruvate dehydrogenase (PDH), could reach as low as zero due to the metabolic flexibility that the iSyn731 model possesses by having alternate enzymes with different cofactor specificities. The max biomass flux range of fumarase (FUM) is found to be (−7.26 to 1.49), compared to the experimentally measured (1.70 to 2.00). Therefore, it appears that under the photoautotrophic condition, the forward direction is kinetically favorable. By restricting the reaction to be irreversible the model predicted a FUM flux range of (0.00 to 1.49) which is close to the experimentally derived one (see Figure 1). However, contrary to MFA measurements these reactions (FUM and ME) are dispensable for in silico biomass production.
iSyn731 Model Testing Using in vivo Gene Essentiality Data
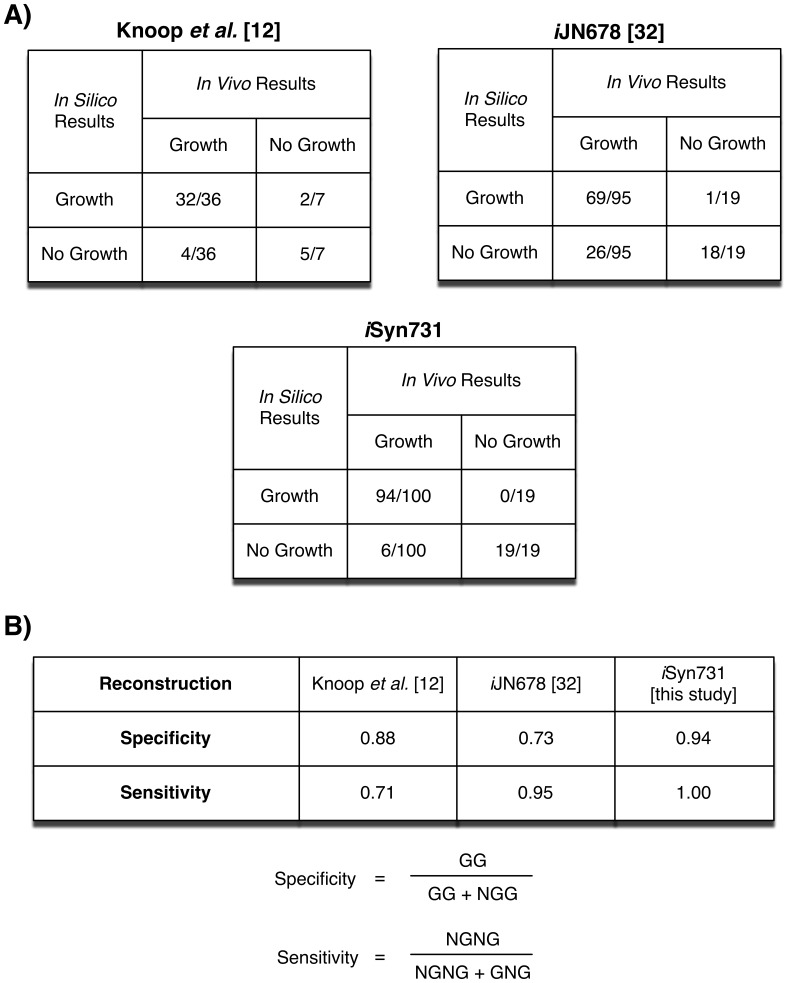
The quality of model iSyn731 for Synechocystis 6803 was tested using experimental data on the viability (or lack thereof) of single gene knockouts. We used the CyanoMutants database [34], [42] that includes in vivo gene essentiality data for 119 genes (i.e., 19 essential and 100 nonessential) with metabolic functions in iSyn731 model. Cases that were flagged with incomplete segregation in the database were omitted in iSyn731 model comparisons. We examined the feasibility of biomass production for the model iSyn731 by comparing the maximum biomass formation upon imposing the gene knockout with the maximum theoretical yield of the wild-type organism. A threshold of 10% of the maximum theoretical yield was used as a cutoff [43]. Comparisons between in vivo and in silico results led to four possible outcomes, as previously delineated by Kumar et al., GG, GNG, NGG and NGNG [43]. Initially, the model correctly predicted 18 out of 19 essential genes (i.e., 18 NGNG and 1 GNG) and 74 out of 100 non-essential genes (i.e., 73 GG and 27 NGG). We next explored the causes of these discrepancies and attempted to mitigate them whenever possible.
The single GNG case corresponds to mutant ΔchlAI exhibiting no growth under aerobic conditions [44]. The ChlAI system is a Mg-protoporphyrin IX monomethylester (MPE) cyclase system that is responsible for forming the isocyclic ring (E-ring) in chlorophylls under aerobic conditions [44]. The model allowed for the BchE and ChlAII systems (alternate cyclase systems) to complement for the loss of the ChlAI system leading to an in silico viable mutant. However, the same literature source [44] suggested that both BchE and ChlAII systems are unlikely to be active under aerobic conditions and thus rescue mutant ΔchlAI. This prompted the introduction of a regulatory restriction in iSyn731 model where only ChlAI reactions were active under aerobic conditions as MPE while ChlAII and BchE system reactions were deactivated. Using these regulatory restrictions resolves the single GNG inconsistency.
Twenty (out of 27) NGG cases were associated with Photosystem I (PSI), Photosystem II (PSII) and other photosynthesis reactions. While reconstructing the model, we assumed that all genes involved in photosynthetic reaction system were essential to the functioning of the overall system. Published literature [45]–[49] suggests that genes involved in photosynthetic reactions form complex interdependencies. We used NCBI COBALT multiple sequence alignment tool [50] to construct a phylogenetic tree of the genes associated with each photosystem along with BLASTp searches to identify putative complementation relationships between genes to explain the inconsistencies between the predicted in silico and in vivo growth. Genes deemed homologous (i.e., lie adjacent in the phylogenetic tree) were linked with “OR” GPR relations implying that the loss of one gene can be complemented by the other. Seven out of twenty NGG cases (i.e., psaD, psaI and psbA2 for PSI and PSII and cpcC2, cpcC1, cpcD, and apcD for other photosynthesis reactions) were resolved by modifying the corresponding GPR using an OR relation [46], [47], [51]–[54]. However, no phylogenetically adjacent or related (or homologous) genes were found for the remaining 13 NGG cases (psaE, psbD2, psbO, psbU, psbV, psb28, psbX, psb27, petE, cpcA, cpcB, apcE, apcF) [48], [49], [51], [54]–[58]. For these cases, the genes were deemed nonessential to the functioning of the reactions in question (i.e., photosynthesis reactions) and thereby the corresponding GPRs were modified to show an OR relation between each of these genes and an ‘unknown gene’, similar to what was previously performed in the refinement of the iMM904 model [59] (see File S5 for detailed information).
The remaining seven NGG cases are associated with a variety of metabolic functions. One such case is the ΔmodBC mutant corresponding to the sole ABC molybdate transporter in the model. Literature evidence [60] revealed that a related cyanobacterium, Anabaena variabilis ATCC 29413, could continue to grow despite the loss of its molybdate ABC transporter due to the presence of another low affinity molybdate transporter or an inducible sulfate transport system that can serve as a low affinity molybdate transporter when required. We found the same gene coding for the sulfate transporter in A. variabilis (cysA) in the iSyn731 model allowing the resolution of the discrepancy by adding a cysA-linked alternate molybdate transporter. Another NGG case is mutant ΔcrtO that cannot produce echinenone (a biomass component) in iSyn731 with no effect on observed growth. Therefore, it appears that iSyn731 cannot capture the flexibility of Synechocystis 6803 metabolism [61] when echinenone production is restricted. The remaining five NGG cases are spread across many metabolic pathways. The ΔctaA mutant eliminates the copper ABC transporter without affecting growth, which alludes to the existence of another unknown mode of copper uptake not present in iSyn731 [62], [63]. The ΔmenG mutant eliminates a reaction for the production of phylloquinone while mutant Δppd affects the production of homogentisate, a precursor for both tocopherols and plastoquinone. Finally, the Δvte3 mutant affects the production of both plastoquinone and α-tocopherol [64] and the viable ΔccmA mutant restricts the production of chorismate (a precursor to aromatic amino acids) and also restricts carboxysome formation [65]–[67]. These six inconsistencies between the model predictions and growth data imply that the cyanobacterium can co-opt another metabolic process to (partially) complement for the gene loss. Unlike the case of the ΔmodBC mutant, we have found no plausible mechanism for the six remaining mutants.
