ABSTRACT
Proteus mirabilis rapidly migrates across surfaces using a periodic developmental process of differentiation alternating between short swimmer cells and elongated hyperflagellated swarmer cells. To undergo this vigorous flagellum-mediated motility, bacteria must generate a substantial proton gradient across their cytoplasmic membranes by using available energy pathways. We sought to identify the link between energy pathways and swarming differentiation by examining the behavior of defined central metabolism mutants. Mutations in the tricarboxylic acid (TCA) cycle (fumC and sdhB mutants) caused altered patterns of swarming periodicity, suggesting an aerobic pathway. Surprisingly, the wild-type strain swarmed on agar containing sodium azide, which poisons aerobic respiration; the fumC TCA cycle mutant, however, was unable to swarm on azide. To identify other contributing energy pathways, we screened transposon mutants for loss of swarming on sodium azide and found insertions in the following genes that involved fumarate metabolism or respiration: hybB, encoding hydrogenase; fumC, encoding fumarase; argH, encoding argininosuccinate lyase (generates fumarate); and a quinone hydroxylase gene. These findings validated the screen and suggested involvement of anaerobic electron transport chain components. Abnormal swarming periodicity of fumC and sdhB mutants was associated with the excretion of reduced acidic fermentation end products. Bacteria lacking SdhB were rescued to wild-type pH and periodicity by providing fumarate, independent of carbon source but dependent on oxygen, while fumC mutants were rescued by glycerol, independent of fumarate only under anaerobic conditions. These findings link multicellular swarming patterns with fumarate metabolism and membrane electron transport using a previously unappreciated configuration of both aerobic and anaerobic respiratory chain components.
IMPORTANCE
Bacterial locomotion and the existence of microbes were the first scientific observations that followed the invention of the microscope. A bacterium can swim through a fluid environment or coordinate motion with a group of bacteria and swarm across a surface. The flagellar motor, which propels the bacterium, is fueled by proton motive force. In contrast to the physiology that governs swimming motility, much less is known about the energy sources required for multicellular swarming on surfaces. In this study, we used Proteus mirabilis as a model organism to study vigorous swarming behavior and genetic and biochemical approaches to define energy pathways and central metabolism that contribute to multicellular motility. We found that swarming bacteria use a complete aerobic tricarboxylic acid (TCA) cycle but do not respire oxygen as the terminal electron acceptor, suggesting that multicellular cooperation during swarming reduces the amount of energy required by individual bacteria to achieve rapid motility.
Introduction
Proteus mirabilis differentiates from a single short rod-shaped swimmer cell into a multinucleate, elongated, and hyperflagellated swarmer cell in response to extreme viscosity or solid surfaces. This periodic developmental process, known as swarming differentiation, requires multicellularity and results in a regular pattern of rapid migration across a surface. Specifically, swarmer cells associate with one another and move in “rafts” (1). As the advancement of the swarm front decreases in velocity (2) and pauses, each polyploid hyperelongated swarmer cell septates and divides into numerous individual swimmer cells in a process known as consolidation. After a period of consolidation, vegetative swimmer cells differentiate into swarmer cells, and this behavior repeats multiple times, resulting in the bull’s-eye appearance of concentric rings of growth on an agar plate (3, 4). This cyclical developmental process and multicellular swarming behavior on solid (1.5% [wt/vol]) agar is characteristic of P. mirabilis and distinguishes it from other bacteria (5). While it is clear that initiation of swarming involves signaling events that lead from surface sensing to upregulation of flhDC and flagellum production (6–9), the molecular and physiological basis for the periodic multicellular swarming behavior remains poorly understood.
P. mirabilis swarmer cells possess an outer membrane composition biochemically distinct from nonswarming bacteria, or swimmer cells. Swarmer cells possess an increased number of long O-antigen side chains on the outer membrane lipopolysaccharide (LPS) (10), which could be important for surface sensing during initiation of swarming (11). Indeed, disruptions in genes responsible for LPS and cell wall biosynthesis affect swarmer cell elongation (8, 11). In contrast, the surface capsular polysaccharide is required for surface migration but not differentiation into elongated swarmer cells (12). Additionally, a mutation in ccmA (curved cell morphology) causes irregularly curved swarmer cells that disrupt alignment into multicellular rafts and produces migration defects (13). Four umo genes, some of which encode membrane proteins, have been identified that upregulate the expression of the flhDC master operon, which in turn controls flagellar biosynthesis, cell division, and swarming (14). However, despite their effect on flhDC expression, the proteins encoded by umo genes localize to the cell membrane or periplasm (14, 15). Swarm cells appear less transcriptionally active than consolidated cells; our study of P. mirabilis gene expression during swarming and consolidation identified umoD, flagellar genes, zapA, amino acid transport genes, peptide transport genes, and cell division genes as upregulated during consolidation (16), perhaps in preparation for the next round of differentiation and swarming.
Numerous differences in physiology occur in P. mirabilis swarmer cells compared to nonswarmer cells. For example, swarmer cells have reduced amino acid uptake and have increased sensitivity to hydrophobic antibacterial agents, suggesting that reorganization of the outer membrane might affect active transport processes and protective properties (10, 17). During swarming, the rates of uptake and incorporation of precursors into DNA, RNA, and protein are also reduced (18). While ATP concentrations remain constant, the rate of oxygen uptake is also reduced during P. mirabilis swarming (18). At the end of the swarm period, during consolidation, macromolecule synthesis and oxygen uptake are restored to levels equivalent to preswarming levels (18). Furthermore, a study of Proteus vulgaris swarming showed that membrane vesicles from swarm cells have reduced rates of NADH, malate, and succinate respiration (19). Interestingly, swarm cells were also found to contain diminished levels of cytochrome b as well as being deficient in cytochrome a and d (19). Since swarming can occur during both anaerobic and aerobic conditions, it has been proposed that the energy required during swarming is generated by fermentation (19). This is surprising, because using fermentation, rather than membrane respiration, would be much less energetically favorable for fueling the flagellar motor.
The proton gradient that is created during respiration contributes to the gradient of both pH and charge across the membrane and collectively determines proton motive force (PMF). It is during aerobic conditions with O2 as a terminal electron acceptor that the largest number of protons can be pumped, thereby producing the largest PMF for the cell. During fermentation, however, bacteria are unable to respire and must generate PMF by reversing the direction of the F1F0 ATPase rotation to consume ATP to pump protons out from the cytoplasm. Consequently, generation of PMF during anaerobic conditions is substantially more difficult for the cell compared to aerobic conditions, because the rotational speed of the flagellum is dependent on the magnitude of the PMF (20), which would be very limited in the absence of membrane respiration during fermentation.
We previously identified two P. mirabilis transposon mutants disrupting genes encoding proteins in the oxidative tricarboxylic acid (TCA) cycle that each displayed an aberrant swarming phenotype (21). These mutations resided in aceE, which encodes pyruvate dehydrogenase, and sdhC, which encodes the cytochrome b-556 subunit of succinate dehydrogenase. Those findings suggested a link between PMF, the TCA cycle, and swarming motility. Surprisingly, however, Western blot analysis using antibodies to the flagellum subunit, FlaA, showed that both mutants produce similar levels of flagellin despite a dramatic difference in the swarming periodicity (21). Pyruvate dehydrogenase converts pyruvate, the end product of glycolysis, to acetyl coenzyme A (acetyl-CoA), which enters a complete oxidative TCA cycle during aerobic conditions. Succinate dehydrogenase functions in the complete TCA cycle and is a component of the aerobic electron transport chain. Because mutations in both genes resulted in aberrant swarming phenotypes, we hypothesized that a complete oxidative TCA cycle is required for swarming. However, previous findings also indicated that aerobic cytochromes are absent in swarmer cells, and fermentation was proposed as an energy source during P. mirabilis swarming (19), despite the notable requirement for PMF to drive flagellar rotation.
In this current study, we used genetic and biochemical approaches to define central metabolic and energy pathways that contribute to multicellular swarming motility. Consistent with previous studies (19), we found that swarming is resistant to sodium azide. However, characterization of swarming for defined central metabolism mutants indicated that the aerobic TCA cycle and fumarate metabolism, independent of fumarate reductase, controls swarming periodicity in P. mirabilis. Here, we present evidence for an intimate connection between energy metabolism and bacterial multicellular behavior that supports a model for anaerobic respiration using a complete oxidative TCA cycle to generate PMF to drive swarming.
