Abstract
Varroa mites (V. destructor) are a major threat to honey bees (Apis melilfera) and beekeeping worldwide and likely lead to colony decline if colonies are not treated. Most treatments involve chemical control of the mites; however, Varroa has evolved resistance to many of these miticides, leaving beekeepers with a limited number of alternatives. A non-chemical control method is highly desirable for numerous reasons including lack of chemical residues and decreased likelihood of resistance. Varroa sensitive hygiene behavior is one of two behaviors identified that are most important for controlling the growth of Varroa populations in bee hives. To identify genes influencing this trait, a study was conducted to map quantitative trait loci (QTL). Individual workers of a backcross family were observed and evaluated for their VSH behavior in a mite-infested observation hive. Bees that uncapped or removed pupae were identified. The genotypes for 1,340 informative single nucleotide polymorphisms were used to construct a high-resolution genetic map and interval mapping was used to analyze the association of the genotypes with the performance of Varroa sensitive hygiene. We identified one major QTL on chromosome 9 (LOD score = 3.21) and a suggestive QTL on chromosome 1 (LOD = 1.95). The QTL confidence interval on chromosome 9 contains the gene ‘no receptor potential A’ and a dopamine receptor. ‘No receptor potential A’ is involved in vision and olfaction in Drosophila, and dopamine signaling has been previously shown to be required for aversive olfactory learning in honey bees, which is probably necessary for identifying mites within brood cells. Further studies on these candidate genes may allow for breeding bees with this trait using marker-assisted selection.
Introduction
Pollination by honey bees (Apis mellifera) is an important part of modern agriculture, and honey bee health has been receiving increased attention recently from the public, beekeepers, and researchers. Honey bees face numerous challenges, including pesticides, pathogens, and parasites (such as Varroa mites, V. destructor) [1]–[3]. Varroa parasitism of honey bees is widely considered to be the greatest threat to beekeeping and has led to substantial colony losses worldwide [4]–[10]. These obligate ectoparasites live in the nest of honey bees and harm individuals and colonies.
The mites require developing honey bees for their reproduction. Mated adult female mites enter brood cells and start laying eggs, one male and up to five female in worker brood cells but an average of 1.3–1.45 new, mature female offspring are produced [11], [12]. The offspring feed on the hemolymph of the developing bee pupa and sibling mites mate with one another. When the adult bee emerges, mature female mites leave the worker cell and enter a phoretic stage while feeding on the hemolymph of adult bees [13]. The cycle is repeated when the female mite enters a new brood cell. When Varroa feed on hemolymph, the bees experience physical and physiological damage, protein levels decrease, and development can be abnormal [14], [15]. One of the worst impacts of Varroa comes from its association with honey bee viruses – mites can vector many honey bee viruses and some viruses can replicate within the mite [16]–[20]. Untreated Varroa-infested colonies usually die after one to four years of mite infestation; however, there have been reports of untreated hives with mites surviving for up to six years [3], [21]–[26].
Although Varroa have been effectively controlled with several miticides, pesticide-resistant populations of mites have appeared [27]–[39]. Miticides have significant drawbacks because they are soluble in the wax combs of the hive and can leave chemical residues in honey and wax, and synergism between chemicals can have negative effects on bee health [2], [34], [40]–[45]. A more sustainable form of control is desirable and the beekeeping industry has already started to benefit from the recent development of stocks that show resistance to mites [46].
A few behavioral traits of bees have been shown to reduce Varroa populations. One important trait is Varroa sensitive hygiene (VSH). Broadly, hygiene in honey bees refers to the act of adult bees removing dead, diseased or parasitized brood from sealed cells [47], [48]. Hygiene has been improved by breeding for bees that effectively remove free-killed brood (FKB). High hygiene bees also remove more Varroa than less hygienic bees [49], [50]. VSH is a form of hygiene in which bees have heightened response to Varroa; greater frequencies of mites are removed by VSH bees than by FKB hygienic bees [51](Danka et al. unpub. obs.). Enhanced mite removal enables VSH bees to effectively slow growth of the Varroa population in a colony [51]–[54]. VSH has a significant heritable component as evidenced by the response of the trait to selection in a USDA breeding program [52], [53], [55]–[57]. Field studies have shown that bees with the VSH trait successfully reduce mite infestations while retaining performance in traits important to beekeepers [58]–[60].
When infested brood is exposed to bees that exhibit high levels of VSH for one week, the mite reproduction decreases [52]. Immature mites may be killed due to uncapping and removal behavior [61]. When an infested pupa is removed from the colony, the adult female mite (and offspring) may be removed along with the pupa. If the adult female mites survive the removal of the host pupae, they usually attach to the bee that is removing the brood [62] but can also roam freely on the comb, where they are exposed to grooming behavior and can be detected and damaged via biting by the bees [49], [63]. It has also been suggested that mites which are removed with pre-pupae and pupae are not likely to produce viable offspring if they invade new brood cells too soon after such events [64]. Thus, the effectiveness of VSH on reducing mite reproduction is due partly to interference with reproduction, and in part to the risks the mite faces once it is out of the safety of the brood cell [65].
Here, we investigate the genetic architecture of VSH. A companion paper takes a similar approach to study mite-grooming behavior, the other behavior that affects mite population growth [66], [67]. The objective of the current study was to use quantitative trait loci (QTL) mapping on a genome-wide scale to look for segregating chromosomal regions for the VSH trait. Current selection relies on colony-level measurements of VSH; for example, observing a reduction in the level of mite infestation in brood or measuring reproductive success of individual mites in brood cells [55], [68]. Identifying the genes involved would assist in the understanding of the genetics and neurobiology of behaviors that confer mite resistance, as well as provide more efficient tools for selective breeding. Here we report progress towards that goal using a high-resolution genetic map integrated with the genomic sequence of the honey bee.
