ABSTRACT
Dermatophytes are a uniquely pathogenic group of fungi that cause most common fungal infections globally. The major cause of athlete’s foot is Trichophyton rubrum, a pathogen of human skin. A recent paper in this journal reported the sequencing and analysis of five additional genome sequences, including that of Trichophyton rubrum. These five join the existing two additional genome sequences to bring the total to seven dermatophyte genome sequences, a notable milestone in the study of these fungi. These additional genomes set the stage for future genome-supported studies on the biology, pathogenicity, and host specificity of this important group of pathogens. To predict how this future might play out, we review the history of Aspergillus genomics since the initial publication of the first three Aspergillus genome sequences in 2005, an event that stimulated important studies of the pathogenic Aspergillus species. From these 7 years of Aspergillus history, we offer some speculation on the future of dermatophyte studies supported by the genome sequences given the similarities, differences, and relative levels of support for studies in these two groups of fungi and the diseases they cause.
Commentary
A recent paper by Martinez et al. in this journal reported the genome sequencing and analysis of five additional dermatophyte species, bringing the total number to seven (1). In this commentary, we will situate this report in the context of current dermatophyte genomics and speculate on the future of the field based on the advances made in Aspergillus genomics after the first three Aspergillus genomes were sequenced in 2005 (2–4).
Dermatophytes are a uniquely pathogenic group of fungi that cause most common fungal infections globally (5). Dermatophytic fungi are contained within three genera, Trichophyton, Epidermophyton, and Microsporum. In the United States alone, millions of individuals seek treatment for dermatophyte infections annually, translating into an economic burden estimated at $400 million per year (6). Moreover, large-scale epidemics have been reported in American troops in conflicts in Europe and an urban childcare center outbreak (7, 8). The knowledge surrounding the mode by which these pathogens cause disease is insufficient, perhaps due to lack of research utilizing modern molecular tools. Due to this deficiency, the development of effective therapeutics has been stunted. Genetic tools have been underutilized in the characterization of these fungi, resulting in a lack of sequenced dermatophyte genomes and their pathogenicity (9).
As noted, seven whole-genome sequences of dermatophyte species have now been generated (see the Broad Institute’s Dermatophyte Comparative Database at http://www.broadinstitute.org/annotation/genome/dermatophyte-comparative/MultiHome.html): the nuclear genome and mitochondrial sequences of Microsporum canis, Microsporum gypseum, Trichophyton equinum, Trichophyton rubrum, and Trichophyton tonsurans (1), as well as the availability of Arthroderma benhamiae and Trichophyton verrucosum genome sequences (10). In their comparative study, Martinez et al. (1) report that the sequenced dermatophytes are enriched relative to other human-associated fungi with four gene families that contribute to their ability to cause disease, an observation that mirrors the original analysis of the first two dermatophyte genomes (10). These include (i) proteases, secreted to degrade skin, that reportedly act as virulence factors; (ii) kinases, including pseudokinases, that are involved in signaling necessary for adapting to the skin niche; (iii) secondary metabolites, compounds that act as toxins, immune system modulators, or signals in the interactions between fungus and host; and (iv) a class of proteins (LysM) that appear to bind and mask cell wall components and carbohydrates, thus avoiding the host’s immune response to the fungi. Overall, these genome sequence identifications are important for revealing genome components that have the potential to further our understanding of the pathogenicity of dermatophytes. The availability of these sequence and analysis data will provide researchers large amounts of useful information that will provide power to studies aimed to decipher and interpret the molecular basis of host colonization, invasion, and specialization.