RESULTS
P. mirabilis central metabolism mutants have altered periods of swarming.
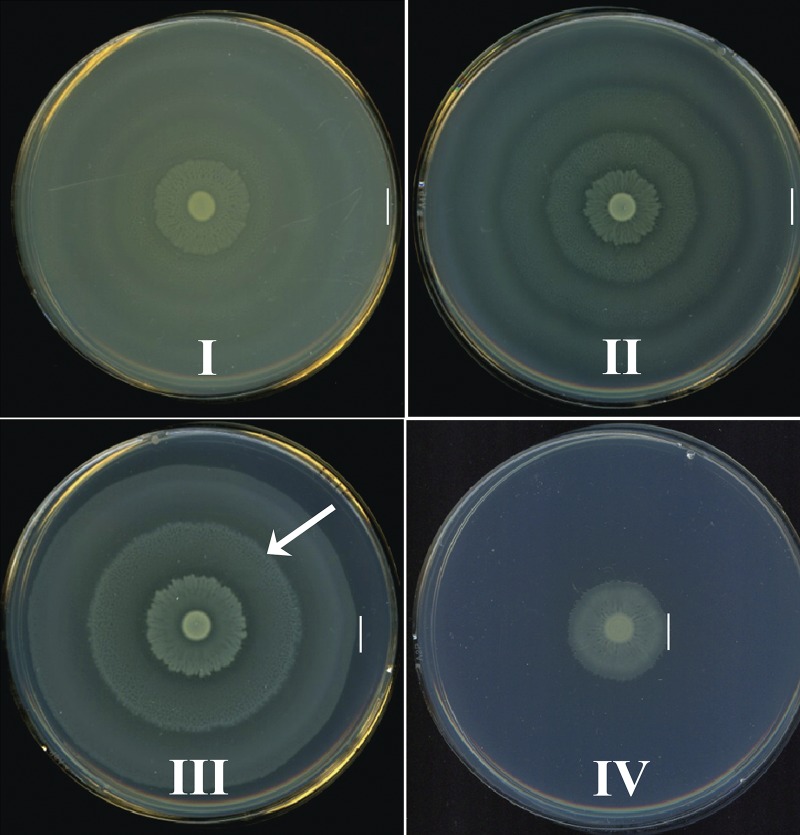
From a previous signature-tagged mutagenesis study (21), we noted that transposon insertions in genes encoding enzymes of the TCA cycle displayed defects in the ability of P. mirabilis HI4320 to swarm but did not affect flagellin production. To better understand the connection between central metabolism and swarming differentiation, we systematically generated defined mutants deficient in specific central pathways and tested their ability to swarm on lysogeny broth (LB) medium solidified with 1.5% (wt/vol) agar. These mutations disrupted genes representing the TCA cycle, both branches of the pentose phosphate pathway (PPP), and glycolysis (Table 1). Remarkably, we found that the swarming phenotypes displayed by wild-type HI4320 and the central metabolism mutants fit into four general classes strictly coinciding with the affected pathway: a normal swarming phenotype as displayed by the wild-type strain (class I) and three distinct classes of altered swarming resulting from disruption of the TCA cycle (class II), the PPP (class III), or glycolysis (class IV) (Fig. 1).
TABLE 1 .
P. mirabilis metabolism mutants examined in this studya
| Pathway | Gene | Description |
|---|---|---|
| Glycolysis | pfkA | Phosphofructokinase |
| pgi | Phosphoglucose isomerase | |
| tpiA | Triosephosphate isomerase | |
| Pentose phosphate | gnd | 6-Phosphogluconate dehydrogenase |
| talB | Transaldolase | |
| Entner-Doudoroff | edd | 6-Phosphogluconate dehydratase |
| TCA cycle | sdhB | Succinate dehydrogenase |
| frdA | Fumarate reductase | |
| fumC | Fumarase | |
| Gluconeogenesis | pckA | Phosphoenolpyruvate carboxykinase |
All mutants were constructed for this study using the TargeTron system as described in Materials and Methods.
FIG 1 .
The swarming patterns of P. mirabilis central pathway mutants fit into four general classes. P. mirabilis wild-type HI4320 and mutants were examined on lysogeny broth (LB) containing 15g/liter agar and 10g/liter NaCl by spotting 5 µl of overnight culture onto the center of a plate and incubating under aerobic conditions at 37°C for 18 h. Swarming phenotype of wild-type HI4320 (class I) and representative images of swarming phenotypes of mutants in the tricarboxylic acid (TCA) cycle (class II), the PPP (class III), and glycolysis (class IV). Class I, which encompasses wild-type HI4320, displays a normal swarming pattern resulting from multicellular migration of cells. For mutants of class II, swarming develops with altered periods or distance between swarming and consolidation cycles resulting in a distinct pattern of rings on the agar surface. Swarming phenotypes of class III display elongated swarming zones with delayed consolidation (arrow). Class IV swarming includes mutants with aberrant phenotypes displaying an overall decreased swarm zone diameter. The vertical lines indicate the outer edge of the swarm.
Class I, wild type, displays a normal swarming pattern resulting from regular periodicity of multicellular migration as observed with wild-type HI4320 (Fig. 1). Class II, TCA cycle, is represented by TCA cycle mutants (fumC and sdhB mutants) that develop shorter periods, as measured by distance, between swarming and consolidation that results in a distinct pattern of rings on the agar surface (Fig. 1). The swarming phenotype of class III mutants in genes for the pentose phosphate pathway (PPP) (gnd and talB mutants) possesses longer periods of swarming following consolidation (Fig. 1, arrow). The elongated swarming zones of class III mutants were all visibly denser, which could possibly result from an overall increase of swarm cell number or a change in swarm cell morphology, than that of class I bacteria. Class IV, the glycolysis mutants (pfkA and tpiA), have a defect that results in decreased net migration and a severely reduced swarm diameter compared to that of wild-type HI4320 or other classes of mutants (Fig. 1).
Central carbon pathways that generate pyruvate are required for swarming differentiation.
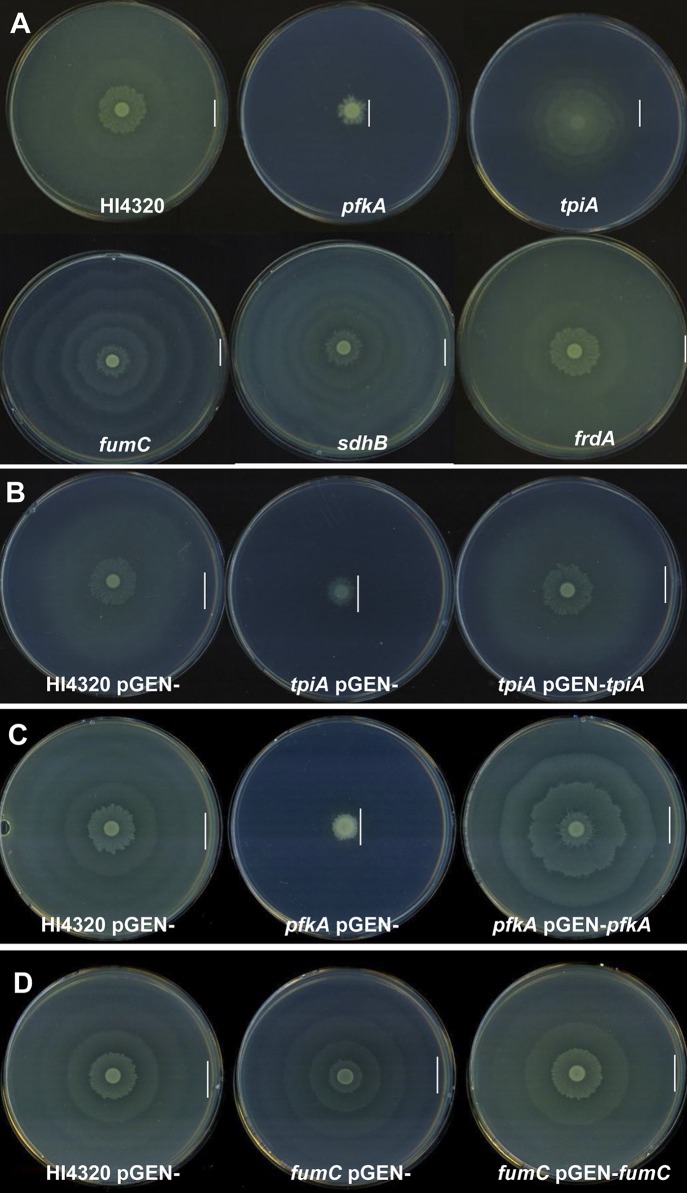
Mutations within pfkA and tpiA that encode glycolytic enzymes resulted in a diminished capacity to migrate; in particular, the diameter of swarming was reduced (class IV) compared to the wild-type HI4320 swarming diameter (Fig. 2A). The defect appeared to result from loss of the respective enzyme, because the crippled swarming phenotype of the tpiA and pfkA mutants was successfully restored to wild-type HI4320 swarming when complemented with the wild-type tpiA or pfkA allele on a plasmid, respectively (Fig. 2B and C).
FIG 2 .
Specific P. mirabilis central metabolism mutants cause aberrant swarming phenotypes. (A) Swarming phenotypes of wild-type HI4320, glycolysis mutants (A) Swarming phenotypes of wild-type HI4320, pfkA (phosphofructokinase) and tpiA (triose phosphate isomerase), glycolysis mutants, fumC (fumarase) and sdhB (succinate dehydrogenase) TCA cycle mutants, frdA (fumarate reductase) the anaerobic TCA cycle mutant. Mutations in pfkA and tpiA result in reduced swarming phenotype diameters, while fumC and sdhB TCA cycle mutants have reduced distances between swarming and consolidation which result in shorter consolidation rafts or rings. The anaerobic TCA cycle mutant has a similar swarming phenotype to that of wild-type HI4320. Complementation of tpiA (B) pfkA (C) and fumC (D) swarming phenotypes. P. mirabilis HI4320 containing pGEN empty vector and each mutant containing pGEN empty vector were compared to mutant-complemented strains. The complemented tpiA (pGEN+tpiA), pfkA (pGEN+pfkA), and fumC (pGEN+fumC) mutants all exhibit a restored swarming phenotype to that of wild-type HI4320. The vertical lines in panels A to D indicate the outer edge of the swarm.
