Abstract
We investigated 15,542 patients with suspected BCR-ABL1- negative myeloproliferative or myelodysplastic/myeloproliferative neoplasm (including 359 chronic myelomonocytic leukemia) by a molecular marker set. JAK2V617F was detected in the suspected categories as follows: polycythemia vera 88.3%, primary myelofibrosis 53.8%, essential thrombocythemia 50.2%, and not further classifiable myeloproliferative neoplasms 38.0%. JAK2 exon 12 mutations were detected in 40.0% JAK2V617F-negative suspected polycythemia vera, MPLW515 mutations in 13.2%JAK2V617F-negative primary myelofibrosis and 7.1% JAK2V617F-negative essential thrombocythemia. TET2 mutations were distributed across all entities but were most frequent in suspected chronic myelomonocytic leukemia (77.8%). CBL mutations were identified in suspected chronic myelomonocytic leukemia (13.9%), primary myelofibrosis (8.0%), and not further classifiable myeloproliferative neoplasm (7.0%). This leads to a stepwise workflow for suspected myeloproliferative neoplasms starting with JAK2V617F and investigating JAK2V617F-negative patients for JAK2 exon 12 or MPL mutations, respectively. In cases in which a myeloproliferative neoplasm cannot be established, analysis for TET2, CBL and EZH2 mutations may be indicated.
Key words: myeloproliferative neoplasms, chronic myelomonocytic leukemia, molecular analyses, mutation screening, diagnostic workflow
Introduction
In recent years, the expansion of the molecular marker panel for the myeloproliferative neoplasms (MPNs) has paved the way to new diagnostic strategies for patients with persistent and otherwise unexplained elevated peripheral blood counts or symptoms such as splenomegaly. Following the detection of the JAK2V617F as the most frequent mutation in the BCR-ABL1-negative MPNs, especially in polycythemia vera (PV), other JAK-STAT pathway activating mutations were detected: mutations in exon 12 of JAK2 in roughly one-third of V617F-negative PV cases, which sometimes present with an apparently isolated erythrocytosis,1,2 and mutations of the MPL gene in V617F-negative essential thrombocythemia (ET) and primary myelofibrosis (PMF).3 Grand et al. identified mutations of the CBL (Casitas B-cell lymphoma) gene in patients with PMF, but not in ET or PV.4 Mutations of TET2 (TET oncogene family member 2) were described in PV, ET, PMF, and in post-PV and post-ET myelofibrosis.5 The EZH2 (histone methyltransferase; enhancer of zeste homolog 2) gene was found to be mutated in 7-13% of patients with myelofibrosis.6,7 Patients with chronic myelomonocytic leukemia (CMML) were observed to have a high frequency of TET2, CBL, and EZH2 mutations. 8-10
Design and Methods
To further evaluate the power and applicability of this new molecular marker set in a routine diagnostic setting, we investigated 15,542 patients with suspicion for a myeloproliferative or myelodysplastic/myeloproliferative neoplasm in whom BCR-ABL1 was ruled out, and developed a stepwise diagnostic workflow based on the results. The study cohort was made up of 7,879 males and 7,663 females, median age 62.9 years (range 0.1-97.5 years). From August 2005 to June 2010, bone marrow as sole sample material (n=5,222), bone marrow in combination with peripheral blood (n=96), or peripheral blood only (n=10,224) were sent from different hematologic centers for diagnosis to the MLL Munich Leukemia Laboratory. In detail, all samples sent with the suspicion for a BCR-ABL1-negative MPN, myelodysplastic/myeloproliferative neoplasm, CMML, or with constellations such as “unexplained leukocytosis” or “unexplained thrombocytosis”, and in whom a diagnosis of chronic myeloid leukemia (CML) had been excluded by genetic analysis, were included in the study. According to clinical symptoms as reported by the referring hematologists, laboratory parameters, and cytomorphological evaluation, indications for molecular diagnostics were as follows: in 739 cases the suspected diagnosis could be narrowed down to PV, in 1,620 cases to ET, and in 324 patients to PMF. In contrast, a total of 11,461 patients had suspicion of an MPN that could not be further subcategorized based on clinical symptoms or laboratory parameters when samples were taken. Isolated erythrocytosis was present in 753 patients. Thirty-six patients had otherwise unexplained thrombosis. Regarding the category of myelodysplastic/myeloproliferative disorders, 250 patients were suspected for an unclassifiable MDS/MPN (MDS/MPNu) and 359 for CMML.11 All patients gave their written informed consent to genetic analysis and scientific studies. The study was approved by the Internal Review Board of the MLL Munich Leukemia Laboratory and was carried out in accordance with the Declaration of Helsinki.
