Abstract
Mutations in an enzyme can result in a neomorphic catalytic activity in cancers. We applied cancer-associated mutations from isocitrate dehydrogenases (IDHs) to homologous residues in the active sites of homoisocitrate dehydrogenases (HIDHs) to derive enzymes that catalyze the conversion of 2-oxoadipate to (R)-2-hydroxyadipate, a critical step for adipic acid production. Thus, we provide a prototypic example of how insights from cancer genome sequencing and functional studies can aid in enzyme redesign.
Keywords: IDH1, IDH2, enzyme design, β-hydroxyacid α-decarboxylase, β-hydroxyacid oxidative decarboxylase, R132H, (R)-2-hydroxyadipate dehydrogenase
New organic synthesis techniques and metabolic engineering methods often rely on the generation of enzymes with novel catalytic activities. Cancer is a process of microevolution that selects for functional alterations that benefit tumor cells, including gain-of-function mutations in metabolic enzymes1. Cancer mutational data therefore represents an until-now unappreciated source of enzyme functional diversity, which is becoming increasingly accessible as cancer sequencing data increases logarithmically with improvements in sequencing technology and as cancer geneticists turn their attention to cellular metabolic enzymes. We hypothesized that insights from mutated enzymes in cancer could guide the redesign of a useful enzyme. To demonstrate this approach, we applied insights from cancer mutations to derive an enzyme that overcomes a hurdle to the clean, bio-based production of adipic acid.
Adipic acid (HOOC(CH2)4COOH, 1,4-butanedicarboxylic acid) is a substrate for the synthesis of nylon-6,6, and is one of the most-produced chemicals worldwide. There is great interest in developing biological methods for adipic acid synthesis to lower the cost of input materials, to obviate the need for fossil fuel substrates, and to reduce the release of pollutants compared to standard catalytic methods of synthesis2–4. Multiple enzymatic transformations that would support one proposed fully biological method5 have recently become technically possible6. However, the enzymatic conversion of 2-oxoadipate to (R)-2-hydroxyadipate, a key step in the proposed pathway, remains a major challenge6.
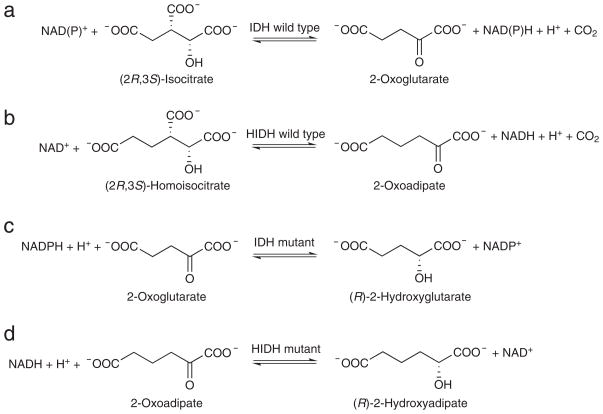
To engineer an enzyme to catalyze the conversion of 2-oxoadipate to (R)-2-hydroxyadipate, termed an (R)-2-hydroxyadipate dehydrogenase, we used insights from gain-of-function IDH mutants found in human cancer. IDHs are ubiquitous throughout life and convert (2R,3S)-isocitrate to 2-oxoglutarate, which is also known as α-ketoglutarate7 (Fig. 1a). All IDHs belong to the enzyme subfamily of metal ion-dependent pyridine-dinucleotide-linked β-hydroxyacid oxidative decarboxylases that act on substrates with the (R) configuration at the C-2 position8 (hereafter, β-hydroxyacid oxidative decarboxylases). HIDHs belong to the same β-hydroxyacid oxidative decarboxylase subfamily as the IDHs, but the HIDHs instead convert (2R,3S)-homoisocitrate, a 6-carbon-backbone analog of (2R,3S)-isocitrate, to 2-oxoadipate in yeast, thermophilic bacteria, and archaea8–13 (Fig. 1b). Recent exome sequencing revealed that missense mutations in NADP+-dependent IDHs, IDH1 and IDH2, occur frequently in the somatic cells of gliomas and other cancers14, 15. The IDH mutations alter active site residues that contact the β-carboxyl of (2R,3S)-isocitrate, including Arg132 of IDH1, Arg140 of IDH2, and Arg172 of IDH216, 17. These mutations cause the IDH enzymes to lose their native oxidative decarboxylase activity and gain a simple oxidoreductase activity to convert 2-oxoglutarate to (R)-2-hydroxyglutarate1, 17 (Fig. 1c). Our goal was to generate a similar oxidoreductase that favored substrates with a 6-carbon backbone. IDH cancer mutants have this desired simple oxidoreductase activity but favor substrates with a 5-carbon backbone, whereas wild-type HIDHs do not have this activity yet act on substrates with a 6-carbon backbone. Here, we applied mutations homologous to those observed in IDHs in cancer to HIDHs to generate a simple oxidoreductase function to convert 2-oxoadipate to (R)-2-hydroxyadipate6, i.e., the desired (R)-2-hydroxyadipate dehydrogenase (Fig. 1d).