In addition to glycolysis, many bacteria employ the Entner-Doudoroff (ED) pathway to catabolize 6-carbon sugars, such as gluconate and sugar acids, into glyceraldehyde-3-phosphate (G3P), ATP, and pyruvate. Another important route for 6-carbon catabolism is the oxidative and nonoxidative PPP, during which NADPH and sugars, such as ribose-5-phosphate, G3P, and fructose-6-phosphate, are interconverted, respectively. Distinct swarming phenotypes were also observed for the PPP (class III); mutations within gnd of the oxidative PPP and talB of the nonoxidative PPP resulted in a swarming phenotype consisting of dense elongated swarming zones following consolidation (Fig. 1, arrow). Interestingly, mutations of the ED pathway (edd mutant) and gluconeogenesis (pckA mutant) result in swarming phenotypes identical to that of the wild-type strain HI4320 (class I). Glycolysis, the ED pathway, and the PPP are all generally involved in the catabolism of 6-carbon sugars to pyruvate, yet only mutations in tpiA, pfkA, talB, and gnd have swarming defects on LB-1.5% agar. Since LB medium lacks a carbohydrate carbon source like glucose, these findings suggest that their swarming defects could result from the inability to produce sufficient pyruvate for energy metabolism or specific macromolecular precursors; DHAP, for cell membranes; fructose-6-P, for peptidoglycan (PfkA); ribose-5-P (TalB), for nucleobases; or NADPH (Gnd), which powers most biosynthetic pathways.
Multicellular swarming requires energy pathways dependent on the TCA cycle.
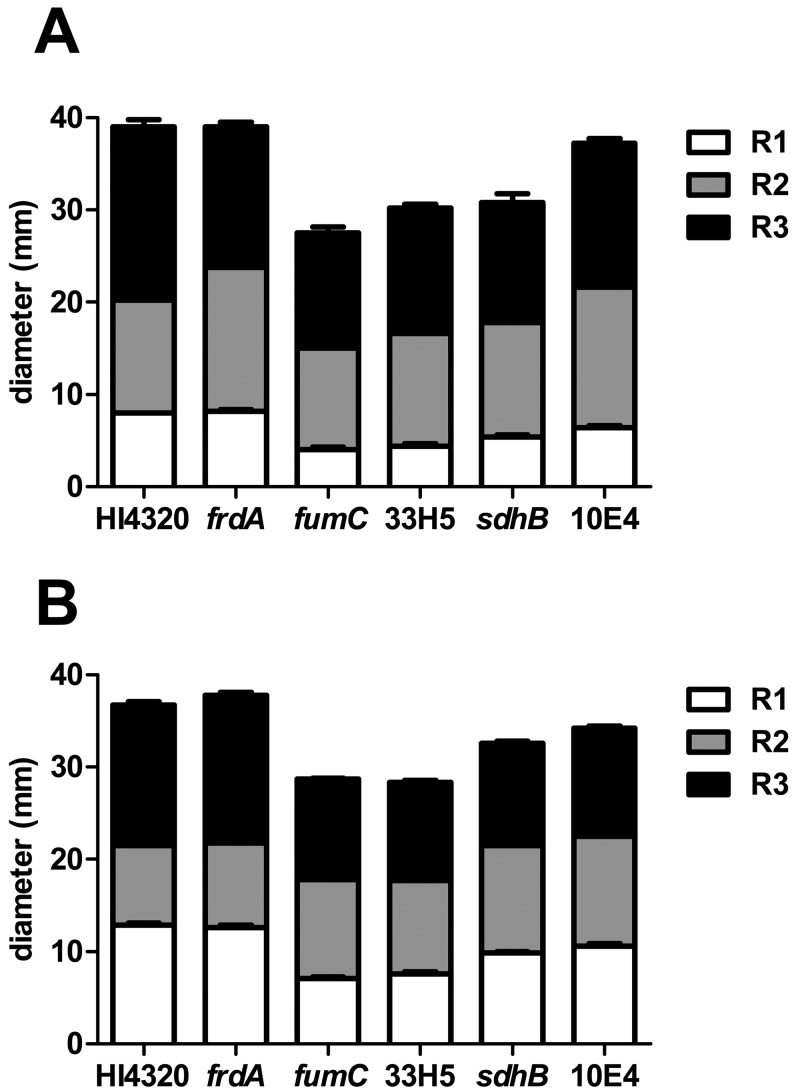
During aerobic respiration, pyruvate generated by glycolysis, the PPP, or the ED pathway is converted to acetyl-CoA by pyruvate dehydrogenase before entering the TCA cycle, where it condenses with oxalacetate to form citrate. Previously, we found that pyruvate dehydrogenase mutants (aceE mutant) produce wild-type levels of flagellin (FlaA) yet are unable to swarm (21), suggesting that an aerobic TCA cycle is required for swarming. The TCA cycle operates during aerobic and anaerobic respiration or fermentation by running in an oxidative cycle (when respiring oxygen) or in an incomplete, reductive, and branched pathway, respectively. Mutants in TCA cycle genes have distinct swarming periodicity phenotypes (class II) compared to wild-type HI4320 (Fig. 2A). The class II phenotype was highly reproducible (see Fig. S1 in the supplemental material) and demonstrated a consistent 50% reduction (4 mm for class II compared to 8 mm for the wild type) in the distance between the consolidation rings that immediately follow the inoculation point compared to the wild type at both 30°C and 37°C (Fig. 3A and B). Bacteria with a mutation within fumarase, the fumC mutant, which functions in both the aerobic and anaerobic TCA cycle, produce shorter distances between swarming and consolidation rafts (Fig. 3). The altered swarming phenotype of the fumC mutant bacteria was restored to wild-type swarming when complemented by the wild-type fumC allele on a plasmid (Fig. 2D). The same aberrant swarming phenotype is observed for the aerobic TCA cycle mutant, the sdhB mutant, which encodes succinate dehydrogenase (Fig. 2A) and was also observed previously for a strain with a mutation in sdhC (21) (see Fig. S1). In contrast, a mutation within frdA, which encodes fumarate reductase, a branched TCA pathway enzyme that is important during anaerobic respiration, causes swarming like in wild-type HI4320 (Fig. 2A). Together, the swarming phenotypes of these three TCA cycle mutants indicate that P. mirabilis swarming is dependent on the complete oxidative pathway since mutation of frdA, which disrupts only the incomplete, reductive (branched) TCA pathway, did not affect swarming periodicity (Fig. 3A and B).
FIG 3 .
P. mirabilis HI4320 oxidative TCA cycle mutants display shorter swarming distances between consolidation rafts in comparison to wild-type HI4320. To quantify the aberrant swarming phenotype, swarm agar plates were inoculated with a 5-µl volume from one overnight culture of wild-type P. mirabilis HI4320 and fumC, sdhB, frdA, 33H5 (fumC), and 10E4 (sdhC) mutants. Plates were incubated for 18 h, and the width of each swarming raft was measured in millimeters. The white-shaded portion of the bar represents the averaged measurements of the first swarm raft (R1), the gray-shaded portion of the bar represents the averaged measurements of the second raft (R2), and the black-shaded portion of the bar represents the averaged measurements of the third raft (R3). In panel A, the bar graph displays the measurements from a total of 5 inoculated agar swarm plates for each strain following incubation at 37°C, and in panel B, measurements were taken from a total of 10 inoculated agar plates of each strain following incubation at 30°C. 33H5 was identified in the present study; in an earlier study (see reference 21), 10E4 was shown to produce aberrant swarming patterns on agar.
Addition of TCA cycle intermediates alters swarming differentiation of P. mirabilis.
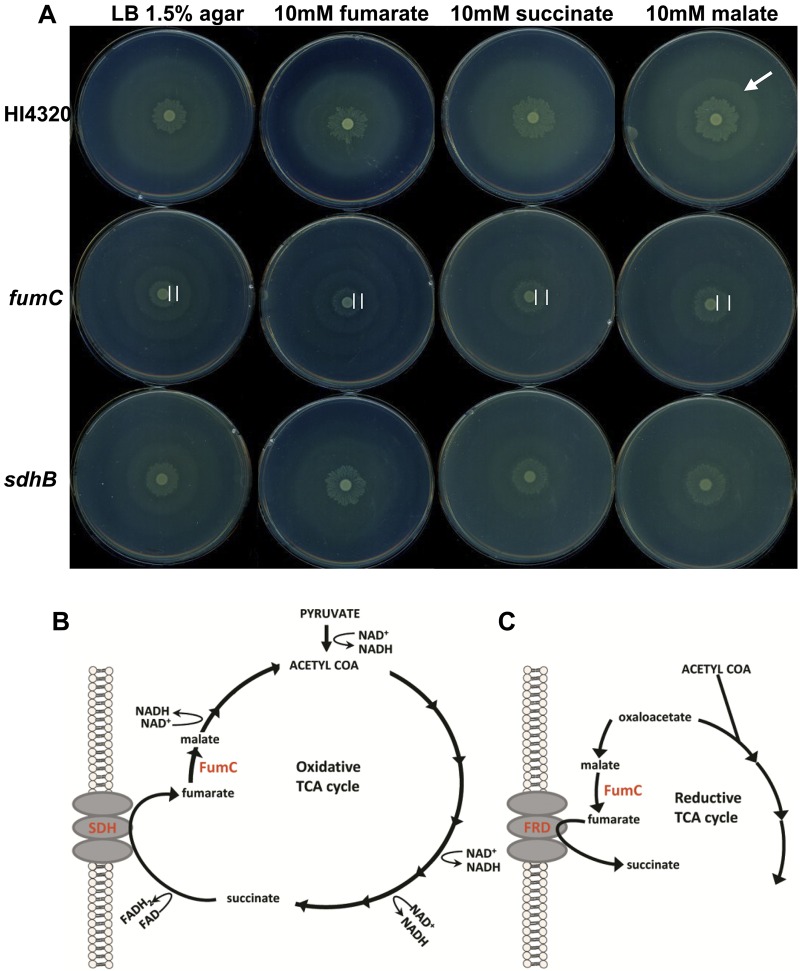
Because disruption of the aerobic oxidative TCA cycle (aceE, sdhB, sdhC, and fumC mutant bacteria) resulted in aberrant swarming phenotypes (class II), we sought to determine whether swarming periodicity of the wild-type parent strain or these TCA cycle mutants could be manipulated or chemically complemented by the addition of exogenously supplied TCA cycle intermediates. We tested P. mirabilis HI4320 and the TCA cycle mutants on agar replete with fumarate, succinate, or malate. The biochemical intermediate for which a specific mutant was deficient in generating was added to the agar to test for restoration of the mutant’s swarming cycles to wild-type periodicity. For example, during the aerobic TCA cycle (Fig. 4B), succinate is oxidized to fumarate by succinate dehydrogenase, SdhB; if the altered swarming phenotype of the sdhB mutant is due to its inability to generate fumarate, then swarming of the sdhB mutant should be restored to the wild-type phenotype by the addition of fumarate. The fumarase mutant (fumC), which is unable to convert fumarate to malate during a complete oxidative TCA cycle (or malate to fumarate in the branched reductive pathway [Fig. 4C]), was also examined on agar containing each of the tested intermediates (Fig. 4A). Indeed, fumarate, succinate, and malate all allow the sdhB mutant to revert to wild-type swarming periodicity, while succinate and malate rescue swarming periodicity in the fumC mutant (Fig. 4A). It was also possible to alter wild-type HI4320 swarming with malate, which caused a more pronounced secondary raft (Fig. 4A, arrow). During the complete oxidative TCA cycle (Fig. 4B), fumarase catalyzes the oxidation of fumarate to malate during aerobic respiration; however, under anaerobic respiration, the TCA cycle operates in a reduced branched pathway during which the intermediate malate is reduced to fumarate by fumarase (Fig. 4C). Collectively, these findings also indicate that swarming differentiation relies on a complete, oxidative TCA cycle because succinate dehydrogenase is part of the aerobic pathway and the fumarase mutant was not affected by the addition of fumarate (Fig. 4A).