After resolving the discrepancies, as described above, iSyn731 correctly predicted all 19 essential genes (i.e., 19 NGNG and 0 GNG) and 94 (out of 100) non-essential genes (i.e., 94 GG and 6 NGG). Figure 2 shows our results and comparisons against two other available Synechocystis 6803 models by Knoop et al. [12] and Nogales et al. [32]. We used the CyanoMutants database [34] to identify 114 genes (i.e., 19 essential and 95 nonessential) having metabolic functions in the iJN678 model by Nogales et al. [32]. Out of 114 genes the iJN678 model correctly predicted 18 essential genes (i.e., 18 NGNG and 1 GNG) and 69 non-essential genes (i.e., 69 GG and 26 NGG). The model by Knoop et al. [12] was tested for 51 mutants but we found that only 43 (i.e., 7 essential and 36 non-essential) of them were reported to have complete segregation [34]. Of these 43, Knoop et al.’s [12] model correctly predicted 5 essential genes (i.e., 5 NGNG and 2 GNG) and 32 nonessential genes (i.e., 32 GG and 4 NGG). The specificity and sensitivity of each of these three models were also calculated and displayed at the bottom of Figure 2.
Figure 2. Comparison of gene essentiality/viability data with predictions by a number of Synechocystis 6803 models.
(A) Tabulated growth (i.e., G) or non-growth (i.e., NG) predictions and experimental data. The first number denotes the number of GG, GNG, NGG and NGNG combinations whereas the second number signifies the number of experimentally observed lethal (or viable) mutants, and (B) Definition and comparison of specificity and sensitivity of all three models. Note that GG denotes both in silico and in vivo growth, NGG represents no growth in silico but in vivo growth. NGNG implies no growth for either in silico or in vivo, whereas GNG marks growth in silico but no growth in vivo.
All 114 genes tested for iJN678 were also present in the iSyn731 model. 26 NGG and one GNG cases present in iJN678 model correspond to NGG and GNG cases that were either fixed or still present in iSyn731 as discussed before. Lethal mutant Δppa is correctly predicted as NGNG in iSyn731 but deemed GNG in Knoop et al. [12] model. This was because ppa in iSyn731 codes for the degradation of both triphosphate into diphosphate and diphosphate to phosphate. Only the latter activity is linked to ppa in the Knoop et al’s model. Out of 4 NGG cases in [12], two involve ΔcmpA and ΔcmpB mutants. Both these genes are involved in the ABC transporter system for bicarbonate from periplasm to cytosol. iSyn731 avoids this inconsistency as it contains an alternate sodium and bicarbonate co-transport system.
Model Comparisons
Synechocystis 6803 model comparisons
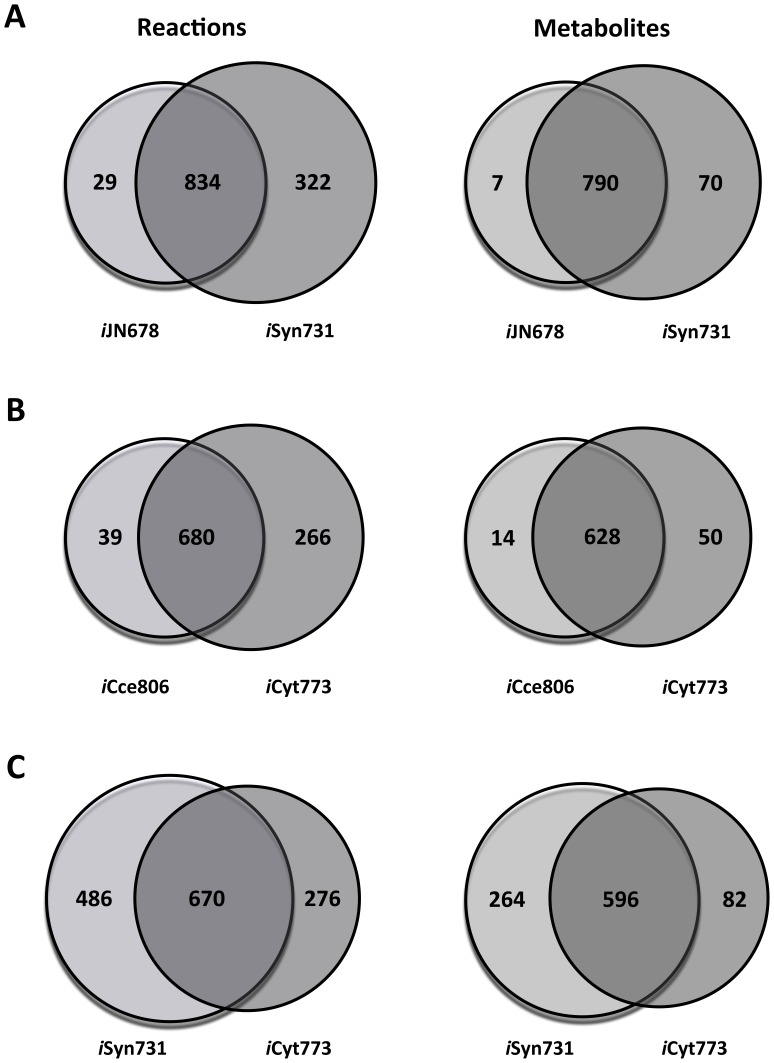
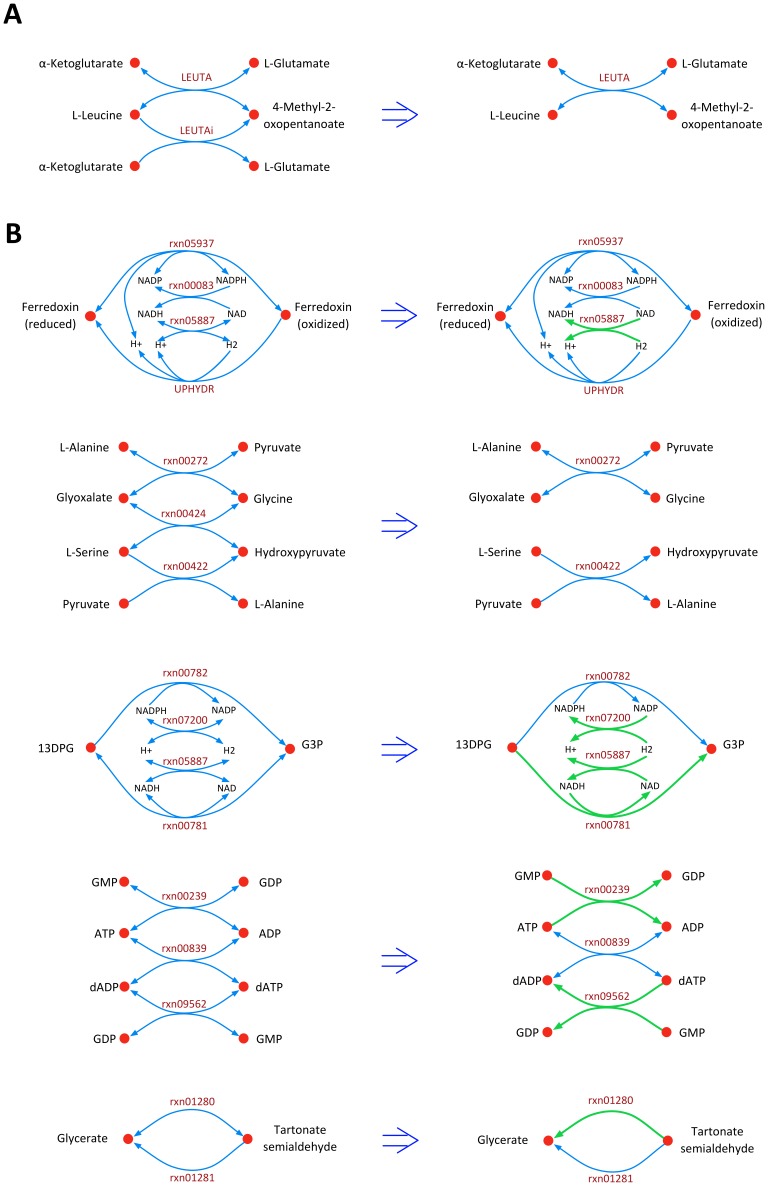
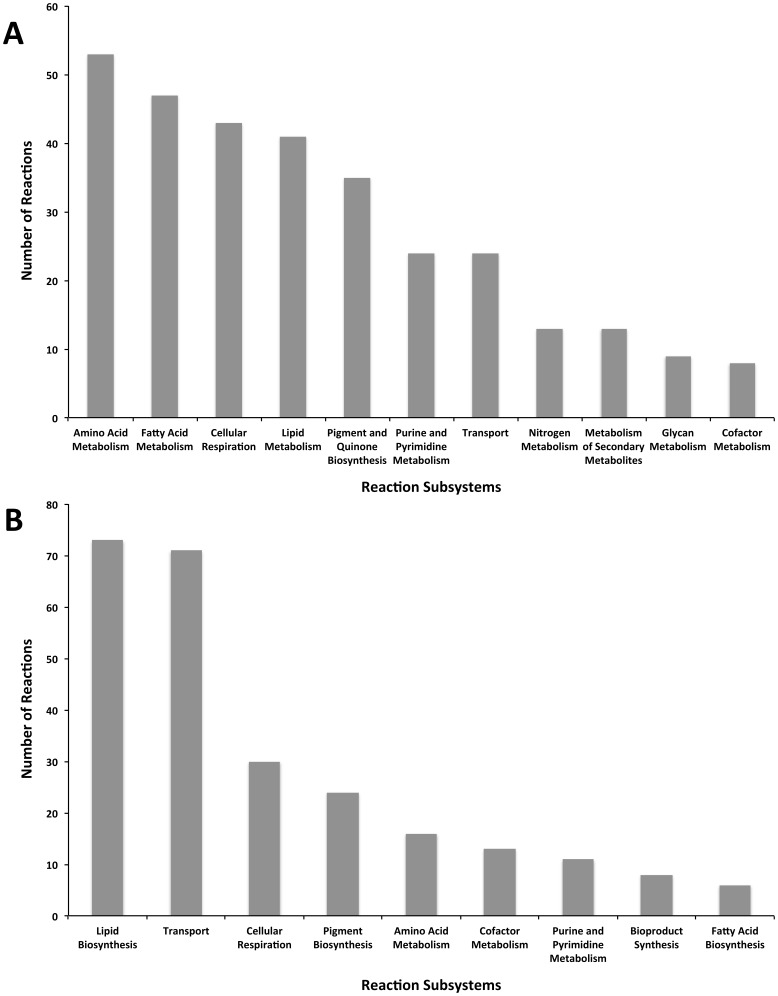
The iSyn731 model integrates the description in the photosystems of the model presented by Nogales et al. [32] and adds additional detail. One notable difference is that iSyn731 uses a separate photon for each reaction center (i.e., PSI and PSII) as they are optimized for different ranges of wavelength [68], whereas iJN678 [32] uses a single photon shared by both photosystem reactions. As many as 322 new reactions (see Figure 3A), are added in iSyn731 distributed across many pathways. Most of the additions are in the lipid and fatty acid metabolism to support the synthesis of measured fatty acids and lipids present in the biomass equation. This list