Abstract
Brucella is an important zoonotic pathogen for which no human vaccine exists. In an infected host, Brucella resides in macrophages but must coordinate expression of multiple virulence factors for successful cell entry and trafficking to acquire this replicative niche. Brucella responds to environmental signals to regulate virulence strategies that circumvent or blunt the host immune response. The Brucella quorum sensing system is a nexus of control for several Brucella virulence factors including flagellar genes and the type IV secretion system. Other sensory transduction systems, such as BvrRS and the newly described LOV-HK, sense environmental factors to control virulence. Here, we examine the contributions of various regulatory systems to Brucella virulence.
Brucella pathogenesis
Brucellosis is a zoonotic disease transmitted to humans by contact with the bodily fluids of infected animals, including non-pasteurized dairy products. Several species from the genus Brucella are etiological agents for human brucellosis, most commonly B. melitensis, which can cause severe chronic disease, and B. abortus and B. suis, which usually cause milder forms of the disease [1]. The natural hosts for these species are cattle and bison for B. abortus; sheep, goats and camels for B. melitensis, and pigs for B. suis.
Brucella has been called a stealthy pathogen [2,3] because it possesses no classical toxins and does not strongly activate innate immunity [2]. Brucella are taken up by phagocytes within the host and establish a replicative niche within macrophages and dendritic cells [4]. Internalization of Brucella occurs through lipid raft mediated macropinocytosis [5,6], after which the Brucella containing vacuole (BCV) moves by altered intracellular trafficking, transiently interacting with early and late endosomes and the lysosome but evading destruction by the lysosome and associating with the endoplasmic reticulum (ER) membranes to form a replicative vacuole [7]. Brucella multiply extensively within the host cell and prevent the death of the cell by inhibiting programmed cell death [8].
Although killed Brucella are taken up by macrophages, they fail to associate with the ER and are rapidly degraded [9], suggesting that de novo protein expression and/or secretion is required for bacterial trafficking during infection. Additionally, vaccination using killed Brucella fails to elicit a protective response [10]. Understanding Brucella gene regulation in response to environmental signals during host cell entry (Figure 1) can provide valuable clues towards production of an effective vaccine and reveal much about the infectious process. Several regulators of global gene expression have been identified in Brucella. Quorum sensing, stringent response, two-component regulatory systems and the recently discovered blue-light responsive LOV-HK protein are implicated in the adaptive response of Brucella to different environments. Here, we discuss different regulatory systems and their contribution to Brucella virulence (Box 1).
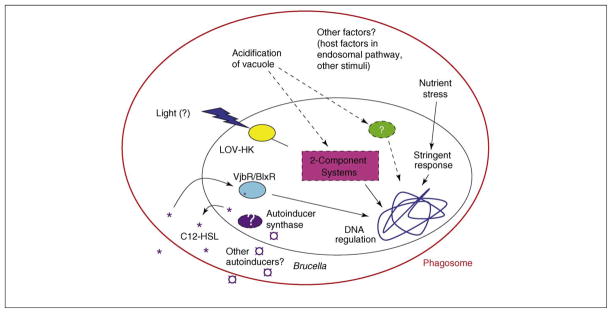
Figure 1.
Environmental signals sensed by Brucella during transition to an intracellular lifestyle. Brucella responds to external factors such as light, decrease in vacuolar pH, nutrient limitation and host factors through multiple regulatory systems. Dashed lines represent unconfirmed pathways, whereas dashed circles represent unidentified proteins.
Box 1. Global regulators involved in Brucella virulence.
Quorum sensing
VjbR
BlxR
Stringent response
Rsh
IHF
Two-component signaling
BvrR-BvrS
PdhS
LOV-HK
Quorum sensing in Brucella
Quorum sensing systems are used by many bacterial species to sense their environment and drive a population-wide transcriptional response. Thus, the use of this regulatory system by the intracellular pathogen Brucella presents a new adaptation of a conserved signaling system.
The fundamental components of a quorum sensing system are an autoinducer signal molecule, synthesized by the autoinducer synthase protein, and a regulatory protein which interacts with the autoinducer to activate transcription of genes in the quorum sensing network. The autoinducer is typically a homoserine lactone, although these molecules differ widely among species [11]. The autoinducer diffuses [11] or is transported through the cell wall [12], allowing the bacterial population to regulate genes coordinately. The regulatory protein contains an N-terminal autoinducer binding domain [13], and a C-terminal DNA binding and dimerization domain [14]. As the concentration of autoinducer increases, the regulatory protein binds to the autoinducer, allowing the regulatory protein to dimerize and bind to an inverted repeat ‘lux box’ sequence upstream of genes to activate their transcription [11,15]. Many bacteria contain more than one autoinducer synthase and quorum sensing regulator pair, which often share considerable overlap in the genes regulated and coordinate expression of these genes under different environmental conditions [11].