FIG 4 .
LB-1.5% agar replete with TCA cycle intermediates alters the interval between swarming and consolidation of P. mirabilis TCA cycle mutants. (A) Wild-type P. mirabilis HI4320 and the fumC and sdhB aerobic TCA cycle mutants were examined on LB-1.5% agar alone or agar containing either 10 mM fumarate, 10 mM succinate, or 10 mM malate. All three TCA cycle intermediates cause the sdhB mutant to revert to a wild-type swarming phenotype, while only succinate and malate cause reversion to the wild-type swarming phenotype for the fumC mutant. The addition of malate causes the formation of a pronounced secondary raft for wild-type HI4320 (arrow). Complete oxidative (B) and incomplete reductive (C) TCA cycle pathways indicating the relevant enzymes, intermediates, and direction of reactions during aerobic and anaerobic processes, respectively.
P. mirabilis swarming is resistant to sodium azide, a poison of aerobic respiration.
Mutation of the oxidative TCA cycle genes fumC and sdhB resulted in shorter periods of swarming. Succinate dehydrogenase can participate in membrane electron transfer during respiration. This predicts that swarming of wild-type strain HI4320 would be affected by adding sodium azide (NaN3) to poison aerobic respiration by irreversibly binding and inhibiting quinones involved in the transfer of electrons to oxygen. Despite the genetic requirement for the complete oxidative (aerobic) TCA cycle, we found that P. mirabilis HI4230 swarms on growth inhibitory concentrations (0.005% [wt/vol]) of NaN3 at 37°C (Fig. 5A). This suggests that P. mirabilis swarming is resistant to sodium azide and does not require aerobic respiration, supporting the notion that anaerobic or fermentation pathways can provide energy for swarming (18, 19).
FIG 5 .
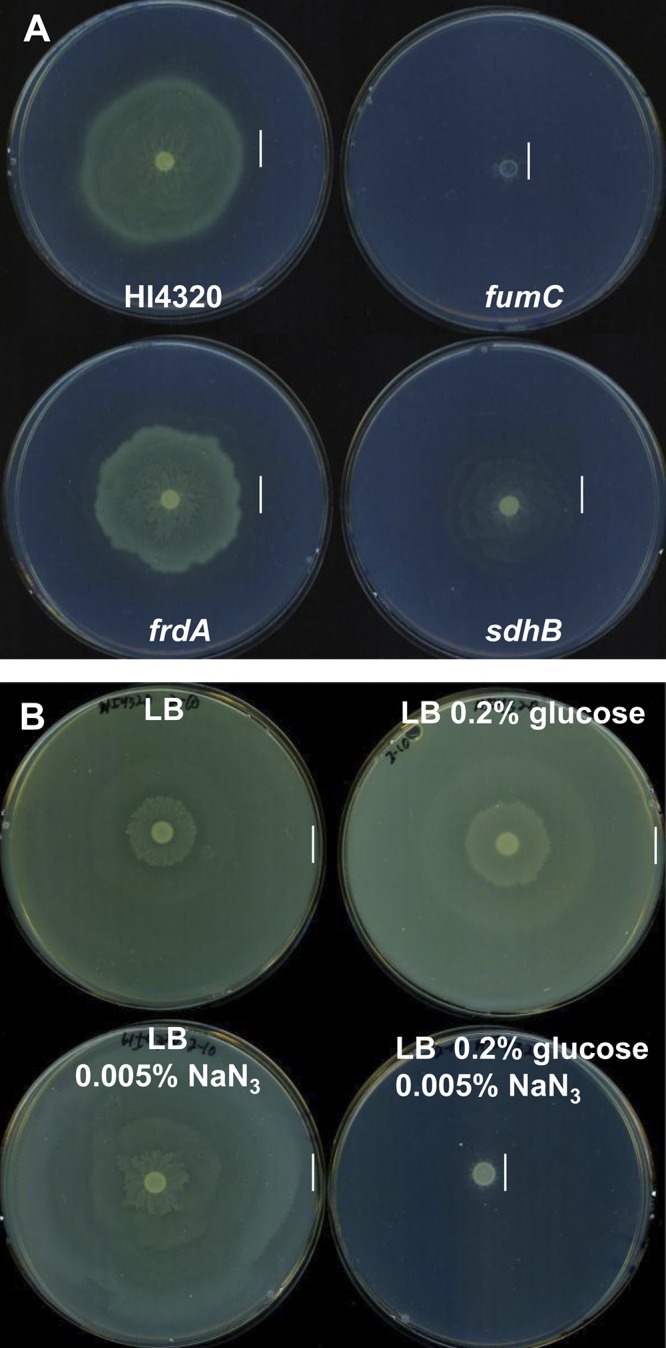
Swarming P. mirabilis HI4320 is resistant to inhibitory sodium azide concentrations. (A) Swarming behavior of wild-type HI4320 and fumC, frdA, and sdhB mutants on LB-1.5% agar containing 0.005% (wt/vol) sodium azide (NaN3) following incubation at 37°C during aerobic conditions. Wild-type P. mirabilis HI4320 and the frdA TCA cycle mutant are resistant to NaN3 and have similar swarming phenotypes. The addition of NaN3 cripples swarming of the fumC mutant, while the sdhB mutant has a reduced swarming phenotype in the presence of NaN3. (B) Glucose inhibits wild-type P. mirabilis swarming on NaN3. P. mirabilis HI4320 was inoculated on LB-1.5% agar alone, agar with 0.02% (wt/vol) glucose, agar containing 0.005% (wt/vol) NaN3, and agar containing 0.005% (wt/vol) NaN3 and 0.02% (wt/vol) glucose. The vertical lines in panels A and B indicate the outer edge of the swarm.
The anaerobic branched TCA pathway mutant was also resistant to NaN3 and had a swarming phenotype identical to that of wild-type HI4320 on azide (Fig. 5A). In the absence of FrdA, a component of the incomplete reductive TCA pathway, either a fermentative pathway or an aerobic pathway that uses succinate dehydrogenase in the complete TCA cycle, would likely operate, because the frdA mutant is unable to assemble an anaerobic respiratory chain using fumarate reductase (Fig. 4C). However, our findings demonstrate that an otherwise aerobic pathway is not using oxygen as a terminal electron acceptor during membrane respiration, because mutation of frdA does not render the bacteria susceptible to sodium azide during swarming. Mutation of fumC, which can function in both the anaerobic and aerobic pathways (Fig. 4B and C), severely inhibits swarming when respiration is poisoned by NaN3, while swarming of the aerobic TCA cycle mutant sdhB was modestly affected by NaN3 (Fig. 5A). Interestingly, swarming of wild-type strain HI4320 was inhibited when 0.02% (wt/vol) glucose was added to LB-1.5% agar containing NaN3 (Fig. 5B), suggesting that in the presence of glucose, which promotes aerobic respiration when oxygen is present (22), P. mirabilis is unable to use its apparently preferred anaerobic-like electron acceptor and energy pathway required for swarming. The activity of azide on swarming was specific for membrane respiration with oxygen rather than acting as an uncoupler, because P. mirabilis was unable to swarm when the proton gradient was collapsed by adding the conventional uncoupler carbonylcyanide m-chlorophenylhydrazone (CCCP) to LB-1.5% agar (data not shown).
Genetic screen to identify genes required for P. mirabilis to swarm on sodium azide.