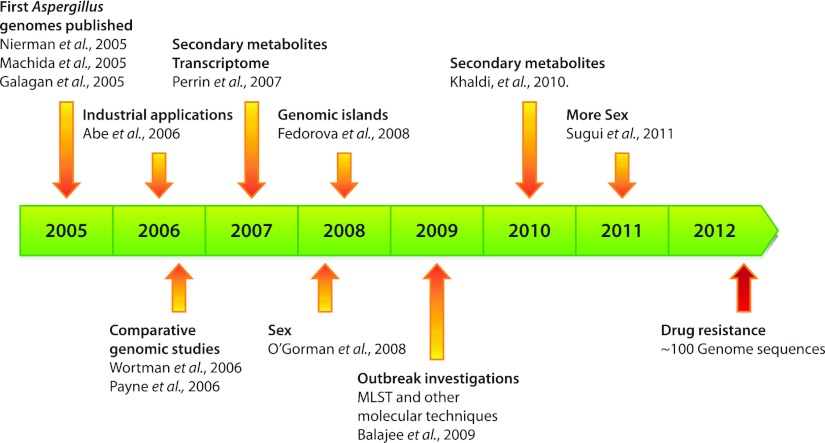
The observations about the dermatophyte genomes are reminiscent of the observations made on the first three Aspergillus genomes that were sequenced and analyzed. This is not surprising given that all dermatophytes and Aspergilli belong to the same phylum, Ascomycota. Characterization and analysis of many virulence-associated traits in Aspergillus species (1) may be useful in the search for such traits in dermatophyte genomes. Additionally, Aspergillus pathogens have been the subject of medically important research, targeting genes associated with replication cycles and secreted enzymes involved in secondary metabolite production. The genome sequences of Aspergillus fumigatus, Aspergillus nidulans, and Aspergillus oryzae were reported in back-to-back Nature papers in 2005 (2–4). Shortly after that publication event, the sequence of Aspergillus flavus was completed (11). A. fumigatus and A. flavus cause invasive aspergillosis in immunocompromised patients, an ability that positions them as the more important fungal pathogens in this group. A. flavus is also an important crop plant pathogen. All of these fungi but A. oryzae are environmental saprophytes whose niche is decaying plant material. A. oryzae, whose genome sequence revealed it to be essentially a derivative of ancestral A. flavus, has experienced centuries of human cultivation as a key ingredient in the production of sake, miso, soy sauce, and other Japanese foods. At the time of the genome sequence publications, all but A. nidulans were presumed to be asexual. As was the case for dermatophytes, the genome analysis of these aspergilli revealed several striking features, including a surprising abundance of secondary metabolite biosynthetic gene clusters, a full set of sexual cycle genes even in the presumed asexual strains, and an abundance of secreted degradative enzymes. The Aspergillus genomes inspired a burst of studies that leveraged these genome sequences, as overviewed in Fig. 1 (12–20). Subsequent post-genome sequence studies have revealed the identity of numerous products of the secondary-metabolite biosynthetic clusters and the roles of some of them in A. fumigatus and A. flavus virulence; have identified conditions for in vitro sexual cycles in A. fumigatus and A. flavus, which has led to genetic analysis studies of these organisms that are now under way; and have supported studies of the roles of many of the secreted proteases and other degradative enzymes in virulence. In addition, multiple strains of A. fumigatus and A. flavus have been sequenced, with the total of A. fumigatus sequences completed or under way approaching 100 (http://gsc.jcvi.org/projects/gsc/a_fumigatus/index.php).
FIG 1 .
Aspergillus genomics timeline. The timeline highlights some of the numerous critical studies since the publication of the first three Aspergillus genome sequences in 2005. The cited papers should be taken as representative, as no rigorous prioritization was imposed in selecting papers to highlight in the timeline.
