ABSTRACT
Group A streptococcus (GAS) causes human pharyngitis and invasive infections and frequently colonizes individuals asymptomatically. Many lines of evidence generated over decades have shown that the hyaluronic acid capsule is a major virulence factor contributing to these infections. While conducting a whole-genome analysis of the in vivo molecular genetic changes that occur in GAS during longitudinal human pharyngeal interaction, we discovered that serotypes M4 and M22 GAS strains lack the hasABC genes necessary for hyaluronic acid capsule biosynthesis. Using targeted PCR, we found that all 491 temporally and geographically diverse disease isolates of these two serotypes studied lack the hasABC genes. Consistent with the lack of capsule synthesis genes, none of the strains produced detectable hyaluronic acid. Despite the lack of a hyaluronic acid capsule, all strains tested multiplied extensively ex vivo in human blood. Thus, counter to the prevailing concept in GAS pathogenesis research, strains of these two serotypes do not require hyaluronic acid to colonize the upper respiratory tract or cause abundant mucosal or invasive human infections. We speculate that serotype M4 and M22 GAS have alternative, compensatory mechanisms that promote virulence.
IMPORTANCE
A century of study of the antiphagocytic hyaluronic acid capsule made by group A streptococcus has led to the concept that it is a major virulence factor contributing to human pharyngeal and invasive infections. However, the discovery that some strains that cause abundant human infections lack hyaluronic acid biosynthetic genes and fail to produce this capsule provides a new stimulus for research designed to understand the group A streptococcus factors contributing to pharyngeal infection and invasive disease episodes.
Observation
Streptococcus pyogenes (group A streptococcus [GAS]) causes significant human morbidity and mortality worldwide. The organism is the most common cause of bacterial pharyngitis and is responsible for severe invasive infections such as necrotizing fasciitis and toxic shock syndrome. GAS strains express a large number of protein virulence factors that contribute to pathogen-host interactions and disease. In addition, GAS strains produce a hyaluronic acid (HA) capsule that has been studied extensively since its discovery more than 100 years ago (1). The capsule is antiphagocytic (2, 3) and is acknowledged to be a key virulence factor contributing to pharyngeal and invasive infections (4–7). GAS capsule production requires two genes (hasA and hasB) for biosynthesis (6). These two genes are located in a three-gene operon that also includes hasC. Genetic inactivation of has genes significantly reduces virulence in multiple animal models of GAS infection (4–6), including pharyngeal colonization of nonhuman primates (7). Decades of study have led to the concept that most GAS strains produce HA capsule, and many investigators believe that it is required for virulence. Here we report that all serotype M4 and M22 strains studied from cases of pharyngitis and invasive disease lack the hasABC capsule gene operon, produce no detectable HA capsule, and multiply extensively ex vivo in human blood.
Serotype M4 and M22 strains lack the hasABC genes.