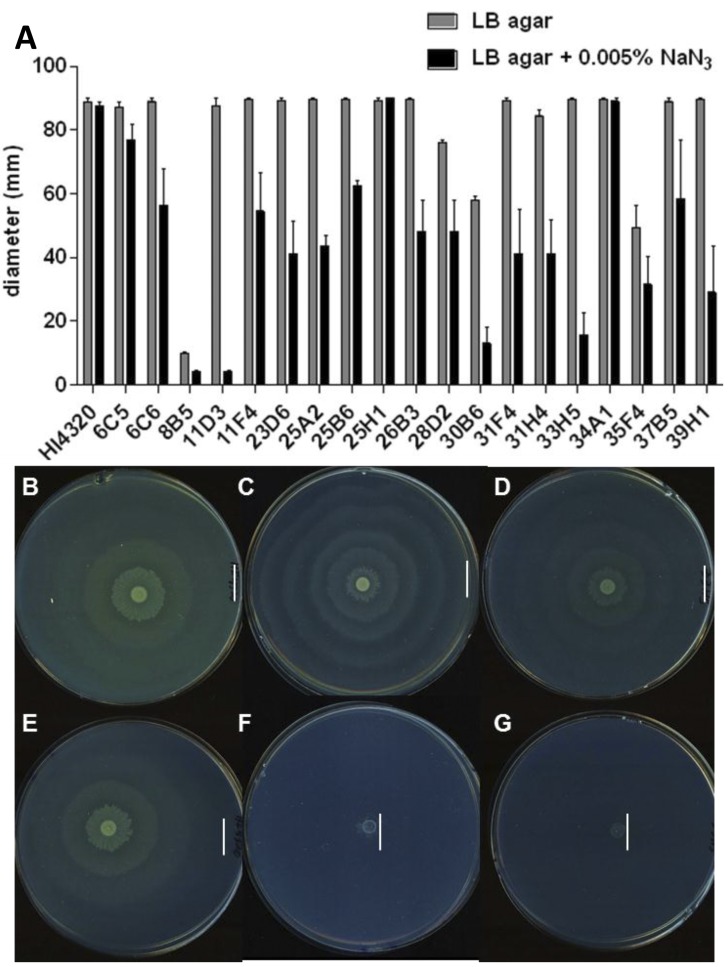
To identify genes that encode or are required for the expression of alternative anaerobic-like respiratory chains, we tested P. mirabilis transposon mutants on LB-1.5% agar containing 0.005% (wt/vol) NaN3 for the loss of swarming only when aerobic respiration is poisoned. Eighteen of 1,920 P. mirabilis transposon mutants consistently demonstrated decreased swarming diameters compared to that of wild-type HI4320 (Fig. 6B and E) on LB-1.5% agar containing NaN3 (Fig. 6A). To eliminate motility-defective mutants, for example, those with disruptions in flagellar genes, all mutants were simultaneously tested for swarming on agar with and without NaN3. Mutants with deficient swarming phenotypes on agar without NaN3 were removed from the screen.
FIG 6 .
Identification of P. mirabilis genes specifically required for swarming on sodium azide. (A) Eighteen out of 1,920 P. mirabilis transposon mutants were sensitive to 0.005% (wt/vol) sodium azide (NaN3) and displayed decreased swarming phenotypes during aerobic conditions at 37°C (black bars). The mutants selected in this screen swarm at 37°C in the absence of NaN3 (gray bars). A previously identified transposon mutant, 8B5, was included as a nonswarming control. Swarming phenotypes of wild-type HI4320 (B), fumC mutant (C), and 33H5 mutant (D) on LB-1.5% agar alone and swarming phenotypes of wild-type HI4320 (E), fumC mutant (F), and 33H5 mutant (G) on agar containing 0.005% (wt/vol) NaN3. The gene disrupted in 33H5 (fumC), has the same swarming phenotypes as the de novo fumC mutant (C). The vertical lines in panels B to G indicate the outer edge of the swarm.
Transposon insertions were identified that disrupted genes encoding proteins involved in energy metabolism that are required during swarming (Table 2). Transposon mutant 37B5 has a disruption in gene argH, which encodes argininosuccinate lyase that catalyzes the breakdown of argininosuccinate to arginine and fumarate (Table 2). Mutant 33H5 has an insertion that disrupts fumC, which encodes the TCA cycle enzyme fumarase (Fig. 6A). Identification of fumarase from these experiments validated our azide swarming screen since the de novo fumC mutant (Fig. 6C) and 33H5 (Fig. 6D) displayed the same aberrant swarming phenotype (see Fig. S2 in the supplemental material), and both are unable to swarm on NaN3, a poison of aerobic respiration (Fig. 6F and G). The transposon insertion in mutant 25H1 (Table 2) disrupts the hybB gene, which encodes hydrogenase-2, an anaerobic cytochrome that catalyzes the oxidation of hydrogen into two protons and two electrons during fermentation. Mutant 11F4 disrupts PMI2646, which encodes a protein that has 75% identity to a quinone hydroxylase of Providencia rettgeri (Table 2). Interestingly, the azide-defective swarming mutant 31H4 has a disruption in rnb, which encodes an exonuclease II. This gene, however, is located approximately 1 kb downstream of genes encoding a predicted electron transport system, RnfABCDGE.
TABLE 2 .
Identification of P. mirabilis genes required for swarming on sodium azide at 37°C
| Mutant | ORF | Gene | Annotation |
|---|---|---|---|
| 37B5 | PMI3240 | argH | Argininosuccinate lyase |
| 33H5 | PMI1296 | fumC | Fumarase |
| 25H1 | PMI0033 | hybB | Hydrogenase 2b cytochrome subunit |
| 25A2 | PMI1807 | fadJ | Fatty acid oxidation complex subunit alpha |
| 11F4 | PMI2646 | Ubiquinone biosynthesis hydroxylase | |
| 31H4 | PMI1311 | rnb | Exoribonuclease II |
| 31F4 | PMI2801 | fkpA | FKBP-type peptidyl-prolyl cis-trans-isomerase |
| 28D2 | PMI2495 | holD | DNA polymerase subunit psi |
| 11D3 | PMI3143 | DNA modification methyltransferase | |
| 6C5 | PMI2669 | pacB | Putative periplasmic member of the SGNH-family of hydrolases |
| 6C6 | PMI1548 | cjrC | TonB-dependent receptor |
| 30B6 | PMI3180 | M23B family outer membrane metalloprotease | |
| 34A1 | PMI1378 | sapA | Peptide transport periplasmic protein |
| 35F4 | PMI3021 | mrcA | Peptidoglycan synthetase |
| 39H1 | PMI2993 | idsD | Hypothetical protein |
| 23D6 | Intergenic | 400-bp 3′ end of acrB, 200-bp 5′ PMI0130/31 biofilm regulator ybaJ/hemolysin modulating protein | |
| 25B6 | PMI1680 | rcsA | Colanic acid capsular biosynthesis activation protein |
| 26B3 | PMI2914 | Hemagglutinin |
In the azide swarming screen of P. mirabilis transposon mutants, genes encoding respiratory enzymes and genes unrelated to energy metabolism were both identified as a result of NaN3 sensitivity (Table 2). Although we expected to identify genes related to alternative respiratory chains or fermentative functions (discussed above), we were not surprised to also find disrupted genes that increased susceptibility to azide but are unrelated to swarming. In addition, we expected to find insertions in genes encoding cell wall machinery, LPS biosynthesis, and capsule production, because an altered permeability barrier might increase susceptibility to sodium azide. Mutants with decreased swarming diameters identified on NaN3 (Fig. 6A) included transposon mutant 25A2 in fadJ, which encodes an oxidation subunit involved in fatty acid metabolism (Table 2); 30B6 in PMI3180, which is adjacent (11 kb downstream) to the LPS O-antigen biosynthetic gene cluster; 25B6 with an insertion in the colonic acid capsular biosynthesis activation protein encoded by rcsA; and 35F4 in mrcA, which encodes a peptidoglycan synthetase (Table 2). Insertions in a hemagglutinin encoded by PMI2914; an FKBP-type peptidyl-prolyl cis-trans-isomerase encoded by fkpA; a periplasmic peptide transport protein; encoded by sapA; and mutant 23D6, in which the transposon was located in an intergenic region between acrB and ybaJ (Table 2) were also found in the screen (Fig. 6A).
Fermentation end products are not excreted during swarming.
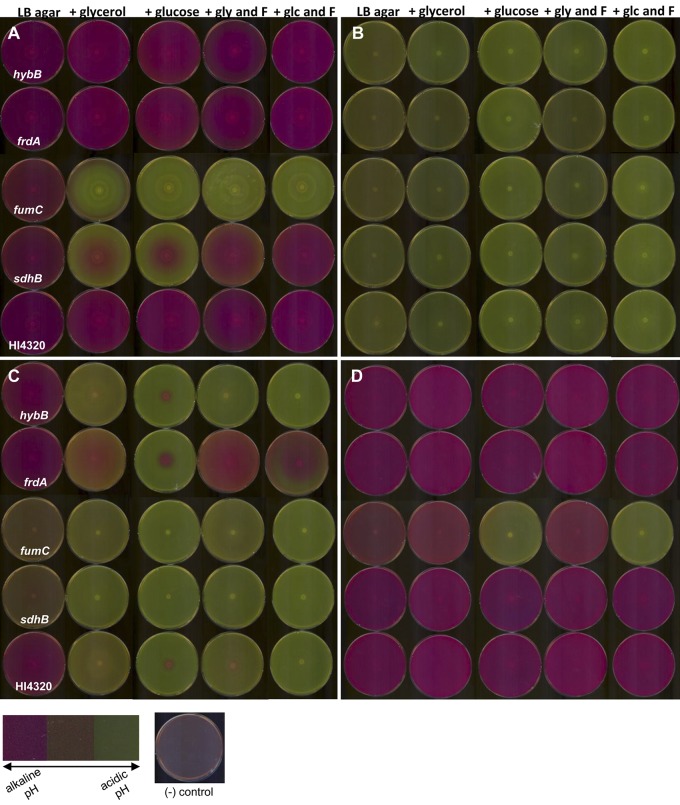
By screening P. mirabilis transposon mutants for loss of swarming on NaN3, a poison of aerobic respiration, we identified hybB and argH, which encode proteins involved in alternative anaerobic processes potentially required to generate energy for P. mirabilis swarming. However, the identification of an insertion in fumC supports our assertion that the enzyme is participating in the oxidative TCA cycle during P. mirabilis swarming. To test whether P. mirabilis swarming is also dependent on fermentation or is using a less canonical respiratory chain in conjunction with the complete aerobic TCA cycle, we used the pH indicator phenol red in LB-1.5% agar to examine swarming of P. mirabilis under aerobic and anaerobic conditions, because excretion of acidic end products is indicative of fermentation. A change in pH was determined by a visible color change of the agar from orange (neutral pH) to pink (alkaline pH) or to yellow (acidic pH). Under aerobic conditions at 30°C and 37°C, wild-type P. mirabilis HI4320 swarming created an alkaline environment (Fig. 7A), raising the surrounding pH, suggesting that the bacteria are not excreting acidic fermentation end products during swarming differentiation.