Analysis for the JAK2V617F (15,363 cases analyzed; sensitivity 1%),12 JAK2 exon 12 (n=2,224; sensitivity 5%),13 and MPLW51514 (n=2678; sensitivity 5%) mutations by different PCR assays followed previous descriptions. The workflow was expanded in the final study period so smaller subsets of patients were investigated for recently detected mutations in genes such as CBL (n=480 investigated), TET2 (n=123), and EZH2 (n=34), sensitivity 10% for each. CBL was analyzed by direct Sanger sequencing of exons 8-9.4 The whole coding region of TET2 was covered with 24 amplicons and also analyzed by Sanger sequencing. EZH2 mutations9 were investigated by high-throughput sequencing (454 technology, Life Sciences, Branford, CT, USA). All mutations that were not obviously damaging (stop or frameshift) were confirmed by data base research, analysis of remission controls and/or buccal swaps to make sure that they are real mutations. CML was excluded in all cases of this study by multiplex RT (reverse transcription)-PCR for various BCR-ABL1 transcripts.15 The diagnosis of CMML was based on cytomorphology. In cases with eosinophilia, the presence of PDGFR rearrangements was excluded according to WHO 2008.11
Results and Discussion
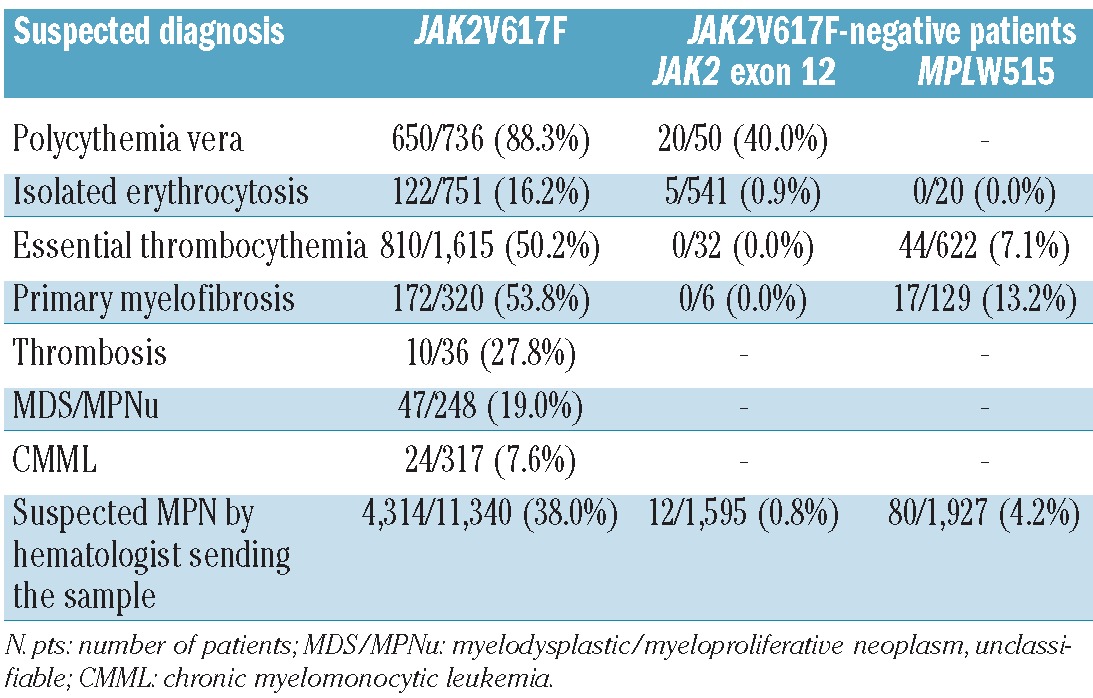
JAK2V617F was detected in 650 of 736 (88.3%) analyzed patients with suspected PV, in 172 of 320 (53.8%) suspected PMF, and in 810 of 1,615 (50.2%) analyzed patients with suspected ET. In addition, 122 of 751 (16.2%) patients with isolated erythrocytosis were JAK2V617F-positive leading to a diagnosis of PV. In patients with suspected not further classifiable MPN, the JAK2V617F was found in 4,314 of 11,340 (38.0%) cases, confirming a diagnosis of an MPN. The frequency of the JAK2V617F was lower in patients with suspected myelodysplastic/myeloproliferative neoplasms (MDS/MPNu 47 of 248, 19.0%; CMML 24 of 317, 7.6%). Patients with unexplained thrombosis were JAK2V617F-positive in 10 of 36 (27.8%) patients. JAK2V617F-negative cases but suspected PV were analyzed for JAK2 exon 12 mutations; these were found in 20 of 50 (40.0%) patients. However, JAK2 exon 12 mutations were rarely detected (5 of 541, 0.9%) in patients with isolated erythrocytosis. JAK2 exon 12 mutations were also found in 12 of 1,595 (0.8%) patients with suspicion for a not further classifiable MPN. No JAK2 exon 12 mutation was identified in 32 cases with ET or 6 PMF, respectively. MPLW515 mutations were detected in 17 of 129 (13.2%) JAK2V617Fnegative PMF and in 44 of 622 (7.1%) JAK2V617F-negative ET. JAK2V617F-negative patients with a suspected not further classifiable MPN but with elevated thrombocyte count showed MPLW515 mutations in 80 of 1,927 (4.2%) leading to narrowing down the diagnosis to ET or PMF (Table 1).