Figure 1. Strategy for enzyme mutagenesis.
(a) IDHs catalyze the NAD(P)+-linked reversible oxidative decarboxylation of (2R,3S)-isocitrate to form 2-oxoglutarate and CO2. (b) HIDHs catalyze the NAD+-linked reversible oxidative decarboxylation of (2R,3S)-homoisocitrate to form 2-oxoadipate and CO2. In the reverse direction, HIDHs catalyze the reductive carboxylation of 2-oxoadipate with CO2 to form (2R,3S)-homoisocitrate. (c) In human cancer, IDH mutants such as HsIDH1-R132H catalyze the non-carboxylating reduction of 2-oxoglutarate to (R)-2-hydroxyglutarate. (d) Analogous HIDH mutants could catalyze the non-carboxylating reduction of 2-oxoadipate to (R)-2-hydroxyadipate, i.e., (R)-2-hydroxyadipate dehydrogenases.
To identify HIDH residues homologous to IDH cancer mutation hotspots, we compared the substrate-bound active site structures of human cytoplasmic NADP+-dependent isocitrate dehydrogenase (HsIDH1)18 and Schizosaccharomyces pombe (Sp) HIDH19 (Fig 2a). Overlay analysis revealed that the critical substrate-binding residues Arg100 and Arg132 contained within α-helix 5 and β-strand 5 of HsIDH1, respectively, were conserved in relative topography to Arg97 and Arg126 of SpHIDH. The structural homology between HsIDH1 and SpHIDH was used as an anchor to align their primary protein sequences with each other, and also to the greater family of β-hydroxyacid oxidative decarboxylases, including isopropylmalate dehydrogenases and tartrate dehydrogenases (Supplementary Results, Supplementary Fig. 1). This alignment differed from a previous alignment of HsIDH1 and the HIDHs that was based on primary sequence alone, which aligned HsIDH1 Arg132 with a gap region in the other β-hydroxyacid oxidative decarboxylases8.
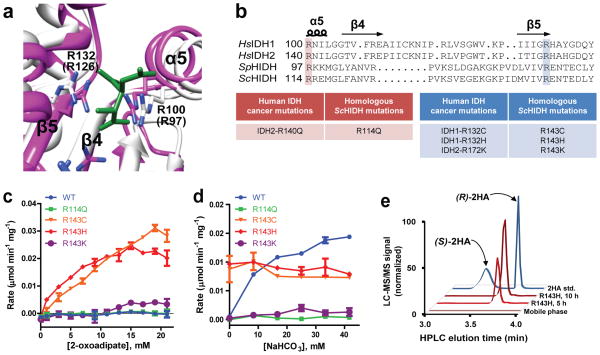
Figure 2. ScHIDH mutants catalyze the NADH-linked production of 2-hydroxyadipate.
(a) Superimposition of the active site for SpHIDH19 (white) onto HsIDH118 (pink; complex with isocitrate in green). Residues Arg100 and Arg132 of HsIDH1 are shown, and corresponding residues for SpHIDH are shown in parentheses. (b) Alignment of HsIDH1, HsIDH2, SpHIDH, and ScHIDH. (c) Initial rate of NADH decrease catalyzed by the indicated ScHIDH mutant at varying concentrations of 2-oxoadipate, and 300 μM NADH. (d) Initial rate of NADH decrease catalyzed by the indicated ScHIDH mutant in the presence of varying concentrations of NaHCO3, as well as 15 mM 2-oxoadipate and 100 μM NADH. (e) LC-MS/MS chromatogram showing (R)-2-hydroxyadipate (Q1/Q3 m/z = 377/161) accumulation in a reaction containing ScHIDH-R143H, 2 mM NADH, 2 mM 2-oxoadipate, and 500 mM HEPES at times indicated. A racemic (R/S)-2-hydroxyadipate standard and mobile phase are shown for reference. Unless otherwise specified, all reactions contained 20 mM MgCl2, 40 ng/μl of the indicated purified enzyme, and 100 mM HEPES, pH 7.3. Data points are mean ± s.d. from n=2 reactions. All plots are representative of three independent experiments.