Target genes in the quorum sensing network
There is evidence that the quorum sensing network in Brucella includes many genes, as do quorum sensing networks in other bacteria [16–20]. Little is known about most of the genes in the quorum sensing network (Table 1), which were identified using microarray [19] and transcriptional reporters [16,19,20]. Many of the genes encode transcriptional regulators, but the target systems regulated by the encoded proteins have not yet been studied. Several of the regulated systems, including the type IV secretion system, flagella and cyclic 1,2-β-glucan synthetase, are known virulence factors, suggesting that further study of the genes regulated by quorum sensing could reveal additional Brucella virulence factors. Much of the research on quorum sensing in Brucella has focused on the type IV secretion system and flagella, which are discussed in greater detail later.
Table 1.
Brucella genes regulated by the quorum sensing network
| Type IV secretion system | Refs |
|---|---|
| Type IV secretion system | [16,19] |
| Effector proteins VceA and VceC | [20] |
| Flagella | |
| Flagellin (FliC) | [20] |
| FliF and FlgE | [16] |
| MotA, MotB, and MotD | [19] |
| FlhB and FlgJ | [19] |
| Transcriptional regulators | |
| OmpR(EnvZ) | [20] |
| Flagellar master regulator (FtcR) | [17] |
| Eight uncharacterized regulators | [19,20] |
| Cell envelope biogenesis | |
| Oxoacyl acyl carrier protein reductase | [19] |
| 1-acyl-SN-glycerol-3-phosphate acyltransferase | [20] |
| Two glycosyltransferases | [19] |
| Bacteroid development protein BacA | [19] |
| Cyclic β 1,2-glucan synthetase (Cgs) | [20] |
| Glucosamine fuctose 6 phosphate aminotransferase | [19] |
| UDP glucose 4 epimerase | [19] |
| GDP mannose 4,6-dehydratase | [19] |
| Signal transduction | |
| Sensory transduction histidine kinase | [19] |
| Two-component system sensor | [19] |
| Other | |
| Biotin synthesis protein BioC | [20] |
| Carbonic anhydrase | [20] |
| Soluble lytic murein transglycosylase | [19] |
| Glutathione S-transferase III | [20] |
| Efflux protein, LysE family, putative | [19] |
| Coproporphyrinogen III oxidase iron | [19] |
| Putative exported protein BopA | [19,20] |
| Osmotically inducible protein C | [19] |
| Proline dehydrogenase | [19] |
| Five hypothetical proteins | [19,20] |
VirB type IV secretion system
The Brucella VirB type IV secretion system [21] is of crucial importance during macrophage infection, being required both during internalization by macropinocytosis [6] and for fusion with the ER to establish the replicative niche [4]. Brucella virB mutants also have an attenuated phenotype in mouse models of infection [22]. Based on these results, it is hypothesized that effectors of the VirB type IV secretion system are required for proper trafficking within the cell, interaction with the ER and survival within a host.
Effector molecules secreted by the VirB system have been elusive. Although reporter fusions to N-terminal portions of several Brucella proteins were secreted in an apparently VirB-dependent manner [23,24], it was subsequently shown that the addition of a Sec secretory tag to the YopP reporter was sufficient to promote VirB-dependent secretion [24]. Therefore, proteins secreted into the periplasm by the Sec-pathway might be non-specifically secreted via VirB. It remains to be determined if this function is required for the fitness of the bacterium or if it is simply an artifact within the experimental systems tested.
Two putative type IV effector proteins, VceA and VceC, were recently identified [20]. These proteins are secreted in a Sec-independent, VirB-dependent manner into the cytosol of infected macrophages. The C-terminus of these proteins in B. suis contains a C-terminal secretion motif seen in proteins secreted by the A. tumefaciens VirB system [25], although this signal is not present in the VceC protein of all Brucella species due to a frameshift [20].
The search for effectors of the VirB system is far from over. The data presented earlier suggest a low specificity of effector recognition by the type IV secretion system in Brucella and the possibility that small periplasmic proteins might leak from the cell via the secretory apparatus. However, the more recently identified proteins contain type IV specific C-terminal secretion signals and are secreted past the membrane of the BCV into the cytosol of the host cell, suggesting that these proteins are type IV secreted effectors.
Flagella
In many bacteria, quorum sensing regulates transition between motility and biofilm formation. Brucella is an apparently non-motile organism; however, it has recently been shown to produce a flagellum [26,27]. Flagellar mutants infect macrophages at wild-type levels but show reduced persistence in a mouse model of infection [26], indicating that the flagellum has an important role in pathogenesis but is not involved in entry into host cells. Whether the flagellum has been adapted to a purpose other than motility, or whether the bacteria are only motile in response to a specific stimulus is currently undetermined. However, the filament protein (FliC) is required for full virulence in the mouse model, which would not be expected if the only role of the flagellum was as a secretory apparatus.