Results
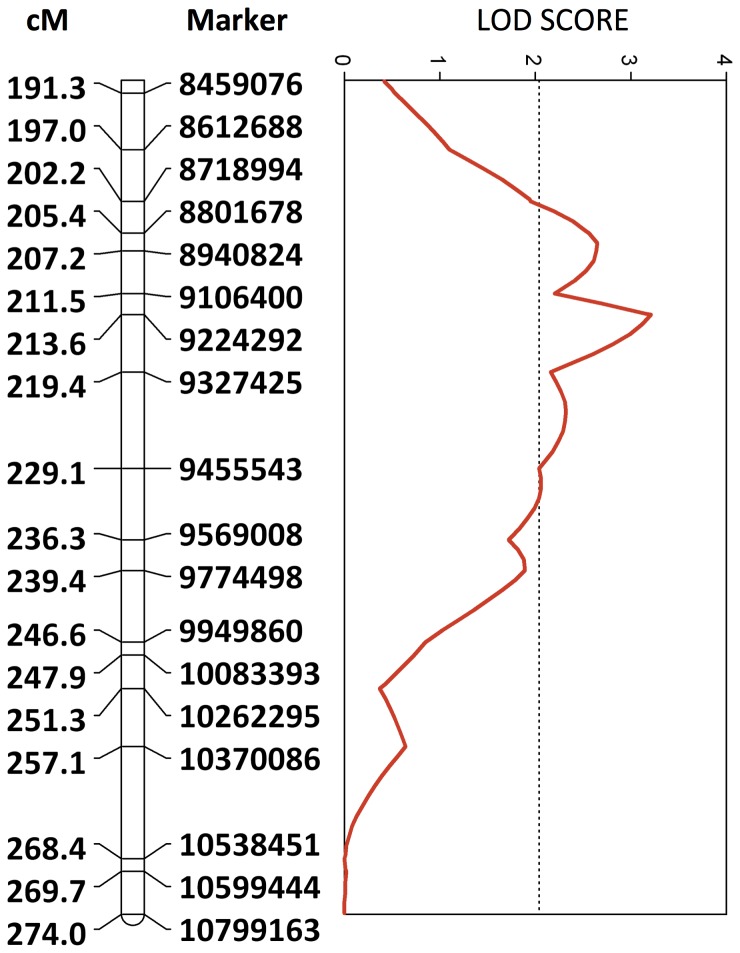
The genotypes for 1,340 informative SNPs were used to construct a high-resolution genetic map and to compare genotypes of individuals that performed VSH behavior (uncapping cells or removing infested pupae, n = 127) to those that did not (n = 111). The use of the Illumina GoldenGate assay provides high call rates and accuracy in calling SNP genotypes [69]. The high average recombination rate across the genome (whole genome: 22.6 cM/Mb, chromosome 9: 35.23 cM/Mb, chromosome 1: 26.144 cM/Mb) was similar to previous estimates [70]–[72]. Interval mapping analysis identified a LOD peak of 3.21 on chromosome 9 (Fig. 1). Permutation tests indicated that this QTL is not significant with the genome-wide threshold for p<0.05, however, it is does surpass the chromosome-wide threshold for p<0.05 (1000 iterations and p<0.05 thresholds: genome-wide = 3.41, chromosome-wide = 2.04) and is above the widely used theoretical threshold of 3.0 [71], [73]–[75]. On average, individuals that were homozygous for the VSH allele were more likely to be individuals who were observed exhibiting VSH behavior. This QTL explains 6.1% of the variance observed and had an effect size of 0.248408. The LOD-1.5 confidence interval spanned about 1.1 Mb of physical distance. There were 63 candidate genes identified in this region (Table S1). Two genes were particularly interesting given the association between general hygienic behavior and odors (Table 1) [76]–[79]: 1) no receptor potential A2, which is associated with vision and olfaction in Drosophila; and 2) dop3, a D2-like dopamine receptor, which has been shown to be involved in aversive olfactory learning and memory in Drosophila [80]–[82], crickets [83], and honey bees [84]–[86].
Figure 1. QTL location on map of chromosome 9.
The physical location in base pairs of SNP probes in the honey bee genome assembly (Amel 4.0) is indicated to the right of the bar. Numbers to the left of the bar are distances in centimorgans (cM). The dotted line indicates the chromosome-wide empirical significance threshold of 0.05 as determined by 1000 permutations of phenotype data.
Table 1. Candidate genes involved in neurological signaling or regulation in QTL region on chromosome 9.
| Honey beegene ID | Drosophilahomolog ID | Predictions from Blast | Putative function |
| GB14619 | CG3620 | similar to no receptor potential A CG3620-PD, isoform D | phosphatidlyinositol phospholipase C activity; vision, olfaction |
| GB14561 | CG33517 | Dop3 D2-like dopamine receptor | aversive olfactory learning |
| GB15650 | similar to dpr6 CG14162-PA | defective proboscis extension response; sensory perception | |
| GB16925 | similar to longitudinals lacking protein,isoform G | putative transcription factor for axon growth and guidance in the CNS and PNS | |
| GB15048 | similar to zinc finger protein 595;longitudinals lacking protein,isoform G-like | putative transcription factor for axon growth and guidance in the CNS and PNS | |
| GB13523 | similar to zinc finger protein 808-like | development of supraesophageal ganglion and ocelli; may promote appendage formation | |
| GB10996 | ATM interactor-like;longitudinals lacking protein,isoforms A/B/D/L | transcription regulation | |
| GB10458 | hypothetical protein LOC724938;longitudinals lacking protein,isoforms A/B/D/L | transcription regulation | |
| GB12094, GB12494 | CG12052 | longitudinals lacking protein,isoforms A/B/D/L | transcription regulation |
| GB17677 | hypothetical protein LOC100578231;longitudinals lacking protein,isoforms A/B/D/L | transcription regulation | |
| GB14763 | similar to zinc finger protein 407-like | transcription regulation | |
| GB14706 | CG7471 | histone deacetylase Rpd3 isoform1Hist_deacetyl superfamily | transcription regulation |
| GB17640 | CG2368 | pipsqueak BTB superfamily | chromatin silencing; olfactory behavior |
| GB12634 | CG12608 | p21-activated protein kinase-interacting protein1-like; WD40 superfamily | signal transduction; pre-mRNA processing, cytoskeleton assembly |
| GB19232 | CG17221 | reticulon-4-interacting protein 1, mitochondrial-likeisoform 1; MDR superfamily; AdoMet_MTases superfamily | mushroom body development |
| GB11986, GB10237 | CG5406 | protein still life, isoform SIF type 1-like, partial;PH-like superfamily;UBQ superfamily,PDZ & RhoGEF superfamilies | signal transduction, regulation of synapse structure and activity |
| GB10808 | CG3894 | neuralized-like protein 2-like isoform 1;neutralized superfamily | signal transduction; myofiber differentiation and maturation |
| GB12006, GB16984 | nicotinic acetylcholine receptorbeta2 subunit and alpha9 subunit;neur_chan_LBD superfamily | neurotransmitter-gated ion-channel ligand binding domain; ion transport | |
| GB12219 | CG3889 | low quality protein: COP9;signalosome complex subunit 1;PCI superfamily | cell differentiation/specification; G-protein pathway suppressor 1 |
| GB12004 | CG2275 | transcription factor AP-1;Jun superfamily;bZIP_1 superfamily | Jun-like transcription factor |
A LOD peak of 1.95 was identified on chromosome 1, however, this QTL is only suggestive since it falls below both the genome-wide and chromosome-wide thresholds for significance (3.41 and 2.5, respectively). The percentage of observed variance explained by this QTL is 3.9% and the effect size is 0.196857. The LOD-1.5 confidence interval spanned approximately 2.0 Mb and contained 37 candidate genes, including a putative odorant receptor, a G-protein coupled receptor, and a protein that is a homolog of synaptic vesicle glycoprotein 2C (Tables 2 and S2).