includes myristic acid (14-carbon saturated fatty acid) and lauric acid (12-carbon saturated fatty acid). iJN678 [32] contained four reactions exhibiting unbounded flux (i.e., two duplicate glycine cleavage reactions and two duplicate leucine transaminase reactions). They form a thermodynamically infeasible cycle (see Figure 4A for leucine transaminase reactions) that was resolved in iSyn731 by eliminating redundant functions. In addition, the glycine cleavage system was recast in detail by abstracting the separate action of the four enzymes (named the T-, P-, L-, and H-proteins) that ultimately catalyze the demethylamination of glycine.
Figure 3. Venn diagram depicting (common and unique) reactions and metabolites between (A) iJN678 [32] and iSyn731, (B) iCce806 [25] and iCyt773, and (C) iSyn731 and iCyt773 models.
Figure 4. Schematics that illustrate the thermodynamically infeasible cycles and subsequent resolution strategies.
(A) Cycles present in iJN678 [32], and (B) Cycles present in iCce805 [25]. Blue colored lines represent the original reaction directionality whereas green ones denote modified directionality to eliminate cycle.
iSyn731 improves upon iJN678 [32] by eliminating lumped reactions whenever a multi-step description is available and expands the range of functions carried out with alternate cofactors. As many as twelve reactions with an enoyl-[acyl-carrier-protein] reductase function were linked with not only NADP but also with the more rare NAD cofactor specificity. Another important difference between iSyn731 and iJN678 [32] is the cellular location of the CO2 fixation (i.e., ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) enzyme). Literature [69], [70] shows that cyanobacteria possess a micro-compartment (i.e., carboxysome) encapsulating RuBisCO and carbonic anhydrase (CA) enzymes. iSyn731 adds carboxysome as a cellular compartment and also all necessary transport reactions [69], [70]. Recently, Zhang and Bryant [33] hypothesized the existence of a functional TCA cycle in most cyanobacterial species using a 2-ketoglutarate to succinate bypassing step. iSyn731 allows for a complete TCA cycle using the bypassing step. In addition, iSyn731 contains an intact heptadecane biosynthesis pathway as recently described [21] unlike earlier Synechocystis 6803 models [12], [29], [30], [32] (see Figure 5A for distribution of unique reactions in iSyn731).
Figure 5. List of added reactions across pathways.
(A) iSyn731 compared to iJN678 [32], and (B) iCyt773 compared to iCce806 [25].
Cyanothece 51142 model comparisons
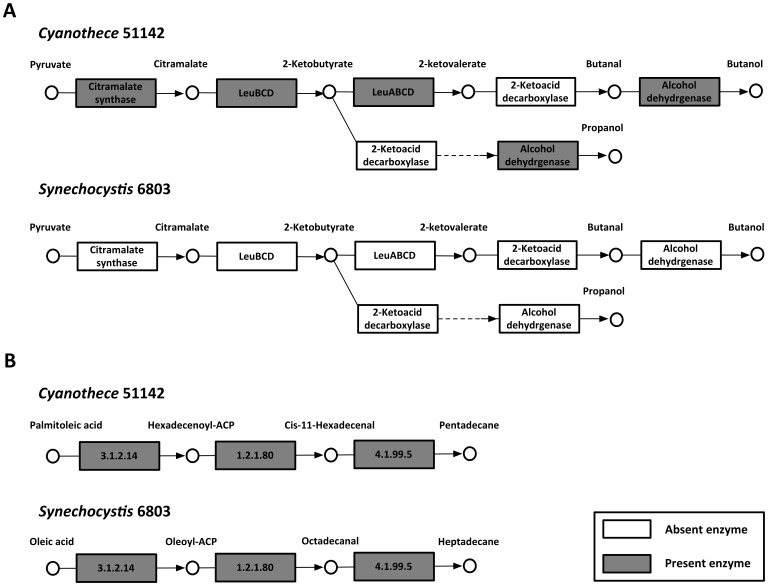
The iCyt773 model for Cyanothece 51142 improves upon the iCce806 model [25]. iCyt773 segregates reactions into the periplasm, thylakoid lumen, carboxysome, and cytoplasm compartments thus introducing an additional 60 transport reactions compared to iCce806 [25]. Unlike iCce806 [25], iCyt773 does not track macromolecule synthesis for DNA, RNA, and proteins to maintain consistency with the Synechocystis 6803 model. This difference accounts for 69 genes present in iCce806 [25] but absent from iCyt773. iCce806 [25] contained 15 reactions which formed five cycles that could carry unbounded metabolic flux (i.e., thermodynamically infeasible cycles). All these cycles were eliminated by restricting reaction directionality and eliminating reactions that were linear combinations of others (coded by the same gene) (see Figure 4B).