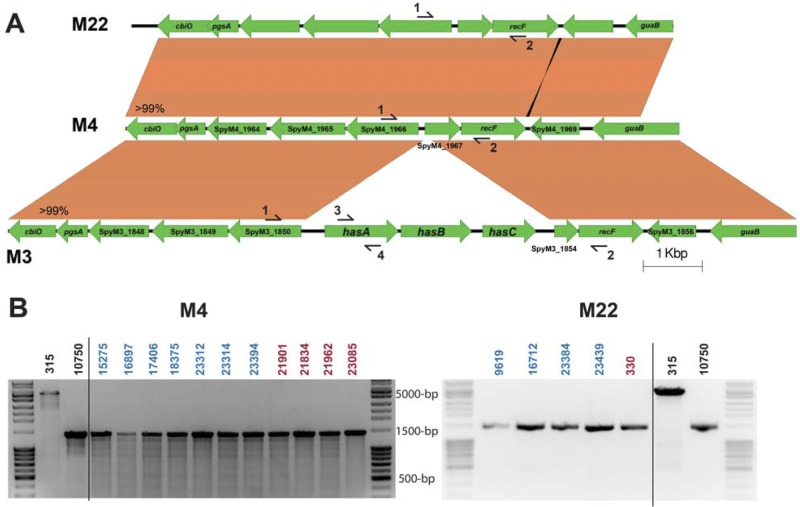
Recent studies reported that inactivating mutations arise in the hasA and hasB capsule biosynthesis genes during longitudinal nonhuman primate (8) and human (our unpublished data) pharyngeal infection. In a study designed to better understand the genetic changes that occur in vivo during human mucosal interaction with GAS, we obtained full-genome sequence data from 23 serotype M4 isolates and 8 serotype M22 isolates cultured sequentially from three and two subjects, respectively. We discovered that the genomes of these serotype M4 strains and a reference M4 genome (strain MGAS10750; accession number NC_008024.1) lack the hasABC operon genes (Fig. 1A). In addition, no hasABC homologues were identified in the MGAS10750 genome using the basic local alignment search tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi). No reference genome sequence is available for a serotype M22 GAS strain. Therefore, we used the MOSAIK assembler to align all genome sequence reads for the eight M22 isolates with the hasABC operon from the serotype M3 reference genome MGAS315, which is highly conserved in all sequenced genomes (see Fig. S1 in the supplemental material). None of these eight strains contained sequences that aligned with the hasABC operon.
FIG 1.
Serotype M4 and M22 GAS (group A streptococcus) strains lack the genes for hyaluronic acid capsule biosynthesis. (A) Schematic of the alignment of the hasABC region and flanking DNA sequence in serotypes M3 (MGAS315), M22 (MGAS23384), and M4 (MGAS10750). Homologous regions were extracted from the reference genomes for serotype M3 and M4 strains. The identical region in serotype M22 was obtained by de novo assembly of short-read genome sequences of strain MGAS23384 using the serotype M4 reference genome as a scaffold and confirmed by Sanger sequencing. The regions flanking the hasABC operon in the reference serotype M3 are >99% identical to the reference genome of serotype M4. Likewise, the serotype M22 sequence is >99% identical to that of the serotype M4 reference sequence. Primer positions for targeted PCR are identified by numbered arrows. (B) Targeted PCR of the hasABC operon using primers 1 and 2 in a subset of serotype M4 (left) and M22 (right) isolates of both pharyngeal (blue) and invasive (red) origin. An approximately 5,000-bp fragment containing hasABC was amplified in strain MGAS315, and a 1,500-bp fragment was amplified from MGAS10750 and strains lacking hasABC.
Microbial capsular polysaccharides vary in sugar residue composition and the glycosidic bonds that link them. No GAS isolate has been described that produces an alternative (i.e., non-HA acid) polysaccharide capsule. It is conceivable that the M4 and M22 GAS strains studied have genes for synthesis of alternative capsules. We next tested for the presence of alternative capsule biosynthesis genes using de novo assembly (Edena) (9) of unmapped genome sequencing reads from each of the 31 serotype M4 and M22 isolates. The resulting assembled contigs were used to query the nonredundant protein sequences (BLASTx) at NCBI. However, no homologues of genes encoding proteins known to participate in capsule production in Streptococcus pneumoniae (10), Streptococcus agalactiae (11), or other microbes were identified. Thus, the genome sequences of these serotype M4 and M22 strains lack genes necessary for production of HA or other known capsule types.
HA capsule biosynthesis genes are absent in a large sample of serotype M4 and M22 GAS strains causing pharyngitis and invasive infections.