Table 2. Candidate genes involved in neurological signaling or regulation in QTL region on chromosome 1.
| Honey beegene ID | Drosophila homolog ID | Predictions from Blast | Putative function |
| GB19123 | CG7497 | prostaglandin E2 receptor EP4 subtype-like | regulation of Rhoprotein signal transduction |
| GB10077 | CG16801 | photoreceptor-specific nuclear receptor | transcription regulation |
| GB16999 | CG31096 | leucine rich repeat G protein coupled receptor | G-protein coupled receptor activity |
| GB18179 | CG15302 | putative odorant receptor 9a | olfaction, G-protein coupled receptor |
| GB10277 | CG4898 | tropomyosin-1; hypothetical protein LOC408583 isoform 1 | muscle contraction; dendrite morphogenesis; lamellipodium assembly |
| GB17608 | CG4898 | tropomyosin-1 | muscle contraction; dendrite morphogenesis; lamellipodium assembly |
| GB17660 | CG4898 | hypothetical protein LOC408583 isoform 1; tropomyosin | muscle contraction; dendrite morphogenesis; lamellipodium assembly |
| GB11694 | hypothetical protein LOC100577365; segmentation polarity homeobox protein engrailed | compartment pattern specification; neuroblast fate determination | |
| GB15566 | CG9015 | segmentation polarity homeobox protein engrailed | compartment pattern specification; neuroblast fate determination |
| GB18087 | CG8759 | nascent polypeptide-associated complex subunit alpha-like isoform 1 | neurogenesis; oogenesis |
| GB14179 | hypothetical protein LOC100577522; defective proboscis extension response, putative | defective proboscis extension response; sensory perception of chemical stimulus |
Discussion
We used genotyping arrays to analyze genotypes for 1340 SNPs in a set of 238 individuals to make a high-density QTL map for VSH-based resistance to Varroa. Six putative QTL influencing hygiene against FKB were previousy identified [87], but we do not see evidence of any of the same QTL in our study. This is despite other studies having shown that FKB hygiene confers some resistance to Varroa [25], [49], and that VSH or VSH-derived bees exhibit high FKB hygiene [65]. Oxley et al. 2010 [87] identified a QTL associated with FKB uncapping behavior on chromosome 9. The nearest marker reported falls outside of the confidence interval for the VSH-related QTL on chromosome 9 we found, and the exact position of the QTL reported for FKB hygiene is uncertain because of low marker density. This suggests that either different QTL are involved in VSH and FKB hygienic behavior, or that differences in the particular populations of bees we tested did not allow us to detect overlap of QTL intervals. In addition, proteomic profiling of honey bee antennae also showed no apparent overlap in peptide signatures between VSH bees and bees with FKB hygiene [88].
Differentially expressed genes between bees exhibiting high and low VSH were identified with microarrays [89]. The high VSH stocks were from the same general population that we used for this QTL study. The microarrays revealed 39 genes that were differentially regulated in the brains of 14-day-old worker bees of low- and high-VSH lines. The results did not fit the hypothesis that differences in VSH behavior were caused by differences in sensitivity to particular olfactory stimuli, although among the 39 genes were three that may be involved in olfaction (a putative odorant binding protein Est65A, arrestin 2 and Antdh homologs). In contrast, the candidate genes we identified in our QTL mapping do show a possible connection between olfactory sensitivity and VSH and show no overlap with the differentially expressed genes in the microarray study. Linkage analyses have at least one advantage over microarray studies for identifying causal variation for traits – they directly tie the inheritance of genomic regions with the trait. Therefore if our QTL are confirmed in independent crosses, we can be confident that genes in the QTL regions are responsible for at least 10% of the differences in the trait measured. The candidate genes in our study may influence the expression of genes identified in the microarray study. It is also possible that although both studies used bees from the same USDA breeding program, different genes were segregating in each study, especially since the studies used different low lines. Brain tissue was used in the microarray study, but that may not be the only relevant tissue for VSH since olfaction starts with the antennae. Additionally, the studies differ in that the array study compared samples from colonies, not individuals, exhibiting different levels of VSH, whereas in our study, the comparisons were made between individuals that were observed performing the behavior.
The candidate genes identified by our study include no receptor potential A (norpA), a putative olfactory receptor, and a dopamine receptor. In addition to these candidates, there were genes in the QTL region with homology to Dmel/dpr, defective proboscis response, which are believed to be involved in chemosensory perception. norpA encodes a phospholipase C that is associated with vision in D. melanogaster [90]–[92], but also has been shown to affect olfaction; mutants defective in norpA exhibited impaired olfactory capabilities [93]. Phospholipase C has been documented in the homogenate of pheromone-sensitive sensilla of the silk moth, Antheraea polyphemus [94], and has been suggested as having a role in olfactory signal transduction in another moth, Spodoptera littoralis [95].
Dopamine (DA) is a catecholamine neurotransmitter and neuromodulator that is involved in behavior, cognition, learning, and memory in both vertebrates and invertebrates. In insects, appetitive learning is reinforced with octopamine and aversive learning is reinforced through a dopamine circuit. In Drosophila, dopamine-receptor-mutant larvae show impaired aversive learning [96], and blocking DA neurons in adults leaves the flies unable to learn to associate an odor stimulus with a punishment [82]. Similarly, in honey bees, the use of DA receptor antagonists blocked aversive learning (exhibited by the extension of a bee’s sting in response to an odor that it was trained to associate with electric shock) [86]. Honey bees have three dopamine receptors (see [97] for review); dop3 is a D2-like dopamine receptor that is widely expressed in the honey bee brain, but shows noticeably different expression from that of dop1 and dop2 [98]. The distribution of dop3 mRNA in cells around the optic and antennal lobes of the honey bee brain also suggests that this D2-like dopamine receptor is involved in processing sensory information [84], [98].
Olfactory cues have been shown to mediate general hygienic behavior [76]–[79], but the role of odor as a stimulus for hygiene by honey bees against Varroa is unclear. Earlier work suggested that the odor of the mite itself is probably not an important cue to A. mellifera [99]. Schöning et al. 2012 [100], however, suggested that bees recognize damaged brood by olfactory cues. The odor profile of brood parasitized by mites with high potential to transmit deformed wing virus (DWV) differed from the odor profile of brood parasitized by mites with low potential to transmit DWV. Hygienic bees preferentially removed pupae infested with mites with a high potential to induce damaging DWV infections, which are more likely to cause deformities and death. Our results support an association between genes involved in olfaction and VSH; however, we cannot rule out the possibility that other (non-olfactory) genes in our QTL regions modulate VSH.
Further work to identify the genes underlying this trait and then utilizing them as diagnostic tools for selective breeding could be valuable for beekeeping. Our mapping study will be followed with studies to analyze differential expression of candidate genes, gene function and association with VSH using gene knockdowns, and sequence differences between alleles. In order to use SNP markers for marker-assisted selection (MAS), it probably will be necessary to have SNP markers within the causal genes because of the high recombination rate of the honey bee genome. MAS may also allow for simultaneous selection and breeding for multiple traits, such as VSH, grooming behavior and physiological resistance to Varroa [67], [101]. If these technical challenges are met and useful markers are developed, MAS may speed selection by targeting sequences of specific genes in potential breeder queens and drones.