The autoinducer
The first component of quorum sensing identified in Brucella was an autoinducer molecule [28]. High performance liquid chromatography (HPLC) of concentrated B. melitensis culture supernatant revealed two fractions with activity in the LasR bioassay system. One fraction contained N-dodecanoyl-DL-homoserine-lactone (C12-HSL) [28], a molecule very similar to the cognate autoinducer of the Pseudomonas aeruginosa LasR quorum sensing system [29]. No molecule was identified for the other fraction; however, it is possible that multiple autoinducers are produced by Brucella. Interestingly, homology searches have failed to identify putative autoinducer synthases in the Brucella genome, so the pathway for synthesis of C12-HSL remains unidentified. Addition of C12-HSL to B. melitensis cultures reduces transcription of the virB operon [28], suggesting that the signal molecule has a negative regulatory role in expression of this important virulence factor.
B. melitensis seems to produce a very low concentration of C12-HSL in vitro compared to the production of autoinducers by other bacteria. One possible explanation for the low recovery of autoinducer is low solubility due to the long acyl chain. However, another intriguing possibility is suggested by a recent report that C12-HSL activates macrophage tumor necrosis factor-α (TNF-α) production [30]. A more sensitive quorum sensing system that allows for lower production of autoinducer might be part of the ‘stealthy’ strategy of the bacterium.
Regulatory proteins
Two putative quorum sensing regulatory proteins have been identified in Brucella: VjbR and BlxR. Both proteins have the classic structure of quorum sensing regulators, with an N-terminal homoserine lactone binding domain and a C-terminal DNA binding domain.
The VjbR regulator is required for expression of the virB and flagellar genes [16,17,19]. Strains containing a vjbR deletion are defective for growth in macrophages and display attenuated virulence in mice [16,19]. However, a vjbR mutant complemented with a vjbR allele containing a deletion in the autoinducer binding domain resulted in expression of virB that was not suppressed by addition of exogenous C12-HSL [18]. This suggests that C12-HSL binds to VjbR to mediate suppression of virB expression. VjbR also regulates expression of the hybrid kinase-response regulator FtcR, and FtcR in turn regulates expression of flagellar components [17]. However, overexpression of FtcR in a vjbR mutant only partially restored expression of the flagellar structural components, suggesting that quorum sensing regulation of flagellar expression could occur through multiple routes [17].
The recently identified BlxR regulatory protein is also required for maximal expression of the virB operon and several flagellar genes, suggesting that there is overlap in the regulatory networks controlled by these two proteins [19]. BlxR does not seem to respond to C12-HSL [18], supporting the idea that multiple signals participate in Brucella quorum sensing. The blxR deletion mutant exhibited a growth defect in macrophages that was similar to the vjbR deletion mutant [19]. However, deletion of blxR did not fully attenuate virulence in a mouse model of infection, unlike the vjbR deletion. Additionally, dissemination of the blxR mutant strain in the mouse model was similar to wild-type B. melitensis, whereas the vjbR mutant was defective for in vivo dissemination [19].
Currently, differences in the regulatory pathways of these proteins have not been elucidated, although the difference in virulence of the mutants indicates that the function of the two proteins is not redundant. Deletion of either quorum sensing regulatory protein produced a similar decrease in expression from transcriptional fusions to genes in the quorum sensing network [19], suggesting that the regulatory networks of the two proteins are convergent. Further evidence for interplay between the two regulatory proteins is the reduction in transcription from both blxR and vjbR promoter fusions in strains lacking expression of one or the other regulatory protein [19]. This suggests that the two regulatory proteins influence expression of each other via positive feedback. However, the VjbR protein did not activate transcription from the blxR promoter in an in vitro assay in Escherichia coli [20], so the observed regulatory effect might be indirect.
With the recent publication of the ‘lux box’-like VjbR recognition sequence comes the first demonstration of direct binding of target promoters by one of the Brucella quorum sensing regulators [20]. The VjbR protein bound to a sequence close to the translational start site in the virB promoter and in the promoters of the newly discovered type IV effector genes vceA and vceC. The response of transcriptional fusions in an E. coli reporter system indicated that numerous genes in the quorum sensing network are directly regulated by VjbR binding.