Table 1.
Results of analyses for JAK2V617F, JAK2 exon 12, and MPLW515 mutations in the different categories of suspicion.
TET2 mutations were distributed across all cohorts with the highest frequency in CMML (14 of 18, 77.8%) that was higher than in suspected PMF (3 of 11, 27.3%), ET (8 of 30, 26.7%), or suspected not further classifiable MPN (16 of 64, 25.0%). CBL mutations showed the highest frequency in suspected CMML (23 of 165, 13.9%). Slightly lower frequencies of CBLmutations were found in suspected PMF (2 of 25, 8.0%) or suspected not further classifiable MPN (14 of 201, 7.0%). No case with CBLmutation was identified in suspected PV (n=32) or ET (n=57). EZH2 mutations were detected in 2 of 9 (22.2%) of patients with suspected PMF and in one of 7 (14.3%) suspected CMML cases, whereas all suspected ET analyzed (n=18) were EZH2 wild-type. Although we investigated only a subset of patients for these novel markers, these results further confirm previous observations that CBLmutations can be found in PMF but do not occur in ET.4 Overall, this study reflects the experience with the above molecular marker panel for a restricted subset of patients rather than providing a complete characterization of molecular mutation profiles of the entities investigated.
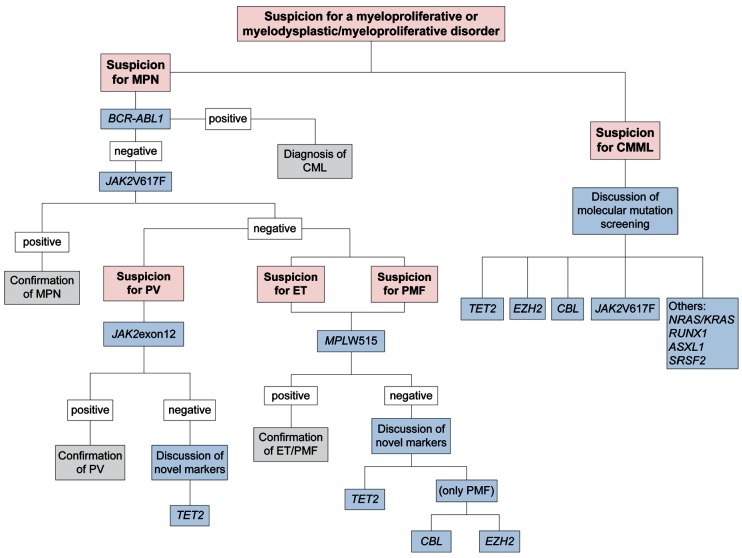
Recently, Tefferi et al. stated that upfront en bloc screening for JAK2V617F, JAK2 exon 12, and MPL mutations was scientifically irrational and economically irresponsible.16 In accordance with this perspective, the results of molecular analysis in our cohort as measured in a routine diagnostic setting emphasize a stepwise molecular diagnostic workflow for patients with suspected myeloproliferative disorders (Figure 1). After exclusion of CML by investigation for the BCR-ABL1 gene fusion, the algorithm starts with JAK2V617F mutation screening. Subsequently, JAK2V617Fnegative patients with suspected PV (e.g. polyglobulia, bone marrow hypercellularity with increase of erythropoiesis, granulopoiesis, and megakaryopoiesis, or reduced erythropoietin levels) or isolated erythrocytosis should be analyzed for a JAK2 exon 12 mutation. LNK mutations were recently suggested as a further marker in PV;17 however, further studies showed that this is not restricted to PV18 and the value of this marker for diagnostics in MPN has to be further evaluated.
Figure 1.
Proposal of a workflow for molecular diagnostics in patients with the suspicion for myeloproliferative or myelodysplastic/myeloproliferative disorders. MPN: myeloproliferative neoplasm; CML: chronic myeloid leukemia; PV: polycythemia vera; ET: essential thrombocythemia; PMF: primary myelofibrosis; CMML: chronic myelomonocytic leukemia.