Materials and Methods
Ethics Statement
No permits were required to conduct the field research or genotyping analyses. The crosses and field research were conducted at the USDA-ARS Honey Bee Breeding, Genetics and Physiology Laboratory in Baton Rouge, LA, which is established and maintained to conduct apicultural research and bee breeding. Genotyping was performed in the Purdue core genomics facility in accordance with university and federal biosafety regulations.
Source of Worker Bees
Queens that produced colonies with either high or low expression of Varroa sensitive hygiene (VSH) were chosen as parents for the production of experimental colonies. The high VSH line was chosen from an ongoing USDA selection program. The high VSH queen produced a colony that removed 85–95% of mite-infested pupae during assays in which a comb of infested capped prepupae was placed into the broodnest for a 1-week period (method as in [68]), while the low VSH queen produced a colony that removed no more than 15% of the mite-infested pupae in similar assays. Fifteen daughter queens from the high VSH line were each mated to a single drone from the low VSH parent using instrumental insemination. The colonies containing F1 workers produced by these queens were evaluated for VSH activity and the colony that had removed the highest percentage of mite-infested pupae (62%, mean of 15 colonies = 40.6%) was used to produce 17 F1 daughter queens. Each F1 daughter queen was backcrossed to a single drone from the high VSH parent, and each colony was evaluated for VSH activity about 7–8 weeks later (mean = 59.0% removal). The colony with the highest removal (83%) of mite-infested pupae, colony A, was used as the source of worker bees that were evaluated for QTL analyses. The drones and queens used to make all generations of crosses were frozen and saved for SNP genotyping. All breeding and behavioral studies were conducted at the USDA, ARS Honey Bee Breeding, Genetics and Physiology Laboratory in Baton Rouge, LA.
Behavioral Studies of Worker Bees
Workers from colony A were classified as hygienic or non-hygienic by direct observation of their behavior when exposed to a comb of highly mite-infested pupae during a 45 minute period. Multiple tests over several weeks were needed to obtain enough workers to perform QTL analyses. Each week for 5 weeks, 300–500 newly emerged workers were individually marked by gluing a small plastic numbered disc to the thorax (E. H. Thorne, Ltd., Lincolnshire, UK); tags were also marked with paint to create enough unique combinations so that each individual bee could be identified. Workers were returned to their colony shortly after being tagged.
Behavioral testing began during the 3rd week when the oldest tagged workers were 15–18 days old, which corresponds to the optimal age for expression of hygienic behavior [102], and continued until 125 workers were identified as non-hygienic and another 125 workers were identified as hygienic (through the 5th week). Each test began when a comb containing mite-infested capped brood was inserted into the center of the broodnest of colony A. Combs were taken from heavily infested colonies and were chosen only if they had >100 square inches of capped brood, 15–20% Varroa infestation levels, and the pupae were predominantly in the white-eyed to pink-eyed stages [55]. The mite-infested comb was left in the center of the broodnest for 15 minutes; afterwards, it was removed carefully with adhering bees to an observation hive kept within a warm room. Two people conducted the behavioral observations, one on each side of the comb. Workers were identified as hygienic if they were observed (1) perforating the wax capping of the cell of a pupa, (2) enlarging the hole of an already perforated cell cap, or (3) removing a pupa from a fully uncapped brood cell. Workers were only sampled if they engaged in these behaviors for >2 minutes, and if the targeted brood cell was infested by Varroa. To determine if a brood cell was infested, a numbered pin was placed next to the brood cell that was manipulated at the moment that each worker bee was sampled. At the end of the test, the remaining worker bees were gently shaken and brushed from the combs, and each manipulated brood cell was examined under a stereomicroscope for the presence of Varroa. Workers were eliminated from the pool of hygienic bees if their hygienic responses were being directed toward cells infested by larvae of the greater wax moth (Galleria mellonella) or the small hive beetle (Athena tumida), or if the pupa was not mite-infested. Non-hygienic workers were identified as workers from the same age cohort as the hygienic workers that did not attempt hygiene during the 15 minutes of direct observation. Most non-hygienic workers were observed standing or walking over brood or engaging in trophallaxis with no attempts to engage in uncapping or removal behavior.
Each hygienic and non-hygienic worker was grabbed from the comb surface using soft forceps and quickly inserted into a plastic vile, which was flash frozen in liquid nitrogen. All samples were stored at −80°C until needed.
Genotyping and QTL Mapping
The DNA of the F1 queen was extracted using the Qiagen DNeasy Blood and Tissue Kit (Qiagen Inc., Valencia, CA) and was sequenced using the ABI SOLiD platform (Life Technologies Corp., Carlsbad, CA). We identified SNPs and designed probes for 1,536 genome-wide SNPs. These probes were used to analyze the genomic DNA of worker bees from the backcross family.
DNA was extracted from 240 individual worker abdomens using Qiagen DNeasy Blood and Tissue Kits. DNA was quantified with a fluorometer (Turner BioSystems, Sunnyvale, CA) and all samples diluted to 50 ng/µl. Genotyping was performed using the Illumina GoldenGate Assay with 250 ng of DNA per individual. Details of the assay can be found at the Illumina website (Illumina, Inc., San Diego, CA, www.illumina.com), but briefly, DNA is fragmented and activated for binding to paramagnetic particles, then hybridized with allele-specific and locus-specific oligonucleotides. The last 3′nucleotide of the allele-specific nucleotide is at the SNP. Extension past the SNP and ligation to the locus-specific oligo follow, giving rise to full-length joined products that serve as templates for PCR with universal primers and dye-labeled allele-specific primers. The dye-labeled PCR products were hybridized to the genotyping array matrix using a complementary address sequence present in the locus-specific primer. The fluorescence signals were read by the BeadArray Reader and analyzed by GenomeStudio software for semi-automated genotype clustering and calling (Illumina, Inc). Probes that had low call rates or were not polymorphic were removed from the data set (216 SNPs).
SNP markers were assembled into linkage groups using JoinMap 4.0 software [103], [104]. The marker orders were obtained by maximum likelihood analysis. Linkage distances between markers were estimated using multipoint analyses and the Kosambi mapping function. Interval mapping was performed with MapQTL 5.0 software [105]. The phenotypes were coded as a binary trait (1 or 0, depending on whether individuals exhibited the behavior). This analysis is effectively interval mapping with the Chi-square statistic. The 1.5-LOD support intervals (which correspond roughly to the 95% confidence intervals) for the QTL positions were determined from the interval mapping LOD values [106] and candidate genes were identified. Sequences for the probes that fall within the 1.5-LOD intervals can be found in Table S3. Genome-wide permutation tests were performed in MapQTL 5.0 to calculate the empirical significance thresholds to identify significant and suggestive QTL [107].
Supporting Information
Complete list of candidate genes for QTL region on chromosome 9.
(DOCX)
Complete list of candidate genes for QTL region on chromosome 1.
(DOCX)
Probe sequences used for genotyping that fall within the 1.5-LOD support interval.