Using the alignment generated above, we identified and produced HIDH mutants that were analogous to human cancer-derived IDH mutants. Given that the enzymatic properties of wild type Saccharomyces cerevisiae (Sc) HIDH (65% amino acid identity with SpHIDH) were well known10–12, we chose to focus our efforts on producing and characterizing mutants of ScHIDH. According to the new alignment and the structural overlay, ScHIDH mutants R114Q, R143C, R143H, and R143K were analogous to the common cancer mutations IDH2-R140Q, IDH1-R132C, IDH1-R132H, and IDH2-R172K (Fig. 2b). These mutants as well as wild type ScHIDH were produced and purified from bacteria (Supplementary Fig. 2).
To investigate whether one or more ScHIDH mutants would convert 2-oxoadipate and NADH to (R)-2-hydroxyadipate and NAD+, specific activity was assessed by measuring the intial rate of NADH decrease catalyzed by the enzymes in reactions containing 2-oxoadipate and NADH. The R143C and R143H mutants, and to a lesser extent the R143K mutant, consumed NADH in a reaction containing 40 ng/μl enzyme, 10 mM 2-oxoadipate, and 300 μM NADH, while no activity was observed for the R114Q mutant or for wild type ScHIDH (Supplementary Fig. 3a). The rate of NADH decrease in the presence of the three Arg143 mutants increased hyperbolically with the concentration of 2-oxoadipate (Fig. 2c) and with the concentration of NADH (Supplementary Fig. 3b). The initial rate of NADH decrease was used to estimate kinetic parameters for the R143H mutant. For the R143H mutant, the substrate concentration for half maximal rate (KM) for 2-oxoadipate was 2.0± 0.34 mM (s.d.) and the KM for NADH was 85 ± 21 uM (s.d.). These constants characterized ScHIDH-R143H to have tight binding to NADH and moderate binding to 2-oxoadipate, comparable to the substrate-binding properties of the WT enzyme10. The maximal reaction rate (Vmax) was 0.041± 0.0021 μmol NADH min−1 mg−1 (s.d.), which equates to a catalytic turnover rate (kcat) of 1.6 min−1. This rate is ~1% of the kcat for WT ScHIDH in the reductive direction10. No activity was observed when ScHIDH-R143H was incubated with 15 mM of either 2-oxoglutarate or 2-oxoheptanedioate, indicating that ScHIDH-R143H is specific for 2-oxoadipate and not other dicarboxylic acids (Supplementary Fig. 3c).
Next, the products of mutant ScHIDH were investigated and compared to wild type ScHIDH-WT. HIDH-WT normally carries out a reductive carboxylation reaction in which CO2 is required as a substrate to carboxylate 2-oxoadipate and form homoisocitrate (see Fig. 1c, from right to left). When CO2 was added to the reaction mix in the form of NaHCO3, ScHIDH-WT demonstrated 2-fold faster NADH consumption than ScHIDH-R143H (Fig. 2d). In contrast to ScHIDH-WT, ScHIDH-R143H and –R143C activity was not stimulated by the addition of NaHCO3. Liquid chromatography tandem mass spectrometry (LC-MS/MS) in negative electrospray ionization mode (-ESI) was undertaken to investigate the products of the wild type and R143H enzymes in the presence of 50 mM NaHCO3 (Supplementary Fig. 4). [3,3,4,4]-2H4-2-hydroxyglutarate was added to samples as an internal standard, and samples were derivatized with diacetyl-L-tartatic acid before binding to the HPLC column (see Supplementary Methods)20, 21. Two Q1/Q3 transitions, each representing a parent ion and a corresponding fragment ion, were monitored for the derivatives of homoisocitrate and of 2-hydroxyadipate. An intense homoisocitrate mass fragmentation pattern was observed in the reaction containing ScHIDH-WT (Q1/Q3 m/z = 421/205 and 421/187). In contrast, the ScHIDH-R143H reaction produced a 2-hydroxyadipate mass fragmentation pattern (Q1/Q3 m/z = 377/161 and 377/143). These results confirmed that ScHIDH-WT performs the expected reductive carboxylation of 2-oxoadipate to produce homoisocitrate and showed that ScHIDH-R143H catalyzes a non-carboxylating reductive reaction to produce 2-hydroxyadipate.