iCyt773 contains 43 unique genes and 266 unique reactions (including transport and alternate cofactor utilizing reactions) as shown in Figure 3B. Figure 5B depicts the distribution of the new reactions across different pathways. Most of the additions are found in lipid and pigment biosynthesis pathways. The iCyt773 model captures in detail the lipid biosynthesis pathway composed of 73 reactions and links as many as 28 biomass precursor lipids (e.g., sulfoquinovosyldiacylglycerols, monogalactosyldiacylglycerols, digalactosyldiacyl-glycerols, and phosphatidylglycerols) directly to the biomass equation. The porphyrin and chlorophyll metabolism and carotenoid biosynthesis pathways were updated to include 24 reactions for the production of accessory pigments such as echinenone, an accessory pigment, and (3Z)-phycocyanobilin, a phycobilin. Accessory pigments donate electrons to chlorophyll rather than directly to photosynthesis. Phycobilins are adapted for many wavelengths not absorbed by chlorophyll thus broadening the spectrum useful for photosynthesis. The variety of pigments in cyanobacteria is well documented [71]–[73] providing so far untapped avenues for engineering increased efficiency in photosynthesis and control of electron transfer processes in biological systems. Another new function in iCyt773 is L-Aspartate Oxidase. L-Aspartate Oxidase allows the deamination of aspartate, forming oxaloacetate a key TCA-cycle metabolite and ammonia. The impact of this addition to iCyt773 is not evident under the photoautotrophic condition but becomes relevant for growth in a medium containing aspartate. iCyt773 also uniquely supports the synthesis of pentadecane as documented by Schirmer et al. [21] and contains an (almost) complete non-fermentative citramalate pathway as suggested by Wu et al. [22].
A number of lumped reactions in iCce806 [25] were recast in detail. For example, pyruvate dehydrogenase (PDH) is a three-enzyme complex that carries out the biotransformation of pyruvate to acetyl-CoA in three steps using five separate cofactors (i.e., TPP, CoA, FAD, lipoate, and NAD). Similar detail was used for lumped steps in the metabolism of glycine, histidine, and serine. All additions to the list of reactions in iCyt773 were corroborated using genome annotations [9] or published literature [20]–[22], [74] with the exception of ten enzymes, whose function in the lipid and pigment biosynthesis pathways was required for biomass production.
A shift in biomass composition was observed under light, dark, and nitrate supplemented (light and dark) conditions. These differences were captured in four separate biomass descriptions present in iCyt773. In addition, we used data from Stockel et al. [75] on the diurnal oscillations for approximately 20% of proteins in Cyanothece 51142 to identify regulatory reaction shutdowns in our metabolic model. File S4 lists the reactions that were inactivated under light and dark conditions, respectively. As expected, the nitrogenase genes cce_0559 and cce_0560, known to be active in the absence of light, exhibited low spectral counts under light conditions. In contrast, photosystem II gene cce_1526, showed no spectral count under dark conditions. Unexpectedly, the data suggested that the Mehler reactions associated gene (cce_2580), known to be active in Synechocystis 6803 [76] and expected to be active in Cyanothece 51142, exhibited lower expression in light than in dark conditions.
iSyn731 and iCyt773 models comparison
Figure 3C illustrates the total number of common and unique reactions and metabolites between iSyn731 and iCyt773 models. The Cyanothece 51142 genome [9], [77] is 1.5 times larger than the one for Synechocystis 6803 [11], nevertheless iCyt773 is smaller than iSyn731 due to differences in the level of detail of annotation and biochemical characterization. As many as 670 reactions and 596 metabolites are shared by both models corresponding to 47% and 63% of the total reactome and metabolome, respectively (see Figure 3C). The higher degree of conservation of metabolites (as opposed to reactions) across the two cyanobacteria suggests that lifestyle adaptations tend to usher new enzymatic activities that most of the time make use of the same metabolite pool without introducing new metabolites. There are 486 reactions that are unique to iSyn731 with no counterpart in iCyt773. These reactions are not preferentially allotted to a handful of specific pathways. Instead they are spread over tens of different pathways. Primary metabolism reactions dispersed throughout fatty acid biosynthesis, lipid metabolism, oxidative phosphorylation, purine and pyrimidine metabolism, transport and exchange reactions account for 295 reactions. Secondary metabolism including chlorophyll and cyanophycin metabolism, folate, terpenoid, phenylpropanoid and flavonoid biosynthesis accounts for the remaining 191 iSyn731-specific reactions. Interestingly, the 276 iCyt773-specific reactions span the same set of diverse pathways implying that the two organisms have adopted unique/divergent biosynthetic capabilities for similar metabolic needs. Fifty-eight span primary metabolism pathways such as purine and pyrimidine metabolism, fatty acid and lipid biosynthesis, amino acid biosynthesis. The remaining 218 reactions describe secondary metabolism such as terpenoid biosynthesis, chlorophyll and cyanophycin biosynthesis, plastoquinone and phyloquinone biosynthesis (see File S6 for detail information). The much larger set of unique iSyn731-specific reactions compared to iCyt773 reflect more complete genome annotation and biochemical characterization rather than augmented metabolic versatility.
A number of distinct differences in metabolism between the two organisms have been accounted for in the two models. For example, iCyt773 does not have the enzyme threonine ammonia-lyase, which catalyzes the conversion of threonine to 2-ketobutyrate and as a consequence lacks the traditional route for isoleucine synthesis. Instead it employs part of the alternative citramalate pathway for isoleucine synthesis with pyruvate and acetyl-CoA as precursors. Follow up literature queries revealed the existence of this alternative pathway in Cyanothece 51142 [22]. Ketobutyrate, an intermediate in the citramalate pathway, can be readily converted to higher alcohols, such as propanol and butanol, via a non-fermentative alcohol production pathway. Using the iCyt773 model, we determined that only 2-ketoacid decarboxylase is missing from these three-step processes. In contrast, iSyn731 was found to have only the traditional route for isoleucine production with the citramalate pathway completely absent (see Figure 6A). In another example, the fermentative 1-butanol pathway is known to be incomplete in both organisms. By querying the developed models we can pinpoint exactly which steps are absent. Specifically, the conversion between 3-hydroxybutanoyl-CoA and butanal is missing in both models. In addition to higher alcohols, higher alkanes (C13 and above) are important biofuel molecules as the main constituents of diesel and jet fuel [21]. Recently reported [21] novel genes involved in the biosynthesis of alkanes in several cyanobacterial strains were incorporated in the models. Metabolic differences in Cyanothece 51142 and Synechocystis 6803 lead to the production of different alkanes (e.g., pentadecane in Cyanothece 51142 and heptadecane in Synechocystis 6803) (see Figure 6B).
Figure 6. Examples of pathways that differ between the two cyanobacteria.
(A) Nonfermentative alcohol production pathway highlighting the present and absent enzymes in Cyanothece 51142 and Synechocystis 6803, and (B) Alkane biosynthesis pathways in Cyanothece 51142 and Synechocystis 6803.