(DOCX)
Acknowledgments
We thank David Dodge for help with the breeding and phenotyping of honey bees used in the study, and Phillip San Miguel and Ann Feil for assistance with the genotyping assays.
Disclaimer: Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Funding Statement
This research was funded by a USDA-NRI award (2008-35302-18803) and the Managed Pollinator Coordinated Agricultural Project (USDA-NIFA 2009-8511805718) to GJH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Evans JD, Schwarz RS (2011) Bees brought to their knees: microbes affecting honey bee health. Trends Microbiol 19: 614–620 doi: 10.1016/j.tim.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 2. Johnson RM, Ellis MD, Mullin CA, Frazier M (2010) Pesticides and honey bee toxicity – USA. Apidologie 41: 312–331 doi: 10.1051/apido/2010018. [Google Scholar]
- 3. Le Conte Y, Ellis M, Ritter W (2010) Varroa mites and honey bee health: can Varroa explain part of the colony losses? Apidologie 41: 353–363 doi: 10.1051/apido/2010017. [Google Scholar]
- 4. Currie RW, Pernal SF, Guzmán-Novoa E (2010) Honey bee colony losses in Canada. J Apic Res 49: 104–106 doi: 10.3896/IBRA.1.49.1.18. [Google Scholar]
- 5. Dahle B (2010) The role of Varroa destructor for honey bee losses in Norway. J Apic Res 49: 124–125 doi: 10.3896/IBRA.1.49.1.26. [Google Scholar]
- 6. Guzmán-Novoa E, Eccles L, Calvete Y, Mcgowan J, Kelly PG, et al. (2010) Varroa destructor is the main culprit for the death and reduced populations of overwintered honey bee (Apis mellifera) colonies in Ontario, Canada. Apidologie 41: 443–450 doi: 10.1051/apido/2009076. [Google Scholar]
- 7. Peterson M, Gray A, Teale A (2010) Colony losses in Scotland in 2004–2006 from a sample survey. J Apic Res 48: 145–146 doi: 10.3896/IBRA.1.48.2.11. [Google Scholar]
- 8. Rosenkranz P, Aumeier P, Ziegelmann B (2010) Biology and control of Varroa destructor . J Invertebr Pathol 103 Supplement: S96–S119 doi: 10.1016/j.jip.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 9. vanEngelsdorp D, Caron D, Hayes J, Underwood RM, Henson M, et al. (2012) A national survey of managed honey bee 2010–11 winter colony losses in the USA: results from the Bee Informed Partnership. J Apic Res 51: 115–124 doi: 10.3896/IBRA.1.51.1.14. [Google Scholar]
- 10. vanEngelsdorp D, Hayes Jr J, Underwood RM, Pettis JS (2010) A survey of honey bee colony losses in the United States, fall 2008 to spring 2009. J Apic Res 49: 7–14 doi: 10.3896/IBRA.1.49.1.03. [Google Scholar]
- 11. Martin SJ (1994) Ontogenesis of the mite Varroa jacobson Oud. in worker brood of the honey bee Apis mellifera L. under natural conditions. Exp Appl Acarol 18: 87–100. [Google Scholar]
- 12. Schultz A (1984) Reproduction and population dynamics of the parasitic mite Varroa jacobsoni Oud. in correlation with the brood cycle of Apis mellifera . Apidologie 5: 401–419. [Google Scholar]
- 13. Sammataro D, Gerson U, Needham G (2000) Parasitic mites of honey bees: life history, implications, and impact. Ann Rev Entom 45: 519–548 doi:10.1146/annurev.ento.45.1.519. [DOI] [PubMed] [Google Scholar]
- 14. Bowen-Walker PL, Gunn A (2001) The effect of the ectoparasitic mite, Varroa destructor on adult worker honeybee (Apis mellifera) emergence weights, water, protein, carbohydrate, and lipid levels. Entomol Exp Appl 101: 207–217 doi: 10.1046/j.1570-7458.2001.00905.x. [Google Scholar]
- 15. Schneider P, Drescher W (1987) Einfluss der parasitierung durch die milbe Varroa jacobsoni Oud. auf das schlupfgewicht, die gewichtsentwicklung, die entwicklung der hypopharynxdrüsen und die lebensdauer von Apis mellifera L. Apidologie 18: 101–110 doi: 10.1051/apido:19870108. [Google Scholar]
- 16. Bowen-Walker PL, Martin SJ, Gunn A (1999) The transmission of deformed wing virus between honeybees (Apis mellifera L.) by the ectoparasitic mite Varroa jacobsoni Oud. J Invertebr Pathol 73: 101–106 doi: 10.1006/jipa.1998.4807. [DOI] [PubMed] [Google Scholar]
- 17. Chen Y, Evans J, Feldlaufer M (2006) Horizontal and vertical transmission of viruses in the honey bee, Apis mellifera . J Invertebr Pathol 92: 152–159 doi: 10.1016/j.jip.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 18. Chen YP, Pettis JS, Collins A, Feldlaufer MF (2006) Prevalence and transmission of honeybee viruses. Appl Environ Microbiol 72: 606–611 doi: 10.1128/aem.72.1.606-611.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gisder S, Aumeier P, Genersch E (2009) Deformed wing virus: replication and viral load in mites (Varroa destructor). J Gen Virol 90: 463–467 doi: 10.1099/vir.0.005579-0. [DOI] [PubMed] [Google Scholar]
- 20. Ongus JR, Peters D, Bonmatin J-M, Bengsch E, Vlak JM, et al. (2004) Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor . J Gen Virol 85: 3747–3755 doi: 10.1099/vir.0.80470-0. [DOI] [PubMed] [Google Scholar]
- 21. Fries I, Imdorf A, Rosenkranz P (2006) Survival of mite infested (Varroa destructor) honey bee (Apis mellifera) colonies in a Nordic climate. Apidologie 37: 564–570 doi: 10.1051/apido:2006031. [Google Scholar]
- 22. Korpela S, Aarhus A, Fries I, Hansen H (1992) Varroa jacobsoni Oud. in cold climates: population growth, winter mortality and influence on the survival of honey bee colonies. J Apic Res 31: 157–164. [Google Scholar]
- 23. Le Conte Y, de Vaublanc G, Crauser D, Jeanne F, Rousselle J-C, et al. (2007) Honey bee colonies that have survived Varroa destructor . Apidologie 38: 566–572 doi: 10.1051/apido:2007040. [Google Scholar]
- 24. Seeley TD (2007) Honey bees of the Arnot Forest: a population of feral colonies persisting with Varroa destructor in the northeastern United States. Apidologie 38: 19–29 doi: 10.1051/apido:2006055. [Google Scholar]
- 25. Spivak M, Reuter GS (2001) Varroa destructor infestation in untreated honey bee (Hymenoptera: Apidae) colonies selected for hygienic behavior. J Econ Entomol 94: 326–331 doi: 10.1603/0022-0493-94.2.326. [DOI] [PubMed] [Google Scholar]