FIG 7 .
Oxidative TCA cycle mutants excrete acidic fermentation end products under aerobic conditions. The hybB, frdA, fumC, and sdhB mutants and wild-type HI4320 were observed on phenol red LB-1.5% agar with and without 0.2% (wt/vol) glycerol (gly) or 0.2% (wt/vol) glucose (glc) as carbon sources and 0.2% (wt/vol) fumarate (F) as an energy source where indicated. Inoculated plates were incubated at 37°C under aerobic conditions (A) and anaerobic conditions (B). In addition, fully developed swarm plates from anaerobic experiments were observed over time following exposure to aerobic conditions. Anaerobic plates were then maintained under aerobic conditions at room temperature and viewed at 6 (C) and 48 (D) h. Following approximately 6 h under aerobic conditions, the phenol red agar of wild-type HI4320 and the hybB and frdA mutants slowly turned alkaline; however, the fumC and sdhB mutants were still visibly acidic. Following approximately 48 h under aerobic conditions, all of the phenol red agar plates returned to alkaline conditions except for the fumC mutant when glucose was present as a carbon source. A color map depicting the color range from alkaline pH (pink) to acidic pH (yellow) is displayed bottom left. To the right of the color map is a reference image of an uninoculated agar plate following incubation under aerobic conditions at 37°C.
Although no exogenous urea was added to the plates, to ensure that the increase in pH was not the result of urease hydrolyzing urea to CO2 and ammonia, P. mirabilis mutations in ureC and argG, required for urease activity and the urea cycle, respectively, were examined on the phenol red agar (see Fig. S3 in the supplemental material). Both the urease mutant and argG mutant bacteria exhibited a similar color change of the agar from orange to pink, indicating alkaline conditions and suggesting that the formation of ammonia by urease was not responsible for the increase in pH by wild-type HI4320 (see Fig. S3).
Swarming of P. mirabilis under anaerobic conditions on phenol red agar, however, resulted in agar turning yellow, indicating an acidic pH (Fig. 7B). These results suggest that under anaerobic conditions, P. mirabilis is swarming using fermentation when a membrane electron acceptor is lacking. It is notable that both argH, which was identified in the screen (Table 2), and argG (see Fig. S3) are arginine auxotrophs, yet only argH had a defect in swarming on sodium azide, suggesting that it is the production of fumarate rather than arginine biosynthesis itself that is important for the energetics that govern swarming.
To better understand how fermentation and respiration produce energy during swarming, carbon source glycerol or glucose, with and without fumarate as an energy source, was added to agar containing phenol red. Under aerobic conditions at 37°C, wild-type HI4320 and sdhB, fumC, frdA, and hybA mutants alkalinized the phenol red agar to turn pink (Fig. 7A). Interestingly, when glycerol or glucose was added to the phenol red agar, the fumC and sdhB mutant bacteria acidified the agar, indicating fermentation (Fig. 7A). Addition of fumarate to the phenol red agar containing either glycerol or glucose rescued the pH of the sdhB mutant to an alkaline pH like that of the wild type (Fig. 7A). Under aerobic conditions, the hybB and frdA mutants remained alkaline under all conditions (Fig. 7A). Under anaerobic conditions at 37°C, wild-type HI4320 and sdhB, fumC, frdA, and hybA mutants were able to ferment and acidify the phenol red agar (Fig. 7B). After anaerobic incubation, plates were exposed to aerobic conditions for approximately 6 h, and wild-type HI4320 and the hybB and frdA mutants slowly turned the medium alkaline; however, the phenol red agar, plated with the fumC and sdhB mutants, remained acidic (Fig. 7C). While swarming of the sdhB mutant could be rescued to wild-type pH by the addition of exogenous fumarate, independent of carbon source and dependent on oxygen, fumC mutant bacteria were rescued to wild-type pH by glycerol, independent of fumarate, under anaerobic conditions (Fig. 7D). Taken together, these findings provide evidence that during swarming, the oxidative TCA cycle is required, because loss of fumarate reductase did not affect swarming under any condition tested, and fumarate was unable to rescue fumC mutant bacteria pH or periodicity during swarming.
DISCUSSION
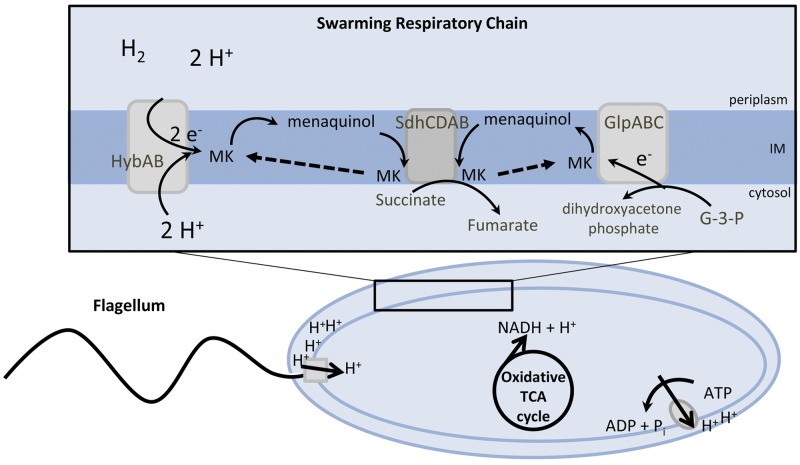
Swarming is a coordinated and multicellular behavior that is dependent upon respiration to generate a proton motive force that drives the rotation of bacterial flagella. Remarkably, P. mirabilis swarm cells are deficient in aerobic cytochromes (19) and swarm on inhibitory growth concentrations of sodium azide, an inhibitor of aerobic respiration. A reduction in membrane cytochromes in swarm cells is surprising, because bacterial propulsion, mediated by rotating flagella, is dependent on proton motive force. Mutations that disrupt genes encoding the TCA cycle enzymes succinate dehydrogenase and fumarase cause an aberrant swarming phenotype; however, disruption of the anaerobic, incomplete reductive TCA cycle enzyme fumarate reductase has no effect on the swarming process. This finding is unexpected, because, in most bacteria, succinate dehydrogenase functions in the complete TCA cycle when oxygen is available as a terminal electron acceptor, while fumarate reductase catalyzes the reverse reaction in the incomplete, reductive TCA cycle in the absence of aerobic respiration. Since we and others (19) observed that swarming is resistant to sodium azide, which inhibits the electron transfer to oxygen, this suggests that P. mirabilis is oxidizing pyruvate using the complete TCA cycle in conjunction with membrane respiration that lacks transfer of electrons to oxygen. Collectively, these findings and our results suggest that P. mirabilis employs an alternative anaerobic-like electron transport chain to reoxidize NAD+ from reduced NADH that is produced during the oxidation of pyruvate to CO2 in the complete TCA cycle and to generate proton motive force to drive flagellar rotation (Fig. 8).
FIG 8 .
Proposed model for P. mirabilis energy metabolism during swarming. The rotation of flagellum and oxidative phosphorylation are dependent on the proton gradient that is generated by membrane respiration to oxidize NADH to NAD+. The majority of NADH is formed during the complete oxidation of pyruvate to CO2 in the TCA cycle. The decreased interval between swarming and consolidation for aerobic TCA cycle mutants suggests that the capacity to reoxidize NAD+ and establish a proton gradient is due to inappropriate fermentation caused by loss of the complete TCA cycle and by the diminished capacity to metabolize fumarate. Extracytoplasmic substrate oxidation of dihydrogen and cytoplasmic reduction of glycerol could maintain the proton gradient and allow for reoxidation of NAD+ in the absence of electron transfer to oxygen. The ability to support motility without respiring oxygen may reflect the reduced energy demand caused by multicellular cooperation during swarming.