Model iCyt773, in contrast to iSyn731, does not have a complete urea cycle as it lacks the enzyme L-arginine aminohydrolase catalyzing the production of urea from L-arginine. Literature sources [77], [78] support this finding and explain the absence of a functional urea cycle as a consequence of the nitrogen-fixation ability of Cyanothece 51142 [79], [80]. Because Cyanothece 51142 can fix nitrogen directly from the atmosphere and produce ammonium via the enzyme nitrogenase, genes corresponding to the activity of L-arginine aminohydrolase and urease (for breaking down urea) become redundant, explaining why they are not present in its genome [80]. In addition to nitrogen metabolism, iCyt773 and iSyn731 models reveal marked differences in anaerobic metabolic capabilities. Unlike iSyn731, iCyt773 includes a L-lactate dehydrogenase activity that enables the complete fermentative lactate production pathway. On the other hand, iSyn731 contains the anaerobic chlorophyll biosynthetic pathway using enzyme protoporphyrin IX cyclase (BchE) that is absent in iCyt773. Other differences in metabolism include lipid and fatty acid synthesis, fructose-6-phosphate shunt and nitrogen fixation. Model iSyn731 traces the location of the double bond for unsaturated fatty acid synthesis pathways, as two separate isomers of unsaturated C18 fatty acids are part of the biomass description. iCyt773 allows for the shunting of fructose-6-phosphate into erythrose-4-phosphate along with acetate and ATP using the fructose-6-phosphate phosphoketolase activity. Finally, both iSyn731 and iCyt773 contain multiple hydrogenases allowing both to produce hydrogen. However, only the latter has a nitrogenase activity that can fix nitrogen while simultaneously producing hydrogen.
Using iSyn731 and iCyt773 to Estimate Production Yields
We tested the recently developed models iSyn731 and iCyt773 by comparing the predicted maximum theoretical product yields with experimentally measured values for two very different metabolic products: isoprene and hydrogen. Isoprene, a volatile hydrocarbon and potential feedstock for biofuel, is mostly produced in plants under heat stress [13]. Cyanobacteria offer promising production alternatives as they can grow to high densities in bioreactors and produce isoprene directly from photosynthesis intermediates [13]. It was reported [13] that Synechocystis 6803 has all but one gene (encoding isoprene synthase) in the methyl-erythritol-4-phosphate (MEP) pathway for isoprene synthesis from dimethylallyl phosphate (DMAPP). Upon cloning the isoprene synthase from kudzu vine (Pueraria montana) into Synechocystis 6803 isoprene production was demonstrated using sunlight and atmospheric CO2 of 4.3×10−4 mole isoprene/mole carbon fixed [81]. We calculated the maximum isoprene yield using iSyn731 to be 3.63×10−5 mole isoprene/mole carbon fixed upon adding the isoprene synthase activity to the model and simulating the conditions described in [41] under maximum biomass production. Similar isoprene yields were obtained with iJN678 [32] while earlier models of Synechocystis 6803 [12], [29]–[31] lack the MEP pathway (partially or completely) and thus do not support isoprene production. The underestimation of the experimentally observed isoprene yield by the model predicted maximum yield may be due to sub-optimal growth of the production strain, differences in the list of measured biomass components, missing isoprene-relevant reactions from the model or more likely a combination of the above factors.
Both Cyanothece 51142 and Synechocystis 6803 produce hydrogen by utilizing nitrogenase and hydrogenase activities, respectively [19]. Under subjective dark conditions [19] whereby (i) stored glycogen acts as a carbon source, (ii) photosynthesis harnesses light energy, and (iii) nitrogenase activity is not restricted, hydrogen production yield for Cyanothece 51142 was measured at 49.67 mole/mole glycogen consumed. Simulating the same conditions in iCyt773 and iCce806 [25] leads to maximum theoretical yields for hydrogen production of 48.43 mole/mole glycogen and 102.4 mole/mole glycogen, respectively. The entire amount of hydrogen produced in iCyt773 is due to the nitrogenase activity. In contrast, the predicted doubling of the maximum hydrogen yield in iCce806 is due to the utilization of the reverse direction of two hydrogen dehydrogenase reactions without any nitrogenase activity. Utilization of the nitrogenase reaction requires the use and recycling of more ATP than simply running the dehydrogenase reactions in reverse. However, it has been reported that hydrogen production in Cyanothece 51142 is primarily mediated by the nitrogenase enzyme [19] in the dark phase. This lends support to the irreversibility of the dehydrogenase reactions (under dark condition) as present in the iCyt773 model. Experimental results for Synechocystis 6803 support up to 4.24 mole/mole glycogen consumed [19], [82] of hydrogen production. iSyn731 predicts a maximum hydrogen theoretical yield of 2.28 mole/mole glycogen consumed while iJN678 [32] yields a value of 2.00 mole/mole glycogen consumed. Again the factors outlined for isoprene production may explain the lower theoretical yields predicted by the two models. The small difference between the model predicted yields is due to the presence of one step lumped biotransformation between isocitrate and oxoglutarate via isocitrate dehydrogenase in iJN678 [32]. iSyn731 describes this biotransformation in two steps (isocitrate → oxalosuccinate → oxoglutarate) [83] generating an additional NADPH and subsequently more hydrogen via the hydrogenase reaction.
Conclusion
In this paper, we expanded upon existing models to develop two genome-scale metabolic models (Synechocystis iSyn731 and Cyanothece iCyt773) for cyanobacterial metabolism by integrating all available knowledge available from public databases and published literature. All metabolite and reaction naming conventions are consistent between the two models allowing for direct comparisons. Systematic gap filling analyses led to the bridging of a number of network gaps in the two models and the elimination of orphan metabolites. Two separate biomass equations as well as two different versions of Cyanothece iCyt773 models were developed for light and dark phases to represent diurnal regulation. The development of two separate models for Cyanothece 51142 (i.e., light and dark) provides the two “end-points” for the future development of dynamic metabolic models capturing the temporal evolution [36], [84]–[86] of fluxes during the transition phases DFBA [87]. Comparisons against available 13C MFA measurements for Synechocystis 6803 [35] revealed that the iSyn731 model upon biomass maximization yields flux ranges that are generally consistent with experimental data. Discrepancies between the two identify metabolic nodes where regulatory constraints are needed in addition to biomass maximization to recapitulate physiological behavior. The ability of iSyn731 to predict the fate of single gene knock-outs was further improved (specificity of 0.94 and sensitivity of 1.00) by reconciling in silico growth predictions with in vivo gene essentiality data [34]. Similar analyses could also be carried out for Cyanothece iCyt773 model once such flux measurements and in vivo gene essentiality data become available.
It is becoming widely accepted that focusing on a single pathway at a time without quantitatively assessing the system-wide implications of genetic manipulations may be responsible for suboptimal production levels. By accounting for both primary and some secondary metabolism pathways, the Cyanothece iCyt773 model can be used to explore in silico the effect of genetic modifications aimed at increased production of useful biofuel molecules. By taking full inventory of Cyanothece 51142 metabolism (as abstracted in iCyt773), and applying available strain optimization techniques [88], [89] optimal gene modifications could be pursued for a variety of targets in coordination with experimental techniques. In particular, the availability of a microaerobic environment in Cyanothece 51142 at certain times during the diurnal cycle can be exploited for the expression of novel pathways that are not usually found in oxygenic cyanobacterial strains that largely maintain an aerobic environment. However, the use of Cyanothece 51142 as a bio-production platform is currently hampered by the inability to efficiently carry out genetic modifications.
By systematically cataloguing the shared (and unique) metabolic content in iSyn731 and iCyt773, successful genetic interventions assessed experimentally for Synechocystis 6803 can be “translated” to Cyanothece 51142. For example, it has been reported [90], [91] that overproduction of fatty alcohols can be achieved in Synechocystis 6803 upon cloning a fatty acyl-CoA reductase (far) from Jojoba (Simmondsia chinensis) and the over-expression of gene slr1609 coding for an acyl-ACP synthetase. By using models iSyn731 and iCyt773 we can infer that in addition to cloning far from Jojoba, over-expression of gene cce_1133 coding for a native acyl-ACP synthetase would be needed to bring about the same overproduction in Cyanothece 51142.
Supporting Information
Synechocystis iSyn731 model along with established GPR, metabolite, gene and protein information.
(XLSX)
Cyanothece iCyt773 model along with established GPR, metabolite, gene and protein information.
(XLSX)
Biomass component measurements and stoichiometry of biomass equation.
(XLSX)
Reactions with diurnal activation/inactivation.
(XLSX)
Comparison of in silico vs. in vivo gene essentiality results for iSyn731 and modifications made in GPR associations.
(XLSX)
Comparison between Synechocystis iSyn731 and iCyt773 models in terms of genes, proteins, reactions and metabolites.
(XLSX)
SBML file of Synechocystis iSyn731 model.