- 26. Villa J, D, Bustamante DM, Dunkley JP, Escobar LA (2008) Changes in Honey Bee (Hymenoptera: Apidae) Colony Swarming and Survival Pre- and Postarrival of Varroa destructor (Mesostigmata: Varroidae) in Louisiana. Ann Entomol Soc Am 101: 867–871 doi: 10.1603/0013-8746(2008)101[867:cihbha]2.0.co;2. [Google Scholar]
- 27. Ellis JD, Delaplane KS, Hood WM (2001) Efficacy of a bottom screen device, Apistan™, and Apilife VAR™, in controlling Varroa destructor . Am Bee J 141: 813–816. [Google Scholar]
- 28. Elzen P, Westervelt D (2004) A scientific note on reversion of fluvalinate resistance to a degree of susceptibility in Varroa destructor . Apidologie 35: 519–520 doi: 10.1051/apido:2004036. [Google Scholar]
- 29. Elzen PJ, Baxter JR, Spivak M, Wilson WT (2000) Control of Varroa jacobsoni Oud. resistant to fluvalinate and amitraz using coumaphos. Apidologie 31: 437–441 doi: 10.1051/apido:2000134. [Google Scholar]
- 30. Elzen PJ, Eischen FA, Baxter JR, Pettis JS, Elzen GW, et al. (1998) Fluvalinate resistance in Varroa jacobsoni from several geographic locations. Am Bee J 138: 674–676. [Google Scholar]
- 31. Elzen PJ, Eischen FA, Baxter JR, Elzen GW, Wilson WT (1999) Detection of resistance in US Varroa jacobsoni Oud. (Mesostigmata: Varroidae) to the acaricide fluvalinate. Apidologie 30: 13–17 doi: 10.1051/apido:19990102. [Google Scholar]
- 32. Elzen PJ, Westervelt D (2002) Detection of coumaphos resistance in Varroa destructor in Florida. Am Bee J 142: 291–292. [Google Scholar]
- 33. Lodesani M, Colombo M, Spreafico M (1995) Ineffectiveness of Apistan® treatment against the mite Varroa jacobsoni Oud in several districts of Lombardy (Italy). Apidologie 26: 67–72 doi: 10.1051/apido:19950109. [Google Scholar]
- 34. Lodesani M, Costa C (2005) Limits of chemotherapy in beekeeping: development of resistance and the problem of residues. Bee World 86: 102–109. [Google Scholar]
- 35. Mathieu L, Faucon J-P (2000) Changes in the response time for Varroa jacobsoni exposed to amitraz. J Apic Res 39: 155–158. [Google Scholar]
- 36. Milani N (1999) The resistance of Varroa jacobsoni Oud. to acaricides. Apidologie 30: 229–234 doi: 10.1051/apido:19990211. [Google Scholar]
- 37. Pettis JS (2004) A scientific note on Varroa destructor resistance to coumaphos in the United States. Apidologie 35: 91–92 doi: 10.1051/apido:2003060. [Google Scholar]
- 38. Sammataro D, Untalan P, Guerrero F, Finley J (2005) The resistance of varroa mites (Acari: Varroidae) to acaricides and the presence of esterase. Int J Acarology 31: 67–74 doi: 10.1080/01647950508684419. [Google Scholar]
- 39.Webster TC, Delaplane KS (2001) Mites of the honey bee. Hamilton, IL: Dadant & Sons Inc. 300 p.
- 40. Haarmann T, Spivak M, Weaver D, Weaver B, Glenn T (2002) Effects of fluvalinate and coumaphos on queen honey bees (Hymenoptera: Apidae) in two commercial queen rearing operations. J Econ Entomol 95: 28–35 doi: 10.1603/0022-0493-95.1.28. [DOI] [PubMed] [Google Scholar]
- 41. Johnson RM, Pollock HS, Berenbaum MR (2009) Synergistic interactions between in-hive miticides in Apis mellifera . J Econ Entomol 102: 474–479 doi: 10.1603/029.102.0202. [DOI] [PubMed] [Google Scholar]
- 42. Lodesani M, Costa C, Serra G, Colombo R, Sabatini AG (2008) Acaricide residues in beeswax after conversion to organic beekeeping methods. Apidologie 39: 324–333 doi: 10.1051/apido:2008012. [Google Scholar]
- 43. Martel A-C, Zeggane S, Aurières C, Drajnudel P, Faucon J-P, et al. (2007) Acaricide residues in honey and wax after treatment of honey bee colonies with Apivar® or Asuntol®50. Apidologie 38: 534–544 doi: 10.1051/apido:2007038. [Google Scholar]
- 44. Wallner K (1999) Varroacides and their residues in bee products. Apidologie 30: 235–248 doi: 10.1051/apido:19990212. [Google Scholar]
- 45. Wu JY, Anelli CM, Sheppard WS (2011) Sub-lethal effects of pesticide residues in brood comb on worker honey bee (Apis mellifera) development and longevity. PLoS ONE 6: e14720 doi: 10.1371/journal.pone.0014720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dietemann V, Pflugfelder J, Anderson D, Charrière J-D, Chejanovsky N, et al. (2012) Varroa destructor: research avenues towards sustainable control. J Apic Res 51: 125–132 doi: 10.3896/ibra.1.51.1.15. [Google Scholar]
- 47. Spivak M, Gilliam M (1998) Hygienic behaviour of honey bees and its application for control of brood diseases and Varroa: Part II. Studies on hygienic behavior since the Rothenbuhler era. Bee World 79: 169–186. [Google Scholar]
- 48. Spivak M, Gilliam M (1998) Hygienic behaviour of honey bees and its application for control of brood diseases and Varroa: Part I. Hygienic behaviour and resistance to American foulbrood. Bee World 79: 124–134. [Google Scholar]
- 49. Boecking O, Spivak M (1999) Behavioral defenses of honey bees against Varroa jacobsoni Oud. Apidologie 30: 141–158 doi: 10.1051/apido:19990205. [Google Scholar]
- 50. Spivak M (1996) Honey bee hygienic behavior and defense against Varroa jacobsoni . Apidologie 27: 245–260 doi: 10.1051/apido:19960407. [Google Scholar]
- 51. Ibrahim A, Spivak M (2006) The relationship between hygienic behavior and suppression of mite reproduction as honey bee (Apis mellifera) mechanisms of resistance to Varroa destructor . Apidologie 37: 31–40 doi: 10.1051/apido:2005052. [Google Scholar]
- 52. Harbo JR, Harris JW (2005) Suppressed mite reproduction linked to the behavior or adult bees. J Apic Res 44: 21–23 doi: 10.3896/IBRA.1.44.1.04. [Google Scholar]
- 53. Harbo JR, Harris JW (2009) Responses to Varroa by honey bees with different levels of Varroa Sensitive Hygiene. J Apic Res 48: 156–161 doi: 10.3896/IBRA.1.48.3.02. [Google Scholar]
- 54. Peng Y-S, Fang Y, Xu S, Ge L (1987) The resistance mechanism of the Asian honey bee, Apis cerana Fabr., to an ectoparasitic mite, Varroa jacobsoni Oudemans. J Invertebr Pathol 49: 54–60 doi: 10.1016/0022-2011(87)90125-x. [Google Scholar]
- 55. Harris JW (2007) Bees with Varroa Sensitive Hygiene preferentially remove mite infested pupae aged ≤ five days post capping. J Apic Res 46: 134–139 doi: 10.3896/IBRA.1.46.3.02. [Google Scholar]