To elucidate potential components of an alternative anaerobic-like electron transport chain, we screened a collection of P. mirabilis transposon mutants to test for the loss of swarming only on sodium azide, a poison of the aerobic respiration chain. We found 18 mutants with a wild-type swarming phenotype on LB-1.5% agar but that displayed reduced swarming diameters on sodium azide; five mutants (33H5, 25H1, 11F4, 37B5, and 25A2) have disruptions in genes having an obvious association with respiration. The identification of fumarase, encoded by fumC, validated the azide swarming screen, because we previously noted that our isogenic fumC mutant was unable to swarm on sodium azide. Fumarase catalyzes the conversion of fumarate to malate during aerobic conditions and malate to fumarate during anaerobic conditions. Since succinate dehydrogenase is operating in the complete TCA cycle, and our findings suggest that during swarming, P. mirabilis is using an anaerobic-like respiratory chain to generate proton motive force, we believe that fumarase is most likely also operating in the complete TCA cycle, converting fumarate to malate. In addition, the enzyme argininosuccinate lyase, encoded by argH, was identified in the azide swarming screen and is required for the production of arginine and fumarate. Since a mutation in argG, which encodes argininosuccinate synthase, is also an arginine auxotroph but does not have a defective swarming phenotype on azide, the activity of argininosuccinate lyase encoded by argH is for production of the TCA cycle intermediate fumarate required for respiration.
hybB, which encodes hydrogenase-2, a known anaerobic respiratory chain component, catalyzes the extracytoplasmic oxidation of dihydrogen into two protons and two electrons, often during fermentation (23). The protons remain in the periplasm of the bacterium, while the electrons are shuttled to a quinone in the electron transport chain of the inner membrane. Simultaneously, two protons located in the bacterial cytoplasm are consumed by hydrogenase-2. The conservation of protons mediated by hydrogenase-2 is consistent with the hypothesis that P. mirabilis swarming is supported by an anaerobic-like respiratory chain, and the identification of hydrogenase-2 in the azide screen suggests that extracytoplasmic energy pathways are important for the physiology of P. mirabilis swarming differentiation (24). Our findings do not address the possibility that P. mirabilis reverses the F1F0 ATP synthase to pump protons during swarming; however, expending ATP generated by substrate-level phosphorylation during fermentations would generate proton motive force (Fig. 8).
An essential component of electron transport chains are hydrophobic quinones that carry electrons and protons during membrane respiration and act as redox mediators linking dehydrogenases and terminal reductases within the electron transport chain. Interestingly, a transposon mutant was identified in the P. mirabilis azide swarming screen that has 75% identity to a quinone hydroxylase of P. rettgeri. Eschereichia coli and related enteric bacteria are known to synthesize three different quinones: ubiquinone, menaquinone, and demethylmenaquinone. During aerobic respiration, ubiquinone is preferred, while during anaerobic conditions, menaquinone is used. Our findings suggest that P. mirabilis could use this predicted quinone hydroxylase to maintain pools of substrates to transfer electrons using the proposed anaerobic-like respiratory chain during swarming.
The fate of NADH, generated from glycolysis and primarily the TCA cycle, is dependent on which terminal electron acceptor is present: oxygen under aerobic conditions or an organic compound under anaerobic conditions. During glycolysis, triose phosphate isomerase, encoded by tpiA, reversibly isomerizes the ketone sugar dihydroxyacetone phosphate (DHAP) to the aldehyde sugar glycerol-3-phosphate (G3P). We have shown that the isogenic tpiA mutant results in a swarming-deficient phenotype, suggesting that both of these glycolysis intermediates, DHAP and G3P, are important during P. mirabilis swarming. Although not identified in the swarming azide screen, another known anaerobic respiratory chain component, glycerol-3-phosphate dehydrogenase, converts G3P to DHAP. It is possible that P. mirabilis utilizes glycerol-3-dehydrogenase to generate energy, because the tpiA mutant is unable to swarm and cannot reversibly isomerize DHAP and G3P. During anaerobic conditions, glycerol can be used as an energy source when fumarate is present as a terminal electron acceptor for fumarate reductase. Our findings show that fumarate reductase is not important for P. mirabilis swarming differentiation; however, future studies may reveal that succinate dehydrogenase fulfills this role.
In respiring cells, NADH and FADH2 are generated during metabolism during oxidation steps of the TCA cycle. To reoxidize these molecules, protons and electrons are shuttled via the electron transport chain within the bacterial cytoplasmic membrane. As electrons are transported through a series of membrane flavoprotein carriers, to a terminal electron acceptor, protons are translocated across the membrane. The proton gradient that is created during respiration contributes to the gradient of both pH and charge across the membrane and collectively determines proton motive force. It is during aerobic conditions with O2 as a terminal electron acceptor that the largest number of protons can be pumped, thereby producing the greatest proton motive force for the cell. During fermentation, however, bacteria are unable to respire and can generate a proton gradient by reversing the rotation of the F1F0 ATPase to consume ATP to pump protons out from the cytoplasm. Consequently, generation of proton motive force during anaerobic conditions is substantially more difficult for the cell compared to that during aerobic conditions (25).
Interestingly, TCA cycle intermediates also can affect patterns formed by motile colonies of E. coli and Salmonella (26, 27). The ability of succinate and fumarate to affect patterns in these bacteria has been suggested to result from the buildup of chemoattractant or chemorepellent (26, 28); however, we propose an alternative explanation: that manipulating concentrations of TCA cycle intermediates affects periodic swarming due to their effect on energy metabolism. This is likely true for P. mirabilis multicellular swarming, since active swarms transplanted onto fresh agar maintain their same periods (3). The connection between energy pathways that generate proton motive force and swarming has broad implications, because surface motility is a behavior shared by many bacteria (28, 29). Our findings suggest that these universal patterns of swarming are dictated by membrane energetics and imply that bacterial cooperation during the development of multicellular swarming decreases the energy required by individual bacteria to travel across a surface.
MATERIALS AND METHODS
Bacterial strains.
P. mirabilis HI4320 was cultured from the urine of a patient presenting with bacteriuria during long-term catheterization (30, 31). P. mirabilis HI4320 mutants (Table 1) were generated using the TargeTron gene knockout system (Sigma). The P. mirabilis tpiA mutant was complemented with a wild copy of the tpiA gene cloned into pGEN-MCS. The ureC (urease) and argG (argininosuccinate synthase) mutants were used as controls for phenol red experiments (32).
Culture media and swarming phenotype testing.
P. mirabilis was routinely cultured in LB medium. Swarming phenotypes of P. mirabilis mutants were compared with the wild-type HI4320 swarming phenotype on LB-1.5% (wt/vol) agar (10.0 g/liter NaCl) by spotting 5 µl of overnight culture onto the center of a plate, incubating under aerobic conditions at 37°C, and examining after 18 h. P. mirabilis HI4230 and TCA cycle mutants were examined under the same conditions on LB-1.5% agar containing a TCA intermediate: 10 mM succinate, 10 mM fumarate, or 10 mM malate. P. mirabilis HI4230, the TCA cycle mutants, and P. mirabilis transposon mutants were also examined on LB-1.5% agar containing inhibitory concentrations of sodium azide, 0.01% (wt/vol) and 0.005% (wt/vol), and incubated under aerobic conditions at 30°C and 37°C. Strain HI4320 was also tested on LB-1.5% agar containing sodium azide and 0.2% (wt/vol) glucose. P. mirabilis HI4320, the ureC mutant, and other metabolism mutants were examined on LB-1.5% agar containing 0.04 g/liter phenol red following incubation under aerobic and anaerobic (BD GasPak EZ Anaerobe) conditions at 30°C. LB-1.5% agar containing phenol red and 0.2% (wt/vol) fumarate-0.2% (wt/vol) glucose, phenol red and 0.2% (wt/vol) glucose alone, phenol red and 0.2% (wt/vol) fumarate-0.2% (wt/vol) glycerol, or phenol red and 0.2% (wt/vol) glycerol alone were tested under aerobic and anaerobic conditions (BD GasPak EZ anaerobe) at 37°C with wild-type P. mirabilis HI4330 and the TCA cycle mutants. For identification of P. mirabilis transposon mutants, arbitrary PCR products were cloned into pCR2.1-TOPO (Invitrogen) and maintained in E. coli TOP10 (Invitrogen). Antibiotics were added as necessary at the following concentrations: kanamycin, 25 µg/ml, and ampicillin, 100 µg/ml.
Sodium azide screen.
Swarming phenotypes of 1,920 P. mirabilis transposon mutants, previously created by us (21), were compared with the wild-type HI4320 swarming phenotype on LB-1.5% agar with and without 0.005% (wt/vol) sodium azide (NaN3), an inhibitor of electron transport. Swarming diameters were measured and documented from triplicate experiments performed on separate days. Transposon mutants unable to swarm on LB-1.5% agar alone were removed from further testing in the NaN3 screen. Arbitrary PCR and molecular cloning into pCR2.1-TOPO (Invitrogen) was carried out as previously described (21, 33) on P. mirabilis transposon mutants with a reduced swarming diameter or inhibited swarming phenotype on LB-1.5% agar containing NaN3 compared to HI4320 following incubation at 37°C under aerobic conditions. For these experiments, transposon mutant 8B5 with a disruption in aceE (pyruvate dehydrogenase), which is swarming deficient, was included as a nonswarming control.
SUPPLEMENTAL MATERIAL
P. mirabilis HI4320 oxidative TCA cycle mutants display aberrant swarming phenotypes in comparison to wild-type HI4320. A 5-µl volume of a liquid suspension from one overnight culture of wild-type P. mirabilis HI4320; fumC, sdhB, and frdA isogenic mutants; and 33H5 and 10E4 transposon mutants was individually spotted in the center of five swarm agar plates and incubated at 30°C for 18 h. Among the replicates, wild-type HI4320 and the frdA reductive TCA cycle mutant have similar swarming phenotypes. Transposon mutant 33H5 has a disruption within fumC which operates in both the reductive and oxidative TCA cycle and has an aberrant swarming phenotype similar to that of the fumC isogenic mutant. Transposon mutant 10E4 has a disruption in sdhC of the oxidative TCA cycle and has the same swarming phenotype as the sdhB mutant, which also operates during oxidative respiration. The swarming phenotypes of the fumC, 33H5, sdhB, and 10E4 mutants are different in comparison to those of wild-type HI4320 and the frdA mutant; an altered pattern of consolidation is visible for those that are required for oxidative respiration. Download Figure S1, TIF file, 2.3 MB.
The P. mirabilis HI4320 de novo fumC mutant and a transposon insertion in fumC are indistinguishable. Inoculation and swarming periodicity of wild-type P. mirabilis HI4320 and fumC and 33H5 mutants, when examined on the same swarm agar plate. (A) wild-type HI4320 and isogenic de novo fumC mutant; (B) wild-type HI4320 and isogenic transposon mutant 33H5; (C) the de novo fumC mutant and 33H5 produce identical phenotypes when inoculated on the same plate. Download Figure S2, TIF file, 3.1 MB.