Sexual reproduction has been suggested as a means to revamp the virulence of fungi via meiotic recombination, which increases the population diversity, and via mating on the human host. These may be associated with antifungal resistance or the rate of pathogenicity of dermatophytes (21). Like the story of sex in A. fumigatus, some dermatophytes, which were once assumed to be asexual, have been demonstrated to possess sexual cycles as well (22). Based on these studies, prediction of unexposed sexual cycles can be assumed from the dermatophytes containing functional sex genes. Recently, identification of the mating type locus (MAT) of five dermatophytes (M. canis, M. gypseum, T. equinum, T. rubrum, and T. tonsurans) with comparable virulence were reported using bioinformatic tools (23). Furthermore, successful mating of T. rubrum with Arthroderma simii suggests that these species have the benefits of sex, including cross-species sexual recombination and adaptation, that may outweigh the efficiencies of an asexual clonal expansion (24). In the Aspergillus species, the APN2 and SLA2 genes, encoding a DNA lyase and cytoskeleton protein, flank the MAT loci (25). However, those MAT genes for dermatophytes are essentially identical and linked on one side of the MAT locus (23). The discovery and characterization of the MAT locus of dermatophytes allows further studies in the pathogenesis to be explored.
The role of LysM proteins was noted for protecting dermatophytes from host immune detection. The importance of these proteins in avoiding detection by the host immune system is supported by the observation that during dermatophyte infection, defective or absent cell-mediated immunity predisposes the host to chronic or recurrent dermatophyte infection (26). Previously, expression of hydrophobin has been demonstrated to inhibit immune recognition in A. fumigatus (27). Dermatophytes A. benhamiae and T. verrucosum, both shown to activate human inflammatory infections, also display moderate expression of a surface hydrophobin gene, suggesting a possible role in immune response functions (10).
Discovery of the abundance of secondary-metabolite biosynthetic clusters in the Aspergillus genomes has led to the identification of the products of many of these clusters and the roles of some of them in virulence. A similar abundance of these clusters has now been noted in the reported dermatophyte genome sequences. For example, melanin, which is an important virulence determinant in Aspergillus (28), was also isolated from dermatophytes (M. canis, M. gypseum, T. equinum, T. rubrum, and T. tonsurans) in vitro and during infection, suggesting a similar role in Aspergillus and dermatophyte pathogenesis (29). Moreover, T. rubrum produces xanthomegnin, a toxin produced by Aspergillus in culture and in the human host (30). Transcriptome analysis revealed differential expression of secondary-metabolite genes during dermatophyte and Aspergillus infections, underscoring their importance in the colonization of tissues and potentially in the manipulation of the host inflammatory response (30). Future studies will undoubtedly leverage the genome sequences of these clusters in dermatophytes to identify their secondary-metabolite products and their potential specific roles in virulence.
Given the recent major progress in the development of broad-scale transcriptional and genome sequence-dependent analyses of dermatophytes (10, 30–32) and a selection of functionally characterized genes (33, 34), full genome sequences will fulfill a critical urgency in the need to develop molecular genetic techniques to study these pathogens. Molecular studies of dermatophyte genomics and pathogenicity have been undertaken in spite of the limited number of sequenced genomes. For example, Vermout and colleagues used RNA silencing as a potential functional genomics tool in M. canis to identify two proteases, SUB3 and DPPIV, coding for subtilisin and dipeptidyl peptidase, respectively (35). Previous studies have also demonstrated the association of increased keratinase with increased disease symptoms in M. canis (36). Several studies have used proteomics to characterize secreted and conidial proteins in T. rubrum, A. benhamiae, and M. canis (10, 37, 38), but these have been limited in number and applicability by the lack of genome sequence. Now that they can be coupled to genome sequences, these and other “omics” methods, such as metabolomics, glycomics, and lipidomics, will be more powerful, and accordingly, will strengthen the understanding and characterization of dermatophyte pathogenesis.
It is clear that the availability of additional dermatophyte genomes will accelerate and enhance molecular studies of these pathogenic fungi. It is therefore most appropriate to celebrate the publication of these new dermatophyte genomes and to note this event as a consequential milestone in the efforts to manage the terrible diseases caused by this group of fungi. The aspergilli and the dermatophytes are closely related, and the new dermatophyte genome sequences reveal features similar to those in the aspergilli. This observation suggests commonality in how these fungi survive and thrive in a mammalian host. Specific features of the dermatophytes and aspergilli diseases—such as invasiveness, fatality, and organ involvement—have resulted in research communities with disproportionate funding support that favored more rapid advancement in the aspergilli than in dermatophytes. Another important factor that favored the aspergilli was the strength of A. nidulans as a model organism and the mature community that had developed around this model prior to the genome sequence publications. However, studies of these two groups of fungi have been, and will continue to be, synergistic, with each community taking lessons from the other. We project that the rate of progress in dermatophyte genomic research will accelerate now in much the same way Aspergillus research accelerated following the publication of the Aspergillus genomes in 2005 and 2006. We look forward to all the exciting and significant findings yet to come.