Role of quorum sensing in pathogenesis
Although both regulatory proteins are required for normal expression of the quorum sensing network in broth [19], expression of virB is suppressed by exogenous autoinducer via the VjbR regulator [18]. This suggests that VjbR acts as a transcriptional activator in the autoinducer-unbound state and that interaction of VjbR with the autoinducer suppresses transcription by changing the conformation of VjbR such that it no longer acts as a transcriptional activator or no longer binds to the promoter. This unusual response to autoinducer accumulation has implications for the role of quorum sensing during infection. Brucella enters the phagocytic cell and traffics to the ER individually, so how does an individual bacterium within a vacuole reach a quorum? The accumulation of signal molecule in the vicinity of the cell is affected by a combination of factors including the local bacterial population density, spatial cell distribution and rate of diffusion of the molecule into the surrounding environment [31]. It is possible that secreted C12-HSL accumulates within the Brucella containing vacuole. However, the highly similar P. aeruginosa LasR autoinducer can rapidly enter mammalian cells [32], suggesting that C12-HSL produced within the vacuole can diffuse out into the cell cytosol. In this case, Brucella would not be exposed to increased concentrations of signal molecule after uptake into the vacuole. The induction of virB observed after entry into the host cell supports this idea. After the vacuole has associated with the ER, Brucella begins replication. Does the population density within the vacuole become high enough to overcome diffusion of the signal molecule, triggering suppression of the target virulence genes in the late stages of cell infection? Immunological response to the type IV secretion system has been documented [33]. Once the intracellular infection has been established, suppression of the virulence factors used for cell entry has the potential to reduce recognition of Brucella by the immune system during chronic intracellular infection. The positive regulatory function of the quorum sensing system is required for full virulence in macrophages [16,19]; however, the role of the suppressor function requires further investigation.
Stringent response to nutrient limitation
A major hurdle in the infection of a host cell by Brucella is the lack of nutrients within the phagosome. Immediately after phagocytosis, Brucella halts synthesis of periplasmic transporters and alters TCA cycle activity as the available extracellular nutrients decrease [34]. A large subset of Brucella genes involved in synthesis of amino acids and nucleotides and metabolism of sugars and nitrogen are dispensable during culture but are required for macrophage infection, highlighting the changing metabolic states that are required for intracellular growth [35,36].
Considering the role that nutrient starvation has in the intracellular replication of Brucella, it is not difficult to consider that same cue having additional roles. The first environment that the bacterium encounters upon phagocytosis is an acidic vacuole deprived of nutrients. Induction of virB expression occurs in response to the acidification of the vacuole and nutrient starvation conditions encountered within the host cell [22,37,38]. Induction under nutrient starved conditions requires the stringent response regulator Rsh and integration host factor (IHF) [39,40]. It has been shown in E. coli that the stringent response induces transcription of IHF [41]. Therefore, the connection can be proposed that nutrient starvation in Brucella induces the stringent response via Rsh, increasing the levels of IHF, which binds to the Brucella virB promoter and induces transcription at neutral pH [39]. Interestingly, deletion of the IHF binding domains in the virB promoter does not alter the response of the promoter to low pH, indicating that other activating factors are involved. The known factors affecting virB promoter transcription are diagramed in Figure 2.
Figure 2.
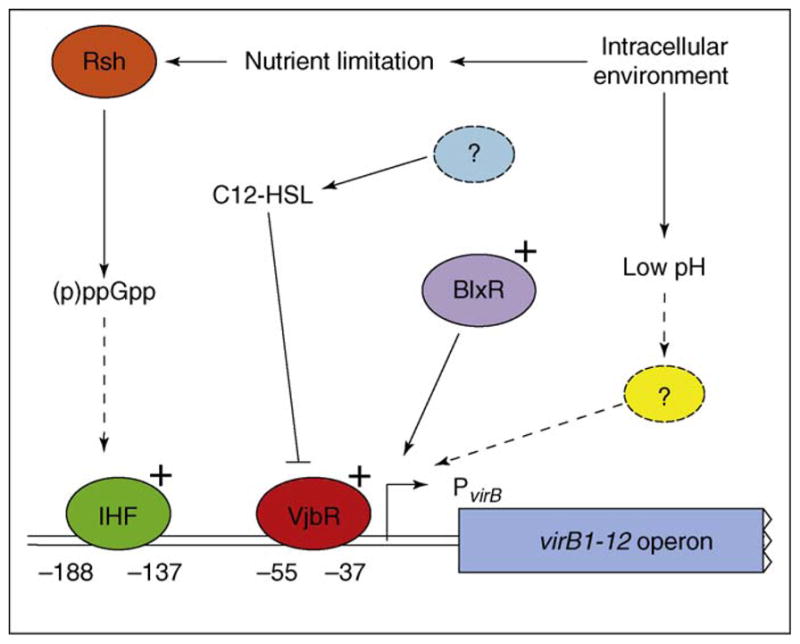
Regulation of the virB operon. Several regulatory systems affect transcription of the virB operon. Positive transcriptional regulators are marked with a ‘+.’ Dashed lines and circles indicate proposed functions that have not been identified to date. Abbreviations: (p)ppGpp, guanosine pentaphosphate.