(XML)
SBML file of Cyanothece 51142 iCyt773 model.
(XML)
Acknowledgments
We acknowledge Thanura Elvitigala and Lawrence Page for contributions during the amino acid and pigment measurements. We also thank Anthony P. Burgard and Alireza Zomorrodi for discussions during the reconstruction process.
Funding Statement
This study was supported by a grant from the United States Department of Energy, Biological and Environmental Research (DOE-BER), grant DE-SC0006870. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Tamagnini P, Axelsson R, Lindberg P, Oxelfelt F, Wunschiers R, et al.. (2002) Hydrogenases and hydrogen metabolism of cyanobacteria. Microbiol Mol Biol Rev 66: 1–20, table of contents. [DOI] [PMC free article] [PubMed]
- 2.Schopf J (2000) The Fossil Record: Tracing the Roots of the Cyanobacterial Lineage. In: B W, Dordrecht PM, editors. The ecology of cyanobacteria: Kluwer Academic Publishers. 13–35.
- 3. Moisander PH, Beinart RA, Hewson I, White AE, Johnson KS, et al. (2010) Unicellular cyanobacterial distributions broaden the oceanic N2 fixation domain. Science 327: 1512–1514. [DOI] [PubMed] [Google Scholar]
- 4. Bryant DA, Frigaard NU (2006) Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol 14: 488–496. [DOI] [PubMed] [Google Scholar]
- 5. Popa R, Weber PK, Pett-Ridge J, Finzi JA, Fallon SJ, et al. (2007) Carbon and nitrogen fixation and metabolite exchange in and between individual cells of Anabaena oscillarioides. Isme Journal 1: 354–360. [DOI] [PubMed] [Google Scholar]
- 6. Ducat DC, Way JC, Silver PA (2011) Engineering cyanobacteria to generate high-value products. Trends Biotechnol 29: 95–103. [DOI] [PubMed] [Google Scholar]
- 7. Savage DF, Way J, Silver PA (2008) Defossiling fuel: How synthetic biology can transform biofuel production. Acs Chemical Biology 3: 13–16. [DOI] [PubMed] [Google Scholar]
- 8. Dismukes GC, Carrieri D, Bennette N, Ananyev GM, Posewitz MC (2008) Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Current Opinion in Biotechnology 19: 235–240. [DOI] [PubMed] [Google Scholar]
- 9. Welsh EA, Liberton M, Stockel J, Loh T, Elvitigala T, et al. (2008) The genome of Cyanothece 51142, a unicellular diazotrophic cyanobacterium important in the marine nitrogen cycle. Proc Natl Acad Sci U S A 105: 15094–15099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zehr JP, Church MJ, Moisander PH (2005) Diversity, distribution and biogeochemical significance of nitrogen-fixing microorganisms in anoxic and suboxic ocean environments. NATO Series book on past and present water column anoxia: Springer. 337–369.
- 11. Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, et al. (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res 3: 109–136. [DOI] [PubMed] [Google Scholar]
- 12. Knoop H, Zilliges Y, Lockau W, Steuer R (2010) The Metabolic Network of Synechocystis sp. PCC 6803: Systemic Properties of Autotrophic Growth. Plant Physiology 154: 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lindberg P, Park S, Melis A (2010) Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metabolic Engineering 12: 70–79. [DOI] [PubMed] [Google Scholar]
- 14. Wu GF, Shen ZY, Wu QY (2001) Possibility to improve the cyanobacterial poly-beta-hydroxybutyrate biosynthesis level. Journal of Chemical Engineering of Japan 34: 1187–1190. [Google Scholar]
- 15. Liu XY, Curtiss R (2009) Nickel-inducible lysis system in Synechocystis sp PCC 6803. Proceedings of the National Academy of Sciences of the United States of America 106: 21550–21554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McHugh K (2005) Hydrogen production methods. Virginia: MPR Associates, Inc.
- 18. Turner J, Sverdrup G, Mann MK, Maness PC, Kroposki B, et al. (2008) Renewable hydrogen production. International Journal of Energy Research 32: 379–407. [Google Scholar]
- 19. Bandyopadhyay A, Stockel J, Min H, Sherman LA, Pakrasi HB (2010) High rates of photobiological H2 production by a cyanobacterium under aerobic conditions. Nat Commun 1: 139. [DOI] [PubMed] [Google Scholar]
- 20. Min H, Sherman LA (2010) Hydrogen production by the unicellular, diazotrophic cyanobacterium Cyanothece sp. strain ATCC 51142 under conditions of continuous light. Appl Environ Microbiol 76: 4293–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schirmer A, Rude MA, Li XZ, Popova E, del Cardayre SB (2010) Microbial Biosynthesis of Alkanes. Science 329: 559–562. [DOI] [PubMed] [Google Scholar]
- 22. Wu B, Zhang BC, Feng XY, Rubens JR, Huang R, et al. (2010) Alternative isoleucine synthesis pathway in cyanobacterial species. Microbiology-Sgm 156: 596–602. [DOI] [PubMed] [Google Scholar]
- 23. Reed JL, Patel TR, Chen KH, Joyce AR, Applebee MK, et al. (2006) Systems approach to refining genome annotation. Proc Natl Acad Sci U S A 103: 17480–17484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Puchalka J, Oberhardt MA, Godinho M, Bielecka A, Regenhardt D, et al. (2008) Genome-scale reconstruction and analysis of the Pseudomonas putida KT2440 metabolic network facilitates applications in biotechnology. PLoS Comput Biol 4: e1000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vu TT, Stolyar SM, Pinchuk GE, Hill EA, Kucek LA, et al. (2012) Genome-scale modeling of light-driven reductant partitioning and carbon fluxes in diazotrophic unicellular cyanobacterium Cyanothece sp. ATCC 51142. PLoS Comput Biol 8: e1002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hong SJ, Lee CG (2007) Evaluation of central metabolism based on a genomic database of Synechocystis PCC6803. Biotechnology and Bioprocess Engineering 12: 165–173. [Google Scholar]
- 27. Shastri AA, Morgan JA (2005) Flux balance analysis of photoautotrophic metabolism. Biotechnology Progress 21: 1617–1626. [DOI] [PubMed] [Google Scholar]
- 28. Yang C, Hua Q, Shimizu K (2002) Metabolic flux analysis in Synechocystis using isotope distribution from 13C-labeled glucose. Metab Eng 4: 202–216. [DOI] [PubMed] [Google Scholar]
- 29. Fu PC (2009) Genome-scale modeling of Synechocystis sp PCC 6803 and prediction of pathway insertion. Journal of Chemical Technology and Biotechnology 84: 473–483. [Google Scholar]
- 30.Montagud A, Navarro E, de Cordoba PF, Urchueguia JF, Patil KR (2010) Reconstruction and analysis of genome-scale metabolic model of a photosynthetic bacterium. Bmc Systems Biology 4: -. [DOI] [PMC free article] [PubMed]
- 31. Montagud A, Zelezniak A, Navarro E, de Cordoba P, Urchueguia JF, et al. (2011) Flux coupling and transcriptional regulation within the metabolic network of the photosynthetic bacterium Synechocystis sp PCC6803. Biotechnology Journal 6: 330–342. [DOI] [PubMed] [Google Scholar]
- 32. Nogales J, Gudmundsson S, Knight EM, Palsson BO, Thiele I (2012) Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis. Proc Natl Acad Sci U S A 109: 2678–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang SY, Bryant DA (2011) The Tricarboxylic Acid Cycle in Cyanobacteria. Science 334: 1551–1553. [DOI] [PubMed] [Google Scholar]
- 34. Nakamura Y, Kaneko T, Miyajima N, Tabata S (1999) Extension of CyanoBase. CyanoMutants: repository of mutant information on Synechocystis sp. strain PCC6803. Nucleic Acids Res 27: 66–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Young JD, Shastri AA, Stephanopoulos G, Morgan JA (2011) Mapping photoautotrophic metabolism with isotopically nonstationary (13)C flux analysis. Metabolic Engineering 13: 656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stockel J, Jacobs JM, Elvitigala TR, Liberton M, Welsh EA, et al. (2011) Diurnal rhythms result in significant changes in the cellular protein complement in the cyanobacterium Cyanothece 51142. PLoS One 6: e16680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kucho K, Okamoto K, Tsuchiya Y, Nomura S, Nango M, et al. (2005) Global analysis of circadian expression in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 187: 2190–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tredici MR, Margheri MC, Philippis RD, Materass R, Bocci F, et al. (1986) Conversion of solar energy into the energy of biomass by culture of marine cyanobacteria. Proceedings of the 1986 International Congress on Renewable Energy Sources 1: 191–199. [Google Scholar]