Footnotes
Citation Rivera ZS, Losada L, Nierman WC. 2012. Back to the future for dermatophyte genomics. mBio 3(6):e00381-12. doi:10.1128/mBio.00381-12.
REFERENCES
- 1. Martinez DA, et al. 2012. Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. mBio 3(5):e00259–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nierman WC, et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156 [DOI] [PubMed] [Google Scholar]
- 3. Machida M, et al. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161 [DOI] [PubMed] [Google Scholar]
- 4. Galagan JE, et al. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115 [DOI] [PubMed] [Google Scholar]
- 5. Weitzman I, Summerbell RC. 1995. The dermatophytes. Clin. Microbiol. Rev. 8:240–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith ES, Fleischer AB, Jr, Feldman SR. 1998. Nondermatologists are more likely than dermatologists to prescribe antifungal/corticosteroid products: an analysis of office visits for cutaneous fungal infections, 1990–1994. J. Am. Acad. Dermatol. 39:43–47 [DOI] [PubMed] [Google Scholar]
- 7. Burzykowski T, et al. 2003. High prevalence of foot diseases in Europe: results of the Achilles Project. Mycoses 46:496–505 [DOI] [PubMed] [Google Scholar]
- 8. Abdel-Rahman SM, Simon S, Wright KJ, Ndjountche L, Gaedigk A. 2006. Tracking Trichophyton tonsurans through a large urban child care center: defining infection prevalence and transmission patterns by molecular strain typing. Pediatrics 118:2365–2373 [DOI] [PubMed] [Google Scholar]
- 9. Grumbt M, Monod M, Staib P. 2011. Genetic advances in dermatophytes. FEMS Microbiol. Lett. 320:79–86 [DOI] [PubMed] [Google Scholar]
- 10. Burmester A, et al. 2011. Comparative and functional genomics provide insights into the pathogenicity of dermatophytic fungi. Genome Biol. 12:R7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Payne G, et al. 2006. Whole genome comparison of Aspergillus flavus and A. oryzae. Med. Mycol. 44:9–12 [DOI] [PubMed] [Google Scholar]
- 12. Abe K, Gomi K, Hasegawa F, Machida M. 2006. Impact of Aspergillus oryzae genomics on industrial production of metabolites. Mycopathologia 162:143–153 [DOI] [PubMed] [Google Scholar]
- 13. Wortman J, et al. 2006. Whole genome comparison of the A. fumigatus family. Med. Mycol. 44:3–7 [DOI] [PubMed] [Google Scholar]
- 14. Perrin RM, et al. 2007. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 3:e50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fedorova ND, et al. 2008. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 4:e1000046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O’Gorman CM, Fuller HT, Dyer PS. 2008. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 457:471–474 [DOI] [PubMed] [Google Scholar]
- 17. Balajee SA, et al. 2009. Sequence-Based Identification of Aspergillus, Fusarium, and Mucorales Species in the Clinical Mycology Laboratory: Where Are We and Where Should We Go from Here?. J. Clin. Microbiol. 47:877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khaldi N, et al. 2010. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 47:736–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fisher MC, Henk DA. 2012. Sex, drugs and recombination: the wild life of Aspergillus. Mol. Ecol. 21:1305–1306 [DOI] [PubMed] [Google Scholar]