Two-component or phosphorelay regulatory systems
Infection is a multi-step process, requiring responses to a complex mixture of environmental cues and antimicrobial defenses. While quorum sensing systems allow a bacterial population to produce a coordinated response to signal molecule accumulation, two-component regulatory systems allow individual bacteria to adjust gene expression in response to environmental stimuli.
Two-component regulation of Brucella virulence factors
Two-component systems are typically composed of a sensor protein that transduces a signal to a response regulator protein. In bacteria, histidine kinases are often responsible for this signal transduction, and are found in two-component regulatory systems involved in biological pathways from osmoregulation to antibiotic production [42]. These histidine kinases phosphorylate aspartyl residues in the REC domains of response regulator proteins [43]. Response regulator proteins are a diverse group with output domains ranging from DNA or protein binding to various enzymatic processes [43]. Predictions indicate more than 20 two-component systems in the Brucella genome, but few have been confirmed or described in detail [43,44].
The BvrR-BvrS two-component regulatory system is required for Brucella pathogenesis and is highly homologous to the ChvI-ChvG regulatory system in Agrobacterium which is important for interaction with the host plant [21]. Transposon mutants of either bvrR or bvrS in Brucella result in pronounced defects in virulence in mouse models, and defects in both entry and trafficking in cellular models of infection [21]. Once inside cells, bvr mutants are unable to prevent phagosome-lysosome fusion and are unable to persist in macrophages [21]. Disruption of the Bvr system leads to changes in the composition of the outer membrane [21,45], resulting in an increased sensitivity to polycations and surfactants [21], possibly due to reduced acylation of lipid A moieties [46]. Despite the importance of this regulatory system in Brucella virulence, little is known about the genes regulated or the environmental signals sensed by the system. BvrR-BvrS has been shown to regulate expression of outer membrane proteins Omp3a and Omp3b [21,45,47]; however, data indicates that these proteins are not essential for Brucella virulence [48,49].
Another two-component system involving the PdhS histidine kinase from B. abortus is involved in the control of cellular division [44]. This system shows homology to the PleC-DivJ system of Caulobacter crescentus [44]. PleC is involved in the formation of asymmetric daughter cells during division and affects the expression of over 100 genes in C. crescentus [50]. Evidence of asymmetric division of Brucella has been observed [50], suggesting that regulation via PdhS could result in the formation of daughter cells that differentially express target genes. Other Brucella two-component systems with effects on virulence could be involved in regulation of nutrient uptake and nitrogen metabolism [51,52]. Recently, a Brucella blue light-sensing histidine kinase has been identified and linked to virulence in a macrophage model [53].
Blue light signaling and virulence in Brucella
Light, which has long been recognized as a signal in plants, has more recently been demonstrated to have a role in the signaling pathways of many bacteria [54,55]. In C. crescentus, a light sensing histidine kinase (LovK) is involved in bacterial attachment, and in Bacillis subtilis a light sensing protein (YtvA) is involved in stress response activation [56,57]. Another light sensing histidine kinase (PST-LOV) that is fused to a response regulator is found in the plant pathogen Pseudomonas syringae [58]. These proteins sense blue light through the utilization of a light, oxygen and voltage (LOV) domain [59]. LOV domains bind the cofactor flavin mononucleotide (FMN), a derivative of riboflavin. Within the LOV domain is an essential cysteine, and upon exposure to blue light a covalent interaction forms between this cysteine and a loosely held flavin mononucleotide [59]. This usually short-lived adduct formation is a signal that affects the structure of the LOV domain. Whole genome BLAST searches of all sequenced bacteria indicate probable LOV domain proteins in 24 separate bacterial species, and the number increases as more bacterial genomes are sequenced [55]. The presence of proteins containing these sensor domains in seemingly unlikely species such as the intracellular pathogen Brucella has piqued interest. Brucella spp. encode a protein (LOV-HK) with an LOV domain at its N-terminus and a histidine kinase domain at its C-terminus. Light increases the enzymatic activity of LOV-HK, and surprisingly, wild-type B. abortus is attenuated in macrophage and mouse models when cultured in the dark in comparison with Brucella that has been exposed to visible light [53].