- 56. Harris JW, Danka RG, Villa JD (2010) Honey bees (Hymenoptera: Apidae) with the trait of varroa sensitive hygiene remove brood with all reproductive stages of varroa mites (Mesostigmata: Varroidae). Ann Entomol Soc Am 103: 146–152 doi: 10.1603/an09138. [Google Scholar]
- 57. Rinderer TE, Harris JW, Hunt GJ, de Guzman LI (2010) Breeding for resistance to Varroa destructor in North America. Apidologie 41: 409–424 doi: 10.1051/apido/2010015. [Google Scholar]
- 58. Danka RG, de Guzman LI, Rinderer TE, Allen Sylvester H, Wagener CM, et al. (2012) Functionality of Varroa-resistant honey bees (Hymenoptera: Apidae) when used in migratory beekeeping for crop pollination. J Econ Entomol 105: 313–321 doi: 10.1603/ec11286. [DOI] [PubMed] [Google Scholar]
- 59. Delaplane KS, Hood WM (1999) Economic threshold for Varroa jacobsoni Oud. in the southeastern USA. Apidologie 30: 383–395 doi: 10.1051/apido:19990504. [Google Scholar]
- 60. Ward K, Danka R, Ward R (2008) Comparative performance of two mite-resistant stocks of honey bees (Hymenoptera: Apidae) in Alabama beekeeping operations. J Econ Entomol 101: 654–659 doi: 10.1603/0022–0493(2008)101[654:cpotms]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 61. Harris JW, Danka RG, Villa JD (2012) Changes in infestation, cell cap condition, and reproductive status of Varroa destructor (Mesostigmata: Varroidae) in brood exposed to honey bees with Varroa sensitive hygiene. Ann Entomol Soc Am 105: 512–518 doi: 10.1603/an11188. [Google Scholar]
- 62. Aumeier P, Rosenkranz P (2001) Scent or movement of Varroa destructor mites does not elicit hygienic behaviour by Africanized and Carniolan honey bees. Apidologie 32: 253–263 doi: 10.1051/apido:2001127. [Google Scholar]
- 63. Thakur RK, Bienefeld K, Keller R (1997) Varroa defense behavior in A. mellifera carnica . Am Bee J 137: 143–148. [Google Scholar]
- 64. Kirrane MJ, De Guzman LI, Rinderer TE, Frake AM, Wagnitz J, et al. (2011) Asynchronous development of honey bee host and Varroa destructor (Mesostigmata: Varroidae) influences reproductive potential of mites. J Econ Entomol 104: 1146–1152 doi: 10.1603/ec11035. [DOI] [PubMed] [Google Scholar]
- 65. Ibrahim A, Reuter GS, Spivak M (2007) Field trial of honey bee colonies bred for mechanisms of resistance against Varroa destructor . Apidologie 38: 67–76 doi: 10.1051/apido:2006065. [Google Scholar]
- 66. Arechavaleta-Velasco ME, Guzmán-Novoa E (2001) Relative effect of four characteristics that restrain the population growth of the mite Varroa destructor in honey bee (Apis mellifera) colonies. Apidologie 32: 157–174 doi: 10.1051/apido:2001121. [Google Scholar]
- 67.Arechavaleta-Velasco ME, Alcala-Escamilla K, Robles-Rios C, Tsuruda JM, Hunt GJ (2012) Fine-scale linkage mapping reveals a small set of candidate genes influencing honey bee grooming behavior in response to Varroa mites. PLoS ONE. doi: 10.1371/journal.pone.0047269. [DOI] [PMC free article] [PubMed]
- 68. Villa JD, Danka RG, Harris JW (2009) Simplified methods of evaluating colonies for levels of Varroa Sensitive Hygiene (VSH). J Apic Res 48: 162–167 doi: 10.3896/IBRA.1.48.3.03. [Google Scholar]
- 69. Fan J-B, Oliphant A, Shen R, Kermani BG, Garcia F, et al. (2003) Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol 68: 69–78 doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- 70. Beye M, Gattermeier I, Hasselmann M, Gempe T, Schioett M, et al. (2006) Exceptionally high levels of recombination across the honey bee genome. Genome Res 16: 1339–1344 doi: 10.1101/gr.5680406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hunt GJ, Page-Jr RE (1995) Linkage map of the honey bee, Apis mellifera, based on RAPD markers. Genetics 139: 1371–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Solignac M, Mougel F, Vautrin D, Monnerot M, Cornuet J-M (2007) A third-generation microsatellite-based linkage map of the honey bee, Apis mellifera, and its comparison with the sequence-based physical map. Genome Biol 8: R66.61–14 doi: 10.1186/gb-2007-8-4-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lander ES, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Page R, Fondrk M, Hunt G, Guzmán-Novoa E, Humphries M, et al. (2000) Genetic dissection of honeybee (Apis mellifera L.) foraging behavior. Journal of Heredity 91: 474–479 doi: 10.1093/jhered/91.6.474. [DOI] [PubMed] [Google Scholar]
- 75. Rueppell O, Pankiw T, Nielsen DI, Fondrk MK, Beye M, et al. (2004) The Genetic Architecture of the Behavioral Ontogeny of Foraging in Honeybee Workers. Genetics 167: 1767–1779 doi: 10.1534/genetics.103.021949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gramacho K, Spivak M (2003) Differences in olfactory sensitivity and behavioral responses among honey bees bred for hygienic behavior. Behavioral Ecology and Sociobiology 54: 472–479 doi: 10.1007/s00265-003-0643-y. [Google Scholar]
- 77. Masterman RM, Ross RR, Mesce KM, Spivak MS (2001) Olfactory and behavioral response thresholds to odors of diseased brood differ between hygienic and non-hygienic honey bees (Apis mellifera L.). J Comp Physiol A Neuroethol Sens Neural Behav Physiol 187: 441–452 doi: 10.1007/s003590100216. [DOI] [PubMed] [Google Scholar]
- 78. Spivak M, Masterman R, Ross R, Mesce KA (2003) Hygienic behavior in the honey bee (Apis mellifera L.) and the modulatory role of octopamine. Journal of Neurobiology 55: 341–354 doi: 10.1002/neu.10219. [DOI] [PubMed] [Google Scholar]
- 79. Swanson J, Torto B, Kells S, Mesce K, Tumlinson J, et al. (2009) Odorants that Induce Hygienic Behavior in Honeybees: Identification of Volatile Compounds in Chalkbrood-Infected Honeybee Larvae. Journal of Chemical Ecology 35: 1108–1116 doi: 10.1007/s10886-009-9683-8. [DOI] [PubMed] [Google Scholar]