- 39. Satish Kumar V, Dasika MS, Maranas CD (2007) Optimization based automated curation of metabolic reconstructions. BMC Bioinformatics 8: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reddy KJ, Haskell JB, Sherman DM, Sherman LA (1993) Unicellular, aerobic nitrogen-fixing cyanobacteria of the genus Cyanothece. J Bacteriol 175: 1284–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bentley FK, Melis A (2012) Diffusion-based process for carbon dioxide uptake and isoprene emission in gaseous/aqueous two-phase photobioreactors by photosynthetic microorganisms. Biotechnol Bioeng 109: 100–109. [DOI] [PubMed] [Google Scholar]
- 42. Nakao M, Okamoto S, Kohara M, Fujishiro T, Fujisawa T, et al. (2010) CyanoBase: the cyanobacteria genome database update 2010. Nucleic Acids Res 38: D379–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kumar VS, Maranas CD (2009) GrowMatch: an automated method for reconciling in silico/in vivo growth predictions. PLoS Comput Biol 5: e1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Minamizaki K, Mizoguchi T, Goto T, Tamiaki H, Fujita Y (2008) Identification of two homologous genes, chlAI and chlAII, that are differentially involved in isocyclic ring formation of chlorophyll a in the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem 283: 2684–2692. [DOI] [PubMed] [Google Scholar]
- 45. Jansson C, Debus RJ, Osiewacz HD, Gurevitz M, McIntosh L (1987) Construction of an Obligate Photoheterotrophic Mutant of the Cyanobacterium Synechocystis 6803 : Inactivation of the psbA Gene Family. Plant Physiol 85: 1021–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chitnis PR, Reilly PA, Nelson N (1989) Insertional Inactivation of the Gene Encoding Subunit-Ii of Photosystem-I from the Cyanobacterium Synechocystis Sp Pcc-6803. Journal of Biological Chemistry 264: 18381–18385. [PubMed] [Google Scholar]
- 47. Nakamoto H (1995) Targeted inactivation of the gene psaI encoding a subunit of photosystem I of the cyanobacterium Synechocystis sp PCC 6803. Plant and Cell Physiology 36: 1579–1587. [PubMed] [Google Scholar]
- 48. Burnap RL, Sherman LA (1991) Deletion Mutagenesis in Synechocystis Sp Pcc6803 Indicates That the Mn-Stabilizing Protein of Photosystem-Ii Is Not Essential for O2 Evolution. Biochemistry 30: 440–446. [DOI] [PubMed] [Google Scholar]
- 49. Shen JR, Ikeuchi M, Inoue Y (1997) Analysis of the psbU gene encoding the 12-kDa extrinsic protein of photosystem II and studies on its role by deletion mutagenesis in Synechocystis sp. PCC 6803. Journal of Biological Chemistry 272: 17821–17826. [DOI] [PubMed] [Google Scholar]
- 50. Papadopoulos JS, Agarwala R (2007) COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23: 1073–1079. [DOI] [PubMed] [Google Scholar]
- 51. Chitnis PR, Reilly PA, Miedel MC, Nelson N (1989) Structure and Targeted Mutagenesis of the Gene Encoding 8-Kda Subunit of Photosystem-I from the Cyanobacterium Synechocystis Sp Pcc-6803. Journal of Biological Chemistry 264: 18374–18380. [PubMed] [Google Scholar]
- 52. Ughy B, Ajlani G (2004) Phycobilisome rod mutants in Synechocystis sp strain PCC6803. Microbiology-Sgm 150: 4147–4156. [DOI] [PubMed] [Google Scholar]
- 53. Delorimier R, Bryant DA, Stevens SE (1990) Genetic-Analysis of a 9 Kda Phycocyanin-Associated Linker Polypeptide. Biochimica Et Biophysica Acta 1019: 29–41. [DOI] [PubMed] [Google Scholar]
- 54. Jallet D, Gwizdala M, Kirilovsky D (2012) ApcD, ApcF and ApcE are not required for the Orange Carotenoid Protein related phycobilisome fluorescence quenching in the cyanobacterium Synechocystis PCC 6803. Biochim Biophys Acta 1817: 1418–1427. [DOI] [PubMed] [Google Scholar]
- 55. Shen JR, Vermaas W, Inoue Y (1995) The Role of Cytochrome C-550 as Studied through Reverse Genetics and Mutant Characterization in Synechocystis Sp Pcc-6803. Journal of Biological Chemistry 270: 6901–6907. [DOI] [PubMed] [Google Scholar]
- 56. Shen JR, Qian M, Inoue Y, Burnap RL (1998) Functional characterization of Synechocystis sp. PCC 6803 Delta psbU and Delta psbV mutants reveals important roles of cytochrome c-550 in cyanobacterial oxygen evolution. Biochemistry 37: 1551–1558. [DOI] [PubMed] [Google Scholar]
- 57. Manna P, Vermaas W (1997) Lumenal proteins involved in respiratory electron transport in the cyanobacterium Synechocystis sp. PCC6803. Plant Molecular Biology 35: 407–416. [DOI] [PubMed] [Google Scholar]
- 58. Shen GZ, Boussiba S, Vermaas WFJ (1993) Synechocystis Sp Pcc-6803 Strains Lacking Photosystem-I and Phycobilisome Function. Plant Cell 5: 1853–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mo ML, Palsson BO, Herrgard MJ (2009) Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst Biol 3: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zahalak M, Pratte B, Werth KJ, Thiel T (2004) Molybdate transport and its effect on nitrogen utilization in the cyanobacterium Anabaena variabilis ATCC 29413. Molecular Microbiology 51: 539–549. [DOI] [PubMed] [Google Scholar]
- 61. Fernandez-Gonzalez B, Sandmann G, Vioque A (1997) A new type of asymmetrically acting beta-carotene ketolase is required for the synthesis of echinenone in the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem 272: 9728–9733. [DOI] [PubMed] [Google Scholar]
- 62. Tottey S, Rich PR, Rondet SA, Robinson NJ (2001) Two Menkes-type atpases supply copper for photosynthesis in Synechocystis PCC 6803. J Biol Chem 276: 19999–20004. [DOI] [PubMed] [Google Scholar]
- 63. Tottey S, Rondet SA, Borrelly GP, Robinson PJ, Rich PR, et al. (2002) A copper metallochaperone for photosynthesis and respiration reveals metal-specific targets, interaction with an importer, and alternative sites for copper acquisition. J Biol Chem 277: 5490–5497. [DOI] [PubMed] [Google Scholar]
- 64. Cheng Z, Sattler S, Maeda H, Sakuragi Y, Bryant DA, et al. (2003) Highly divergent methyltransferases catalyze a conserved reaction in tocopherol and plastoquinone synthesis in cyanobacteria and photosynthetic eukaryotes. Plant Cell 15: 2343–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sakuragi Y, Zybailov B, Shen G, Jones AD, Chitnis PR, et al. (2002) Insertional inactivation of the menG gene, encoding 2-phytyl-1,4-naphthoquinone methyltransferase of Synechocystis sp. PCC 6803, results in the incorporation of 2-phytyl-1,4-naphthoquinone into the A(1) site and alteration of the equilibrium constant between A(1) and F(X) in photosystem I. Biochemistry 41: 394–405. [DOI] [PubMed] [Google Scholar]
- 66. Dahnhardt D, Falk J, Appel J, van der Kooij TA, Schulz-Friedrich R, et al. (2002) The hydroxyphenylpyruvate dioxygenase from Synechocystis sp. PCC 6803 is not required for plastoquinone biosynthesis. FEBS Lett 523: 177–181. [DOI] [PubMed] [Google Scholar]
- 67. Ogawa T, Marco E, Orus MI (1994) A gene (ccmA) required for carboxysome formation in the cyanobacterium Synechocystis sp. strain PCC6803. J Bacteriol 176: 2374–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taiz L, Zeiger E (2002) Plant Physilogy. Massachusetts: Sinauer Associates, Inc., Publishers.