Unlike similar LOV-HK proteins in other bacteria, B. melitensis LOV-HK is slow to return to a pre-stimulated state when removed from light in an in vitro assay [53]. This suggests that activation of the B. melitensis LOV-HK protein by light could produce a longer acting downstream signal. Disruption of the B. abortus LOV-HK gene abolished the light-modulated virulence and reduced bacterial proliferation in light-exposed macrophages to a level similar to that observed during infections conducted in the dark [53]. Moreover, mutagenesis of the conserved cysteine in the LOV domain abolished the light response and demonstrated that cysteinyl-flavin bond formation in the LOV domain is necessary for light-mediated virulence [53]. These results provide a clear link between LOV-HK and a light-induced increase in virulence. Light as a signal for virulence invites speculation. Could sensing ambient light through LOV-HK signal Brucella to regulate genes necessary for survival in the environment outside the host? Does the light signal Brucella perceives through LOV-HK originate from an internal host redox source? The answers to these and other questions will reveal the nature and purpose of LOV-HK signaling in Brucella.
Conclusion
Bacterial virulence factors must be tightly regulated during the infectious process, both to optimize the functioning of the virulence factors in promoting survival and thwarting host defenses, and to minimize the chance that the host will produce an effective immune response against these proteins. Brucella establishes an intracellular niche and persists chronically within the host, thus, control of virulence factor expression is particularly important in Brucella pathogenesis. Brucella regulates the expression of multiple virulence factors in response to signal molecule accumulation via the quorum sensing system, and responds to starvation conditions by upregulating the type IV secretion system via the stringent response. Multiple two-component systems contribute to gene regulation, including alteration of outer membrane composition mediated by BvrR-BvrS, asymmetric cell division potentially mediated by PdhS, and increased virulence in response to visible light mediated by LOV-HK. Examination of the kinetics of the regulatory networks controlling virulence factor expression and the environmental signals sensed by these systems will offer insight into the adaptive mechanisms of Brucella pathogenicity. Additionally, these regulatory networks might reveal further pathogenic mechanisms used by an organism in which few virulence factors have been characterized to date.
Acknowledgments
Work in the authors’ lab is supported by National Institutes of Health/NIAID GLRCE for Biodefense and Emerging Infectious Disease Research Program Grant 1U54-AI-057153, National Institutes of Health Grants 1R01AI073558 and R43AI064959-02, and Binational Agricultural Research and Development Grant US-3829-06 R.
References
- 1.Pappas G, et al. Brucellosis. N Engl J Med. 2005;352:2325–2336. doi: 10.1056/NEJMra050570. [DOI] [PubMed] [Google Scholar]
- 2.Barquero-Calvo E, et al. Brucella abortus uses a stealthy strategy to avoid activation of the innate immune system during the onset of infection. PLoS One. 2007;2:e631. doi: 10.1371/journal.pone.0000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohler S, et al. What is the nature of the replicative niche of a stealthy bug named Brucella? Trends Microbiol. 2003;11:215–219. doi: 10.1016/s0966-842x(03)00078-7. [DOI] [PubMed] [Google Scholar]
- 4.Celli J, et al. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J Exp Med. 2003;198:545–556. doi: 10.1084/jem.20030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S, et al. Membrane sorting during swimming internalization of Brucella is required for phagosome trafficking decisions. Microb Pathog. 2002;33:225–237. doi: 10.1006/mpat.2002.0531. [DOI] [PubMed] [Google Scholar]
- 6.Watarai M, et al. Modulation of Brucella-induced macropinocytosis by lipid rafts mediates intracellular replication. Cell Microbiol. 2002;4:341–355. doi: 10.1046/j.1462-5822.2002.00195.x. [DOI] [PubMed] [Google Scholar]
- 7.Rajashekara G, et al. Brucella: functional genomics and host-pathogen interactions. Anim Health Res Rev. 2006;7:1–11. doi: 10.1017/S146625230700117X. [DOI] [PubMed] [Google Scholar]
- 8.He Y, et al. Brucella melitensis triggers time-dependent modulation of apoptosis and down-regulation of mitochondrion-associated gene expression in mouse macrophages. Infect Immun. 2006;74:5035–5046. doi: 10.1128/IAI.01998-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Starr T, et al. Brucella intracellular replication requires trafficking through the late endosomal/lysosomal compartment. Traffic. 2008;9:678–694. doi: 10.1111/j.1600-0854.2008.00718.x. [DOI] [PubMed] [Google Scholar]