Since HIDHs are stereospecific for (R)-hydroxyacids as their substrates/products8, we predicted that the 2-hydroxyadipate produced by the ScHIDH mutants would be the (R) enantiomer of 2-hydroxyadipate. To confirm this, quantitative LC-MS/MS was performed using a 2-hydroxyadipate standard, and derivation with diacetyl-L-tartaric anhydride allowed the (R)- and (S)- enantiomers to be discriminated based on their elution time from the chiral HPLC column. (R)-2-hydroxyadipate stochiometrically increased as NADH was consumed in reactions containing R143H, R143C, and R143K mutants (Supplementary Fig. 5). (S)-2-hydroxyadipate was undetectable in those reactions as shown in Fig. 2e. Even assuming that the (S) enantiomer was at the lower limit of quantification (0.84 μM), these results indicated that the (R) enantiomer was produced in enantiomeric excess of >99.1% for the R143H mutant. This is consistent with the conversion of NADH, H+, and 2-oxoadipate to NAD+ and (R)-2-hydroxyadipate. Thus, ScHIDH-R143H, -R143C, and -R143K mutants are (R)-2-hydroxyadipate dehydrogenases.
We next investigated whether the new activity introduced by cancer-analogous mutations to ScHIDH was generalizable to an HIDH from a distantly related species, the thermophilic bacterium Thermus thermophilus9 (Supplementary Fig. 6). Arg118 of TtHIDH was analogous to Arg132 of HsIDH1 based on structure-informed alignment. Interestingly, the WT enzyme from this species did display some HCO3-independent activity that resulted in stoichiometric (R)-2-hydroxyadipate production, though this activity was significantly increased in the TtHIDH-R118H mutant. However, unlike ScHIDH-R143H, TtHIDH-R118H consumed NADH at a 3-fold faster rate in the presence of 15mM 2-oxoglutarate than in the presence of 15mM 2-oxoadipate. Because of this substrate promiscuity, TtHIDH-R118H is likely not an ideal (R)-2-hydroxyadipate dehydrogenase for bio-based adipate production. Nevertheless, the finding that mutation of conserved residues conferred a change-of-function not only to HsIDH1 and HsIDH2, but also to distantly related HIDHs, illuminated a conserved feature of the subfamily of β-hydroxyacid oxidative decarboxylases that act on (R)-hydroxy substrates to be redesigned into simple oxidoreductases.
We showed here that redesigned HIDH enzymes catalyze the novel NADH-dependent conversion of 2-oxoadipate to (R)-2-hydroxyadipate. These results defined a potentially generalizable rule that mutation of the arginine residues homologous to Arg132 of IDH1 (Arg143 of ScHIDH, Arg118 of TtHIDH) can introduce a change-of-function to enzymes of this subfamily. The redesigned enzymes had excellent enantioselectivity and moderate substrate binding properties. This establishment of desired activity for HIDH mutants represents first generation enzymes that can be further improved in catalytic turnover rate by directed evolution methods. The redesigned enzymes may be useful for bio-based adipic acid production, for other metabolic engineering applications, and for the in vitro synthesis of chiral compounds. Metabolic enzymes running at high catalytic rates, such as malic enzyme and malate dehydrogenase, are known to produce off-target metabolites at low levels due to mechanistic promiscuity errors22. In a similar manner, IDHs catalyze off-target 2-hydroxyglutarate production at low levels23. Active site IDH mutations in cancer alter substrate binding properties, structural conformational equilibrium, binding affinity for competitive inhibitors, and the rate of 2-hydroxyglutarate-producing catalysis to apparently favor 2-hydroxyglutarate production1, 23. Though our approach to enzyme redesign was agnostic to chemical mechanism, the homology between HIDHs and IDHs suggests that analagous mechanistic changes are at work in the redesigned HIDH mutants.
Our strategy for enzyme redesign was to apply functional mutations observed in cancer cells to homologous enzymes. Though documented gain-of-function activities in metabolic enzymes resulting from cancer mutations are extremely rare so far, these results suggested that sequencing cancer genomes can provide a powerful source of functional mutation diversity to mine for additional novel enzyme activities. Even the numerous rarely-occurring mutations that are of marginal interest from a molecular pathology standpoint due to their rarity could still provide valuable insights for enzyme redesign. While computational design and directed evolution are certain to remain the major avenues to novel enzyme design, this study demonstrated that prospecting for alterations from human disease, and in particular cancer, can help to rapidly address a specific problem in enzyme design.