- 20. Sugui JA, et al. 2011. Identification and characterization of an Aspergillus fumigatus “supermater” pair. mBio 2(6):e00234–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsueh YP, Heitman J. 2008. Orchestration of sexual reproduction and virulence by the fungal mating-type locus. Curr. Opin. Microbiol. 11:517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heitman J. 2010. Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host Microbe 8:86–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li W, Metin B, White TC, Heitman J. 2010. Organization and evolutionary trajectory of the mating type (MAT) locus in dermatophyte and dimorphic fungal pathogens. Eukaryot. Cell 9:46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anzawa K, Kawasaki M, Mochizuki T, Ishizaki H. 2010. Successful mating of Trichophyton rubrum with Arthroderma simii. Med. Mycol. 48:629–634 [DOI] [PubMed] [Google Scholar]
- 25. Fraser JA, et al. 2007. Evolution of the mating type locus: insights gained from the dimorphic primary fungal pathogens histoplasma capsulatum, Coccidioides immitis, and Coccidioides posadasii. Eukaryot. Cell 6:622–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Almeida SR. 2008. Immunology of dermatophytosis. Mycopathologia 166:277–283 [DOI] [PubMed] [Google Scholar]
- 27. Aimanianda V, et al. 2009. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460:1117–1121 [DOI] [PubMed] [Google Scholar]
- 28. Youngchim S, Morris-Jones R, Hay RJ, Hamilton AJ. 2004. Production of melanin by Aspergillus fumigatus. J. Med. Microbiol. 53:175–181 [DOI] [PubMed] [Google Scholar]
- 29. Youngchim S, Pornsuwan S, Nosanchuk JD, Dankai W, Vanittanakom N. 2011. Melanogenesis in dermatophyte species in vitro and during infection. Microbiology 157:2348–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Staib P, et al. 2010. Differential gene expression in the pathogenic dermatophyte Arthroderma benhamiae in vitro versus during infection. Microbiology 156:884–895 [DOI] [PubMed] [Google Scholar]
- 31. Giddey K, et al. 2007. Comprehensive analysis of proteins secreted by Trichophyton rubrum and Trichophyton violaceum under in vitro conditions. J. Proteome Res. 6:3081–3092 [DOI] [PubMed] [Google Scholar]
- 32. Liu T, et al. 2007. The use of global transcriptional analysis to reveal the biological and cellular events involved in distinct development phases of Trichophyton rubrum conidial germination. BMC Genomics 8:100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamada T, Makimura K, Abe S. 2006. Isolation, characterization, and disruption of dnr1, the AreA/Nit-2-like nitrogen regulatory gene of the zoophilic dermatophyte, Microsporum canis. Med. Mycol. 44:243–252 [DOI] [PubMed] [Google Scholar]
- 34. Fachin AL, Ferreira-Nozawa MS, Maccheroni W, Jr, Martinez-Rossi NM. 2006. Role of the ABC transporter TruMDR2 in terbinafine, 4-nitroquinoline N-oxide and ethidium bromide susceptibility in Trichophyton rubrum. J. Med. Microbiol. 55:1093–1099 [DOI] [PubMed] [Google Scholar]
- 35. Vermout S, et al. 2007. RNA silencing in the dermatophyte Microsporum canis. FEMS Microbiol. Lett. 275:38–45 [DOI] [PubMed] [Google Scholar]
- 36. Viani FC, Dos Santos JI, Paula CR, Larson CE, Gambale W. 2001. Production of extracellular enzymes by Microsporum canis and their role in its virulence. Med. Mycol. 39:463–468 [DOI] [PubMed] [Google Scholar]
- 37. Sriranganadane D, et al. 2011. Identification of novel secreted proteases during extracellular proteolysis by dermatophytes at acidic pH. Proteomics 11:4422–4433 [DOI] [PubMed] [Google Scholar]
- 38. Leng W, et al. 2008. Proteomic profile of dormant Trichophyton rubrum conidia. BMC Genomics 9:303 [DOI] [PMC free article] [PubMed] [Google Scholar]