- 10.Harms JS, et al. Evaluation of recombinant invasive, non-pathogenic Eschericia coli as a vaccine vector against the intracellular pathogen. Brucella J Immune Based Ther Vaccines. 2009;7:1. doi: 10.1186/1476-8518-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn AK, Stabb EV. Beyond quorum sensing: the complexities of prokaryotic parliamentary procedures. Anal Bioanal Chem. 2007;387:391–398. doi: 10.1007/s00216-006-0730-9. [DOI] [PubMed] [Google Scholar]
- 12.Pearson JP, et al. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol. 1999;181:1203–1210. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanzelka BL, Greenberg EP. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J Bacteriol. 1995;177:815–817. doi: 10.1128/jb.177.3.815-817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi SH, Greenberg EP. Genetic dissection of DNA binding and luminescence gene activation by the Vibrio fischeri LuxR protein. J Bacteriol. 1992;174:4064–4069. doi: 10.1128/jb.174.12.4064-4069.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antunes LC, et al. A mutational analysis defines Vibrio fischeri LuxR binding sites. J Bacteriol. 2008;190:4392–4397. doi: 10.1128/JB.01443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delrue RM, et al. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell Microbiol. 2005;7:1151–1161. doi: 10.1111/j.1462-5822.2005.00543.x. [DOI] [PubMed] [Google Scholar]
- 17.Leonard S, et al. FtcR is a new master regulator of the flagellar system of Brucella melitensis 16 M with homologs in Rhizobiaceae. J Bacteriol. 2007;189:131–141. doi: 10.1128/JB.00712-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uzureau S, et al. Mutations of the quorum sensing-dependent regulator VjbR lead to drastic surface modifications in Brucella melitensis. J Bacteriol. 2007;189:6035–6047. doi: 10.1128/JB.00265-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rambow-Larsen AA, et al. Putative quorum-sensing regulator BlxR of Brucella melitensis regulates virulence factors including the type IV secretion system and flagella. J Bacteriol. 2008;190:3274–3282. doi: 10.1128/JB.01915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Jong MF, et al. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol Microbiol. 2008;70:1378–1396. doi: 10.1111/j.1365-2958.2008.06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seleem MN, et al. Brucella: A pathogen without classic virulence genes. Vet Microbiol. 2007;129:1–14. doi: 10.1016/j.vetmic.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 22.Sieira R, et al. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J Bacteriol. 2000;182:4849–4855. doi: 10.1128/jb.182.17.4849-4855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchesini MI, et al. N-terminal-capturing screening system for the isolation of Brucella abortus genes encoding surface exposed and secreted proteins. Microb Pathog. 2004;37:95–105. doi: 10.1016/j.micpath.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Lavigne JP, et al. Identification of a new virulence factor, BvfA, in Brucella suis. Infect Immun. 2005;73:5524–5529. doi: 10.1128/IAI.73.9.5524-5529.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vergunst AC, et al. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci U S A. 2005;102:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fretin D, et al. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell Microbiol. 2005;7:687–698. doi: 10.1111/j.1462-5822.2005.00502.x. [DOI] [PubMed] [Google Scholar]
- 27.Abdallah AI, et al. Type III secretion homologs are present in Brucella melitensis, B. ovis, and B. suis biovars 1, 2, and 3. Curr Microbiol. 2003;46:241–245. doi: 10.1007/s00284-002-3789-3. [DOI] [PubMed] [Google Scholar]
- 28.Taminiau B, et al. Identification of a quorum-sensing signal molecule in the facultative intracellular pathogen Brucella melitensis. Infect Immun. 2002;70:3004–3011. doi: 10.1128/IAI.70.6.3004-3011.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson JP, et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci U S A. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomi K, et al. Mouse and human cell activation by N-dodecanoyl-DL-homoserine lactone, a Chromobacterium violaceum autoinducer. Infect Immun. 2006;74:7029–7031. doi: 10.1128/IAI.00038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hense BA, et al. Does efficiency sensing unify diffusion and quorum sensing? Nat Rev Microbiol. 2007;5:230–239. doi: 10.1038/nrmicro1600. [DOI] [PubMed] [Google Scholar]
- 32.Ritchie AJ, et al. The immunomodulatory Pseudomonas aeruginosa signalling molecule N-(3-oxododecanoyl)-L-homoserine lactone enters mammalian cells in an unregulated fashion. Immunol Cell Biol. 2007;85:596–602. doi: 10.1038/sj.icb.7100090. [DOI] [PubMed] [Google Scholar]
- 33.Rolan HG, et al. VirB12 is a serological marker of Brucella infection in experimental and natural hosts. Clin Vaccine Immunol. 2008;15:208–214. doi: 10.1128/CVI.00374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamontagne J, et al. Intracellular adaptation of Brucella abortus. J Proteome Res. 2009;8:1594–1609. doi: 10.1021/pr800978p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ficht TA. Intracellular survival of Brucella: defining the link with persistence. Vet Microbiol. 2003;92:213–223. doi: 10.1016/s0378-1135(02)00367-x. [DOI] [PubMed] [Google Scholar]