P. mirabilis swarming is alkaline when examined on swarm agar containing phenol red. A visible color change of agar containing phenol red is observed when wild-type P. mirabilis HI4320 (A), P. mirabilis ureC mutant (B), and P. miribilis argG mutant (C) are spotted and grown aerobically. The color change from orange to pink indicates that swarming of wild-type HI4320 produces alkaline conditions. The ureC and argG mutants also exhibited the same color change on phenol red agar, indicating alkaline conditions and verifying that urease is not causing the change in pH. Download Figure S3, TIF file, 1.6 MB.
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service grants AI43363 and AI59722 from the National Institutes of Health.
Footnotes
Citation Alteri CJ, Himpsl SD, Engstrom MD, Mobley HLT. 2012. Anaerobic respiration using a complete oxidative TCA cycle drives multicellular swarming in Proteus mirabilis. mBio 3(6):e00365-12. doi:10.1128/mBio.00365-12.
REFERENCES
- 1. Jones BV, Young R, Mahenthiralingam E, Stickler DJ. 2004. Ultrastructure of Proteus Mirabilis swarmer cell rafts and role of swarming in catheter-associated urinary tract infection. Infect. Immun. 72:3941–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Douglas CW. 1979. Measurement of proteus cell motility during swarming. J. Med. Microbiol. 12:195–199 [DOI] [PubMed] [Google Scholar]
- 3. Matsuyama T, et al. 2000. Dynamic aspects of the structured cell population in a swarming colony of Proteus mirabilis. J. Bacteriol. 182:385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rauprich O, et al. 1996. Periodic phenomena in Proteus mirabilis swarm colony development. J. Bacteriol. 178:6525–6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rather PN. 2005. Swarmer cell differentiation in Proteus mirabilis. Environ. Microbiol. 7:1065–1073 [DOI] [PubMed] [Google Scholar]
- 6. Morgenstein RM, Rather PN. 2012. Role of the Umo proteins and the Rcs phosphorelay in the swarming motility of the wild type and an O-antigen (waaL) mutant of Proteus mirabilis. J. Bacteriol. 194:669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morgenstein RM, Szostek B, Rather PN. 2010. Regulation of gene expression during swarmer cell differentiation in Proteus mirabilis. FEMS Microbiol. Rev. 34:753–763 [DOI] [PubMed] [Google Scholar]
- 8. Belas R, Goldman M, Ashliman K. 1995. Genetic analysis of Proteus mirabilis mutants defective in swarmer cell elongation. J. Bacteriol. 177:823–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Belas R, Suvanasuthi R. 2005. The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein. J. Bacteriol. 187:6789–6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Armitage JP, Smith DG, Rowbury RJ. 1979. Alterations in the cell envelope composition of Proteus mirabilis during the development of swarmer cells. Biochim. Biophys. Acta 584:389–397 [DOI] [PubMed] [Google Scholar]
- 11. Morgenstein RM, Clemmer KM, Rather PN. 2010. Loss of the waaL O-antigen ligase prevents surface activation of the flagellar gene cascade in Proteus mirabilis. J. Bacteriol. 192:3213–3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gygi D, et al. 1995. A cell-surface polysaccharide that facilitates rapid population migration by differentiated swarm cells of Proteus mirabilis. Mol. Microbiol. 17:1167–1175 [DOI] [PubMed] [Google Scholar]
- 13. Hay NA, Tipper DJ, Gygi D, Hughes C. 1999. A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J. Bacteriol. 181:2008–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dufour A, Furness RB, Hughes C. 1998. Novel genes that upregulate the Proteus mirabilis flhDC master operon controlling flagellar biogenesis and swarming. Mol. Microbiol. 29:741–751 [DOI] [PubMed] [Google Scholar]
- 15. Cusick K, Lee YY, Youchak B, Belas R. 2012. Perturbation of FliL interferes with Proteus mirabilis swarmer cell gene expression and differentiation. J. Bacteriol. 194:437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pearson MM, Rasko DA, Smith SN, Mobley HL. 2010. Transcriptome of swarming Proteus mirabilis. Infect. Immun. 78:2834–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Armitage JP, Rowbury RJ, Smith DG. 1975. Indirect evidence for cell wall and membrane differences between filamentous swarming cells and short non-swarming cells of Proteus mirabilis. J. Gen. Microbiol. 89:199–202 [DOI] [PubMed] [Google Scholar]
- 18. Armitage JP. 1981. Changes in metabolic activity of Proteus mirabilis during swarming. J. Gen. Microbiol. 125:445–450 [DOI] [PubMed] [Google Scholar]
- 19. Falkinham JO, III, Hoffman PS. 1984. Unique developmental characteristics of the swarm and short cells of Proteus vulgaris and Proteus mirabilis. J. Bacteriol. 158:1037–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gabel CV, Berg HC. 2003. The speed of the flagellar rotary motor of Escherichia coli varies linearly with protonmotive force. Proc. Natl. Acad. Sci. U. S. A. 100:8748–8751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Himpsl SD, Lockatell CV, Hebel JR, Johnson DE, Mobley HL. 2008. Identification of virulence determinants in uropathogenic Proteus mirabilis using signature-tagged mutagenesis. J. Med. Microbiol. 57(Pt 9):1068–1078 [DOI] [PubMed] [Google Scholar]
- 22. Reams SG, Clark DP. 1988. Glucose repression of anaerobic genes of Escherichia coli is independent of cyclic AMP. FEMS Microbiol. Lett. 56:231–236 [Google Scholar]
- 23. Adams MW, Mortenson LE, Chen J-S. 1981. Hydrogenase. Biochim. Biophys. Acta 594:105–176 [DOI] [PubMed] [Google Scholar]
- 24. Hooper AB, DiSpirito AA. 1985. In bacteria which grow on simple reductants, generation of a proton gradient involves extracytoplasmic oxidation of substrate. Microbiol. Rev. 49:140–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hochachka PW, Mommsen TP. 1983. Protons and anaerobiosis. Science 219:1391–1397 [DOI] [PubMed] [Google Scholar]
- 26. Budrene EO, Berg HC. 1991. Complex patterns formed by motile cells of Escherichia coli. Nature 349:630–633 [DOI] [PubMed] [Google Scholar]
- 27. Budrene EO, Berg HC. 1995. Dynamics of formation of symmetrical patterns by chemotactic bacteria. Nature 376:49–53 [DOI] [PubMed] [Google Scholar]
- 28. Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57:249–273 [DOI] [PubMed] [Google Scholar]
- 29. Kearns DB. 2010. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8:634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mobley HL, Warren JW. 1987. Urease-positive bacteriuria and obstruction of long-term urinary catheters. J. Clin. Microbiol. 25:2216–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Warren JW, Tenney JH, Hoopes JM, Muncie HL, Anthony WC. 1982. A prospective microbiologic study of bacteriuria in patients with chronic indwelling urethral catheters. J. Infect. Dis. 146:719–723 [DOI] [PubMed] [Google Scholar]
- 32. Johnson DE, et al. 1993. Contribution of proteus Mirabilis urease to persistence, urolithiasis, and acute pyelonephritis in a mouse model of ascending urinary tract infection. Infect. Immun. 61:2748–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burall LS, et al. 2004. Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold. Infect. Immun. 72:2922–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
P. mirabilis HI4320 oxidative TCA cycle mutants display aberrant swarming phenotypes in comparison to wild-type HI4320. A 5-µl volume of a liquid suspension from one overnight culture of wild-type P. mirabilis HI4320; fumC, sdhB, and frdA isogenic mutants; and 33H5 and 10E4 transposon mutants was individually spotted in the center of five swarm agar plates and incubated at 30°C for 18 h. Among the replicates, wild-type HI4320 and the frdA reductive TCA cycle mutant have similar swarming phenotypes. Transposon mutant 33H5 has a disruption within fumC which operates in both the reductive and oxidative TCA cycle and has an aberrant swarming phenotype similar to that of the fumC isogenic mutant. Transposon mutant 10E4 has a disruption in sdhC of the oxidative TCA cycle and has the same swarming phenotype as the sdhB mutant, which also operates during oxidative respiration. The swarming phenotypes of the fumC, 33H5, sdhB, and 10E4 mutants are different in comparison to those of wild-type HI4320 and the frdA mutant; an altered pattern of consolidation is visible for those that are required for oxidative respiration. Download Figure S1, TIF file, 2.3 MB.
The P. mirabilis HI4320 de novo fumC mutant and a transposon insertion in fumC are indistinguishable. Inoculation and swarming periodicity of wild-type P. mirabilis HI4320 and fumC and 33H5 mutants, when examined on the same swarm agar plate. (A) wild-type HI4320 and isogenic de novo fumC mutant; (B) wild-type HI4320 and isogenic transposon mutant 33H5; (C) the de novo fumC mutant and 33H5 produce identical phenotypes when inoculated on the same plate. Download Figure S2, TIF file, 3.1 MB.
P. mirabilis swarming is alkaline when examined on swarm agar containing phenol red. A visible color change of agar containing phenol red is observed when wild-type P. mirabilis HI4320 (A), P. mirabilis ureC mutant (B), and P. miribilis argG mutant (C) are spotted and grown aerobically. The color change from orange to pink indicates that swarming of wild-type HI4320 produces alkaline conditions. The ureC and argG mutants also exhibited the same color change on phenol red agar, indicating alkaline conditions and verifying that urease is not causing the change in pH. Download Figure S3, TIF file, 1.6 MB.