- 69. Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM (2008) Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat Rev Microbiol 6: 681–691. [DOI] [PubMed] [Google Scholar]
- 70. Badger MR, Price GD (2003) CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot 54: 609–622. [DOI] [PubMed] [Google Scholar]
- 71. Paerl HW (1984) Cyanobacterial Carotenoids - Their Roles in Maintaining Optimal Photosynthetic Production among Aquatic Bloom Forming Genera. Oecologia 61: 143–149. [DOI] [PubMed] [Google Scholar]
- 72. Glazer AN (1977) Structure and molecular organization of the photosynthetic accessory pigments of cyanobacteria and red algae. Mol Cell Biochem 18: 125–140. [DOI] [PubMed] [Google Scholar]
- 73. Poutanen EL, Nikkila K (2001) Carotenoid pigments as tracers of cyanobacterial blooms in recent and postglacial sediments of the Baltic Sea. Ambio 30: 179–183. [PubMed] [Google Scholar]
- 74. Collins MD, Jones D (1981) Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev 45: 316–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Stockel J, Jacobs JM, Elvitigala TR, Liberton M, Welsh EA, et al. (2011) Diurnal rhythms result in significant changes in the cellular protein complement in the cyanobacterium Cyanothece 51142. PLoS One 6: e16680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Allahverdiyeva Y, Ermakova M, Eisenhut M, Zhang P, Richaud P, et al. (2011) Interplay between flavodiiron proteins and photorespiration in Synechocystis sp. PCC 6803. J Biol Chem 286: 24007–24014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bandyopadhyay A, Elvitigala T, Welsh E, Stockel J, Liberton M, et al.. (2011) Novel metabolic attributes of the genus cyanothece, comprising a group of unicellular nitrogen-fixing Cyanothece. Mbio 2. [DOI] [PMC free article] [PubMed]
- 78. Quintero MJ, Muro-Pastor AM, Herrero A, Flores E (2000) Arginine catabolism in the cyanobacterium Synechocystis sp. Strain PCC 6803 involves the urea cycle and arginase pathway. J Bacteriol 182: 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Solomon CM, Collier JL, Berg GM, Glibert PM (2010) Role of urea in microbial metabolism in aquatic systems: a biochemical and molecular review. Aquatic Microbial Ecology 59: 67–88. [Google Scholar]
- 80. Tripp HJ, Bench SR, Turk KA, Foster RA, Desany BA, et al. (2010) Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature 464: 90–94. [DOI] [PubMed] [Google Scholar]
- 81.Connor MR, Atsumi S (2010) Synthetic Biology Guides Biofuel Production. Journal of Biomedicine and Biotechnology. [DOI] [PMC free article] [PubMed]
- 82. Antal TK, Lindblad P (2005) Production of H2 by sulphur-deprived cells of the unicellular cyanobacteria Gloeocapsa alpicola and Synechocystis sp. PCC 6803 during dark incubation with methane or at various extracellular pH. J Appl Microbiol 98: 114–120. [DOI] [PubMed] [Google Scholar]
- 83. Muro-Pastor MI, Reyes JC, Florencio FJ (1996) The NADP+-isocitrate dehydrogenase gene (icd) is nitrogen regulated in cyanobacteria. J Bacteriol 178: 4070–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Stoeckel J, Welsh EA, Liberton M, Kunnvakkam R, Aurora R, et al. (2008) Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes. Proceedings of the National Academy of Sciences of the United States of America 105: 6156–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jensen PA, Lutz KA, Papin JA (2011) TIGER: Toolbox for integrating genome-scale metabolic models, expression data, and transcriptional regulatory networks. Bmc Systems Biology 5. [DOI] [PMC free article] [PubMed]
- 86.Colijn C, Brandes A, Zucker J, Lun DS, Weiner B, et al.. (2009) Interpreting Expression Data with Metabolic Flux Models: Predicting Mycobacterium tuberculosis Mycolic Acid Production. Plos Computational Biology 5. [DOI] [PMC free article] [PubMed]
- 87. Mahadevan RE, Edwards JS, Doyle FJ (2003) Dynamic Flux Analysis of diauxic growth in Escherichia coli . Biophysical Journal 83: 1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim J, Reed JL (2010) OptORF: Optimal metabolic and regulatory perturbations for metabolic engineering of microbial strains. Bmc Systems Biology 4. [DOI] [PMC free article] [PubMed]
- 89.Ranganathan S, Suthers PF, Maranas CD (2010) OptForce: An Optimization Procedure for Identifying All Genetic Manipulations Leading to Targeted Overproductions. Plos Computational Biology 6. [DOI] [PMC free article] [PubMed]
- 90. Gao Q, Wang W, Zhao H, Lu X (2012) Effects of fatty acid activation on photosynthetic production of fatty acid-based biofuels in Synechocystis sp. PCC6803. Biotechnol Biofuels 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tan X, Yao L, Gao Q, Wang W, Qi F, et al. (2011) Photosynthesis driven conversion of carbon dioxide to fatty alcohols and hydrocarbons in cyanobacteria. Metab Eng 13: 169–176. [DOI] [PubMed] [Google Scholar]
- 92.Allen MM (1968) Simple Conditions for Growth of Unicellular Blue-Green Algae on Plates. Journal of Phycology 4: 1–&. [DOI] [PubMed]
- 93. Reddy KJ, Haskell JB, Sherman DM, Sherman LA (1993) Unicellular, Aerobic Nitrogen-Fixing Cyanobacteria of the Genus Cyanothece. Journal of Bacteriology 175: 1284–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394. [Google Scholar]
- 95. Lichtenthaler HK (1987) Chlorophylls and Carotenoids - Pigments of Photosynthetic Biomembranes. Methods in Enzymology 148: 350–382. [Google Scholar]
- 96. Steiger S, Schafer L, Sandmann G (1999) High-light-dependent upregulation of carotenoids and their antioxidative properties in the cyanobacterium Synechocystis PCC 6803. Journal of Photochemistry and Photobiology B-Biology 52: 14–18. [Google Scholar]
- 97. Arnon DI, Mcswain BD, Tsujimot.Hy, Wada K (1974) Photochemical Activity and Components of Membrane Preparations from Blue-Green-Algae.1. Coexistence of 2 Photosystems in Relation to Chlorophyll Alpha and Removal of Phycocyanin. Biochimica Et Biophysica Acta 357: 231–245. [DOI] [PubMed] [Google Scholar]
- 98. Mortazavi AW, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcripts by RNA-Seq. Nature Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 99. Varma A, Palsson BO (1994) Metabolic Flux Balancing - Basic Concepts, Scientific and Practical Use. Bio-Technology 12: 994–998. [Google Scholar]
- 100.Kumar VS, Ferry JG, Maranas CD (2011) Metabolic reconstruction of the archaeon methanogen Methanosarcina Acetivorans. Bmc Systems Biology 5. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Synechocystis iSyn731 model along with established GPR, metabolite, gene and protein information.
(XLSX)
Cyanothece iCyt773 model along with established GPR, metabolite, gene and protein information.
(XLSX)
Biomass component measurements and stoichiometry of biomass equation.
(XLSX)
Reactions with diurnal activation/inactivation.
(XLSX)
Comparison of in silico vs. in vivo gene essentiality results for iSyn731 and modifications made in GPR associations.
(XLSX)
Comparison between Synechocystis iSyn731 and iCyt773 models in terms of genes, proteins, reactions and metabolites.
(XLSX)
SBML file of Synechocystis iSyn731 model.
(XML)
SBML file of Cyanothece 51142 iCyt773 model.
(XML)