- 36.Kohler S, et al. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc Natl Acad Sci U S A. 2002;99:15711–15716. doi: 10.1073/pnas.232454299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boschiroli ML, et al. The Brucella suis virB operon is induced intracellularly in macrophages. Proc Natl Acad Sci U S A. 2002;99:1544–1549. doi: 10.1073/pnas.032514299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouot B, et al. Production of the type IV secretion system differs among Brucella species as revealed with VirB5- and VirB8-specific antisera. Infect Immun. 2003;71:1075–1082. doi: 10.1128/IAI.71.3.1075-1082.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sieira R, et al. Integration host factor is involved in transcriptional regulation of the Brucella abortus virB operon. Mol Microbiol. 2004;54:808–822. doi: 10.1111/j.1365-2958.2004.04316.x. [DOI] [PubMed] [Google Scholar]
- 40.Dozot M, et al. The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB. Cell Microbiol. 2006;8:1791–1802. doi: 10.1111/j.1462-5822.2006.00749.x. [DOI] [PubMed] [Google Scholar]
- 41.Ali Azam T, et al. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karniol B, Vierstra RD. The HWE histidine kinases, a new family of bacterial two-component sensor kinases with potentially diverse roles in environmental signaling. J Bacteriol. 2004;186:445–453. doi: 10.1128/JB.186.2.445-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hallez R, et al. The asymmetric distribution of the essential histidine kinase PdhS indicates a differentiation event in Brucella abortus. EMBO J. 2007;26:1444–1455. doi: 10.1038/sj.emboj.7601577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamontagne J, et al. Extensive cell envelope modulation is associated with virulence in Brucella abortus. J Proteome Res. 2007;6:1519–1529. doi: 10.1021/pr060636a. [DOI] [PubMed] [Google Scholar]
- 46.Manterola L, et al. The lipopolysaccharide of Brucella abortus BvrS/BvrR mutants contains lipid A modifications and has higher affinity for bactericidal cationic peptides. J Bacteriol. 2005;187:5631–5639. doi: 10.1128/JB.187.16.5631-5639.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez-Goni I, et al. Regulation of Brucella virulence by the two-component system BvrR/BvrS. Vet Microbiol. 2002;90:329–339. doi: 10.1016/s0378-1135(02)00218-3. [DOI] [PubMed] [Google Scholar]
- 48.Edmonds MD, et al. Attenuation of a Brucella abortus mutant lacking a major 25 kDa outer membrane protein in cattle. Am J Vet Res. 2001;62:1461–1466. doi: 10.2460/ajvr.2001.62.1461. [DOI] [PubMed] [Google Scholar]
- 49.Manterola L, et al. BvrR/BvrS-controlled outer membrane proteins Omp3a and Omp3b are not essential for Brucella abortus Virulence. Infect Immun. 2007;75:4867–4874. doi: 10.1128/IAI.00439-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hallez R, et al. Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol. 2004;12:361–365. doi: 10.1016/j.tim.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 51.Dorrell N, et al. Identification, cloning and initial characterisation of FeuPQ in Brucella suis: a new sub-family of two-component regulatory systems. FEMS Microbiol Lett. 1998;162:143–150. doi: 10.1111/j.1574-6968.1998.tb12991.x. [DOI] [PubMed] [Google Scholar]
- 52.Dorrell N, et al. Investigation into the role of the response regulator NtrC in the metabolism and virulence of Brucella suis. Microb Pathog. 1999;27:1–11. doi: 10.1006/mpat.1999.0278. [DOI] [PubMed] [Google Scholar]
- 53.Swartz TE, et al. Blue-light-activated histidine kinases: two-component sensors in bacteria. Science. 2007;317:1090–1093. doi: 10.1126/science.1144306. [DOI] [PubMed] [Google Scholar]
- 54.Briggs WR. The LOV domain: a chromophore module servicing multiple photoreceptors. J Biomed Sci. 2007;14:499–504. doi: 10.1007/s11373-007-9162-6. [DOI] [PubMed] [Google Scholar]
- 55.Losi A. The bacterial counterparts of plant phototropins. Photochem Photobiol Sci. 2004;3:566–574. doi: 10.1039/b400728j. [DOI] [PubMed] [Google Scholar]
- 56.Purcell EB, et al. A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci U S A. 2007;104:18241–18246. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Avila-Perez M, et al. Blue light activates the σB-dependent stress response of Bacillus subtilis via YtvA. J Bacteriol. 2006;188:6411–6414. doi: 10.1128/JB.00716-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao Z, et al. A blue light inducible two-component signal transduction system in the plant pathogen Pseudomonas syringae pv. tomato. Biophys J. 2008;94:897–905. doi: 10.1529/biophysj.107.108977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Losi A. Flavin-based blue-light photosensors: a photobiophysics update. Photochem Photobiol. 2007;83:1283–1300. doi: 10.1111/j.1751-1097.2007.00196.x. [DOI] [PubMed] [Google Scholar]