- 80. Busto GU, Cervantes-Sandoval I, Davis RL (2010) Olfactory learning in Drosophila . Physiol 25: 338–346 doi: 10.1152/physiol.00026.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schroll C, Riemensperger T, Bucher D, Ehmer J, Völler T, et al. (2006) Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr Biol 16: 1741–1747 doi: 10.1016/j.cub.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 82. Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, et al. (2003) Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila . J Neurosci 23: 10495–10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Unoki S, Matsumoto Y, Mizunami M (2005) Participation of octopaminergic reward system and dopaminergic punishment system in insect olfactory learning revealed by pharmacological study. Eur J Neurosci 22: 1409–1416 doi: 10.1111/j.1460-9568.2005.04318.x. [DOI] [PubMed] [Google Scholar]
- 84. Beggs KT, Mercer AR (2009) Dopamine receptor activation by honey bee queen pheromone. Curr Biol 19: 1206–1209 doi: 10.1016/j.cub.2009.05.051. [DOI] [PubMed] [Google Scholar]
- 85. Sandoz J-C (2011) Behavioural and neurophysiological study of olfactory perception and learning in honeybees. Front Syst Neurosci 5: 1–20 doi: 10.3389/fnsys.2011.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Vergoz V, Roussel E, Sandoz J-C, Giurfa M (2007) Aversive learning in honeybees revealed by the olfactory conditioning of the sting extension reflex. PLoS ONE 2: e288 doi: 10.1371/journal.pone.0000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Oxley PR, Spivak M, Oldroyd BP (2010) Six quantitative trait loci influence task thresholds for hygienic behaviour in honeybees (Apis mellifera). Mol Ecol 19: 1452–1461 doi: 10.1111/j.1365-294X.2010.04569.x. [DOI] [PubMed] [Google Scholar]
- 88. Parker R, Guarna M, Melathopoulos A, Moon K-M, White R, et al. (2012) Correlation of proteome-wide changes with social immunity behaviors provides insight into resistance to the parasitic mite, Varroa destructor, in the honey bee (Apis mellifera). Genome Biol 13: R81 doi: 10.1186/gb-2012-139-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Le Conte Y, Alaux C, Martin JF, Harbo JR, Harris JW, et al. (2011) Social immunity in honeybees (Apis mellifera): transcriptome analysis of varroa-hygienic behaviour. Insect Mol Biol 20: 399–408 doi: 10.1111/j.1365-2583.2011.01074.x. [DOI] [PubMed] [Google Scholar]
- 90. Meyertholen EP, Stein PJ, Williams MA, Ostroy SE (1987) Studies of the Drosophila norpA phototransduction mutant. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 161: 793–798 doi: 10.1007/bf00610221. [DOI] [PubMed] [Google Scholar]
- 91.Pak WL (1979) Study of photoreceptor function using Drosophila mutants. Neurogenetics: genetic approaches to the nervous system.
- 92. Pollock VP, Radford JC, Pyne S, Hasan G, Dow JAT, et al. (2003) norpA and itpr mutants reveal roles for phospholipase C and inositol (1,4,5)- trisphosphate receptor in Drosophila melanogaster renal function. J Exp Biol 206: 901–911 doi: 10.1242/jeb.00189. [DOI] [PubMed] [Google Scholar]
- 93. Riesgo-Escovar J, Raha D, Carlson JR (1995) Requirement for a phospholipase C in odor response: overlap between olfaction and vision in Drosophila . Proc Natl Acad Sci U S A 92: 2864–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Maida R, Redkozubov A, Ziegelberger G (2000) Identification of PLC[beta] and PKC in pheromone receptor neurons of Antheraea polyphemus . Neuroreport 11: 1773–1776. [DOI] [PubMed] [Google Scholar]
- 95. Chouquet B, Lucas P, Bozzolan F, Solvar M, Maïbèche-Coisné M, et al. (2010) Molecular characterization of a phospholipase C β potentially involved in moth olfactory transduction. Chem Senses 35: 363–373 doi: 10.1093/chemse/bjq024. [DOI] [PubMed] [Google Scholar]
- 96. Selcho M, Pauls D, Han K-A, Stocker RF, Thum AS (2009) The role of dopamine in Drosophila larval classical olfactory conditioning. PLoS ONE 4: e5897 doi: 10.1371/journal.pone.0005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mustard JA, Vergoz V, Mesce KA, Klukas KA, Beggs KT, et al.. (2012) Dopamine signaling in the bee. In: Galizia CG, Eisenhardt D, Giurfa M, editors. Honeybee neurobiology and behavior. Netherlands: Springer. 199–209.
- 98. Beggs KT, Hamilton IS, Kurshan PT, Mustard JA, Mercer AR (2005) Characterization of a D2-like dopamine receptor (AmDOP3) in honey bee, Apis mellifera . Insect Biochem Mol Biol 35: 873–882 doi: 10.1016/j.ibmb.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 99. Boecking O, Drescher W (1994) Rating of signals which trigger Apis mellifera bees to remove mite-infested brood. Apidologie 25: 433–512 doi: 10.1051/apido:19940501. [Google Scholar]
- 100. Schöning C, Gisder S, Geiselhardt S, Kretschmann I, Bienefeld K, et al. (2012) Evidence for damage-dependent hygienic behaviour towards Varroa destructor-parasitised brood in the western honey bee, Apis mellifera . J Exp Biol 215: 264–271 doi: 10.1242/jeb.062562. [DOI] [PubMed] [Google Scholar]
- 101. Behrens D, Huang Q, Geßner C, Rosenkranz P, Frey E, et al. (2011) Three QTL in the honey bee Apis mellifera L. suppress reproduction of the parasitic mite Varroa destructor . Ecol Evol 1: 451–458 doi: 10.1002/ece3.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Arathi HS, Burns I, Spivak M (2000) Ethology of hygienic behaviour in the honey bee Apis mellifera L. (Hymenoptera: Apidae): behavioural repertoire of hygienic bees. Ethology 106: 365–379 doi: 10.1046/j.1439-0310.2000.00556.x. [Google Scholar]
- 103. Jansen J, de Jong AG, van Ooijen JW (2001) Constructing dense genetic linkage maps. Theor Appl Genet 102: 1113–1122 doi: 10.1007/s001220000489. [Google Scholar]
- 104.van Ooijen JW (2006) JoinMap® 4, Software for the calculation of genetic linkage maps in experimental populations. Wageningen, Netherlands: Kyazma, B. V.
- 105.van Ooijen JW (2004) MapQTL® 5, Software for the mapping of quantitative trait loci in experimental populations. Wageningen, Netherlands: Kyazma, B. V.
- 106. Dupuis J, Siegmund D (1999) Statistical methods for mapping quantitative trait loci from a dense set of markers. Genetics 151: 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete list of candidate genes for QTL region on chromosome 9.
(DOCX)
Complete list of candidate genes for QTL region on chromosome 1.
(DOCX)
Probe sequences used for genotyping that fall within the 1.5-LOD support interval.
(DOCX)