Supplementary Material
Acknowledgments
This work is supported by NIH grant R01 CA1403160 (H.Y.). We thank Paul F. Cook for an HIDH expression plasmid, Eric Spana for S. cerevisiae genomic DNA, Ping Fan for technical assistance, as well as Irwin Fridovich and Ines Batinic-Haberle for feedback on the manuscript.
Abbreviations
- IDH
isocitrate dehydrogenase
- HIDH
homoisocitrate dehydrogenase
- Hs
Homo sapiens
- Sp
Schizosaccharomyces pombe
- Sc
Saccharomyces cerevisiae
- Tt
Thermus thermophilus
Footnotes
Competing financial interests
The authors are listed on a patent that is pending related to the mutated enzymes discussed in the text. This intellectual property is managed by Duke University.
Author contributions
Z.J.R. and B.D.C., performed experiments and wrote the manuscript; I.S., performed LC-MS/MS analysis; D.D.B. and J.H.S., provided reagents; H.Y., contributed reagents, funding, and critical feedback on the manuscript.
Additional information
Supplementary information is available online at http://www.nature.com/naturechemicalbiology/. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html. Correspondence and requests for materials should be addressed to H.Y.
References
- 1.Dang L, et al. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niu W, Draths KM, Frost JW. Biotechnol Prog. 2002;18:201–211. doi: 10.1021/bp010179x. [DOI] [PubMed] [Google Scholar]
- 3.Noack H, Georgiev V, Blomberg MR, Siegbahn PE, Johansson AJ. Inorg Chem. 2011;50:1194–1202. doi: 10.1021/ic101405u. [DOI] [PubMed] [Google Scholar]
- 4.Sato K, Aoki M, Noyori R. Science. 1998;281:1646–1647. doi: 10.1126/science.281.5383.1646. [DOI] [PubMed] [Google Scholar]
- 5.Burgard AP, Pharkya P, Osterhout RE. 2010/0330626. US patent. 2010
- 6.Parthasarathy A, Pierik AJ, Kahnt J, Zelder O, Buckel W. Biochemistry. 2011;50:3540–3550. doi: 10.1021/bi1020056. [DOI] [PubMed] [Google Scholar]
- 7.Uhr ML, Thompson VW, Cleland WW. J Biol Chem. 1974;249:2920–2927. [PubMed] [Google Scholar]
- 8.Aktas DF, Cook PF. Biochemistry. 2009;48:3565–3577. doi: 10.1021/bi8022976. [DOI] [PubMed] [Google Scholar]
- 9.Miyazaki J, Kobashi N, Nishiyama M, Yamane H. J Biol Chem. 2003;278:1864–1871. doi: 10.1074/jbc.M205133200. [DOI] [PubMed] [Google Scholar]
- 10.Lin Y, et al. Biochemistry. 2007;46:890–898. doi: 10.1021/bi062067q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin Y, et al. Biochemistry. 2008;47:4169–4180. doi: 10.1021/bi702361j. [DOI] [PubMed] [Google Scholar]
- 12.Lin Y, West AH, Cook PF. Biochemistry. 2009;48:7305–7312. doi: 10.1021/bi900175z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen R, Jeong SS. Protein Sci. 2000;9:2344–2353. doi: 10.1110/ps.9.12.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsons DW, et al. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mardis ER, et al. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan H, et al. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward PS, et al. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, et al. J Biol Chem. 2004;279:33946–33957. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- 19.Bulfer SL, Hendershot JM, Trievel RC. Proteins. 2012;80:661–666. doi: 10.1002/prot.23231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Struys EA, Jansen EE, Verhoeven NM, Jakobs C. Clin Chem. 2004;50:1391–1395. doi: 10.1373/clinchem.2004.033399. [DOI] [PubMed] [Google Scholar]
- 21.Jin G, et al. PLoS One. 2011;6:e16812. doi: 10.1371/journal.pone.0016812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kranendijk M, Struys EA, Salomons GS, Van der Knaap MS, Jakobs C. J Inherit Metab Dis. 2012;35:571–587. doi: 10.1007/s10545-012-9462-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pietrak B, et al. Biochemistry. 2011;50:4804–4812. doi: 10.1021/bi200499m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.