When there is suspicion for a JAK2V617F-negative PMF or ET, patients should be investigated for an MPLW515 mutation. In case the diagnosis of an MPN cannot be established with any of these three markers, screening for a TET2 mutation may be discussed for all still unexplained symptomatic patients. Samples with suspected PMF could be investigated for CBL or EZH2 mutations in cases in which none of the above markers is positive. In cases of suspicion for CMML, a comprehensive molecular analysis including JAK2V617F, NRAS, RUNX1, KRAS, TET2, CBL, EZH2, ASXL1, and SRSF219 should be performed, as this is a genetically very complex disease with significant frequencies of mutations in the abovementioned genes.9,20 The exact sequence of the respective analyses should be defined in future studies. These step-wise procedures can guide the workflow in the laboratory and establish the correct diagnosis in patients with suspected myeloproliferative disorders or CMML in a more efficient and economic way
Footnotes
Authorship and Disclosures: The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Scott LM, Tong W, Levine RL, Scott MA, Beer PA, Stratton MR, et al. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med. 2007;356(5):459-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cross NC.Genetic and epigenetic complexity in myeloproliferative neoplasms Hematology Am Soc Hematol Educ Program. 2011;2011208-14 [DOI] [PubMed] [Google Scholar]
- 3.Pikman Y, Lee BH, Mercher T, McDowell E, Ebert BL, Gozo M, et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006;3(7):e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grand FH, Hidalgo-Curtis CE, Ernst T, Zoi K, Zoi C, McGuire C, et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood. 2009;113(24):6182-92 [DOI] [PubMed] [Google Scholar]
- 5.Tefferi A, Pardanani A, Lim KH, Abdel-Wahab O, Lasho TL, Patel J, et al. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia. 2009;23(5):905-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42(8):722-6 [DOI] [PubMed] [Google Scholar]
- 7.Abdel-Wahab O, Pardanani A, Patel J, Wadleigh M, Lasho T, Heguy A, et al. Concomitant analysis of EZH2 and ASXL1 mutations in myelofibrosis, chronic myelomonocytic leukemia and blast-phase myeloproliferative neoplasms. Leukemia. 2011;25(7):1200-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohlmann A, Grossmann V, Klein HU, Schindela S, Weiss T, Kazak B, et al. Nextgeneration sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J Clin Oncol. 2010;28(24):3858-65 [DOI] [PubMed] [Google Scholar]
- 9.Grossmann V, Kohlmann A, Eder C, Haferlach C, Kern W, Cross NC, et al. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia. 2011;25(5):877-9 [DOI] [PubMed] [Google Scholar]
- 10.Jankowska AM, Makishima H, Tiu RV, Szpurka H, Huang Y, Traina F, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 2011;118(14):3932-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swerdlow S, Campo E, Lee Harris N, Jaffe E, Pileri S, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th Lyon: IARC Press; 2008 [Google Scholar]
- 12.Schnittger S, Bacher U, Kern W, Schröder M, Haferlach T, Schoch C. Report on two novel nucleotide exchanges in the JAK2 pseudokinase domain: D620E and E627E. Leukemia. 2006;20(12):2195-7 [DOI] [PubMed] [Google Scholar]
- 13.Schnittger S, Bacher U, Haferlach C, Geer T, Müller P, Mittermüller J, et al. Detection of JAK2 exon 12 mutations in 15 patients with JAK2V617F negative polycythemia vera. Haematologica. 2009;94(3):414-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnittger S, Bacher U, Haferlach C, Dengler R, Kröber A, Kern W, et al. Detection of an MPLW515 mutation in a case with features of both essential thrombocythemia and refractory anemia with ringed sideroblasts and thrombocytosis. Leukemia. 2008;22(2):453-5 [DOI] [PubMed] [Google Scholar]
- 15.Cross NC, Melo JV, Feng L, Goldman JM. An optimized multiplex polymerase chain reaction (PCR) for detection of BCR-ABL fusion mRNAs in haematological disorders. Leukemia. 1994;8(1):186-9 [PubMed] [Google Scholar]
- 16.Tefferi A, Noel P, Hanson CA. Uses and abuses of JAK2 and MPL mutation tests in myeloproliferative neoplasms a paper from the 2010 William Beaumont hospital symposium on molecular pathology. J Mol Diagn. 2011;13(5):461-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lasho TL, Pardanani A, Tefferi A. LNK mutations in JAK2 mutation-negative erythrocytosis. N Engl J Med. 2010;363(12):1189-90 [DOI] [PubMed] [Google Scholar]
- 18.Pardanani A, Lasho T, Finke C, Oh ST, Gotlib J, Tefferi A. LNK mutation studies in blast-phase myeloproliferative neoplasms, and in chronic-phase disease with TET2, IDH, JAK2 or MPL mutations. Leukemia 2010;24(10):1713-8 [DOI] [PubMed] [Google Scholar]
- 19.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011;478(7367):64-9 [DOI] [PubMed] [Google Scholar]
- 20.Meggendorfer M, Roller A, Haferlach T, Eder C, Dicker F, Grossmann V, et al. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML). Blood. 2012. August 23 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]