The sequential GAS isolates examined originated from five epidemiologically independent subjects. However, this relatively small sample of isolates may not represent the breadth of diversity present in serotype M4 and M22 GAS strains. Strains of these two serotypes are frequent causes of pharyngitis and invasive GAS disease (12). For example, serotype M4 and M22 GAS combined caused approximately 14% of pharyngitis cases in a recent study of cases in Ontario, Canada (13). To determine if a lack of HA capsule genes was a common feature of these strains, we performed targeted PCR of the hasABC operon in 477 serotype M4 and M22 strains from diverse localities (see Tables S1 and S2 in the supplemental material). The DNA sequence flanking the hasABC operon in all sequenced GAS strains is highly conserved (Fig. 1A). Thus, we designed primers to amplify the region containing hasABC or the intervening sequence in strains lacking hasABC (Fig. 1A). We studied 401 serotype M4 and 76 serotype M22 pharyngitis GAS isolates from Ontario, Canada, and Houston, TX, recovered between 2002 and 2010. In addition, targeted PCR was performed on 11 invasive serotype M4 and 3 invasive serotype M22 GAS strains from the United States (see Tables S1 and S2 in the supplemental material). As assessed by PCR, none of the strains studied had the hasABC genes (Fig. 1B).
It is possible that the HA capsule biosynthesis genes are located in a different region of the genome in these strains. Therefore, next we performed targeted PCR of the hasA gene (Fig. 1A) (14). Consistent with the hasABC operon results, none of the M4 or M22 strains yielded an amplification product (data not shown). Moreover, serotype M4 and M22 strains did not produce detectable HA as assayed with a commercially available kit (Table 1). Given the geographic and temporal diversity of the strains examined, our data suggest that most or all serotype M4 and M22 GAS strains circulating in the regions studied lack the genes necessary for HA capsule biosynthesis and do not make an HA capsule.
TABLE 1 .
GAS strains used for hyaluronic acid and bactericidal assays
| MGASa strain | Serotype | Year | Infection type/Source | HA (ng/ml) | Reference |
|---|---|---|---|---|---|
| Serotype M3 | |||||
| 315 | M3 | 1991 | Toxic shock-like | 22,143 | 23 |
| 315ΔhasA | M3 | None | This study | ||
| Serotype M4 | |||||
| 10750 | M4 | 2001 | Pharyngitis | None | 24 |
| 15275 | M4 | 2002 | Pharyngitis | None | 13 |
| 16897 | M4 | 2007 | Pharyngitis | None | 13 |
| 17406 | M4 | 2008 | Pharyngitis | None | 13 |
| 18375 | M4 | 2009 | Pharyngitis | None | 13 |
| 21834 | M4 | 2009 | Bacteremia | None | This study |
| 21901 | M4 | 2009 | Invasive | None | This study |
| 21962 | M4 | 2010 | Soft tissue | None | This study |
| 23085 | M4 | 2010 | Soft tissue | None | This study |
| 23312 | M4 | Unknown | Pharyngeal | None | 21 |
| 23314 | M4 | Unknown | Pharyngeal | None | 21 |
| 23394 | M4 | Unknown | Pharyngeal | None | 21 |
| Serotype M22 | |||||
| 330 | M22 | 1991 | Invasive | None | 23 |
| 9619 | M22 | 2005 | Pharyngeal | None | 22 |
| 16712 | M22 | 2006 | Pharyngeal | None | 22 |
| 23384 | M22 | Unknown | Pharyngeal | None | 21 |
| 23439 | M22 | Unknown | Pharyngeal | None | 21 |
Musser group A streptococcus.
Serotype M4 GAS strains grow extensively in human blood despite failure to make an HA capsule.
The antiphagocytic function of the GAS HA capsule is generally believed to be an essential virulence factor for invasive infections in mice (4, 6). However, many of the strains we studied had been cultured from patients with invasive infections, and M4 and M22 strains have been reported in epidemiological studies of sterile-site infections (12, 15). Therefore, we hypothesized that these GAS strains would persist in human blood in the absence of capsule. Consistent with the hypothesis, all serotype M4 strains tested grew extensively in donor blood ex vivo (Fig. 2).
FIG 2.
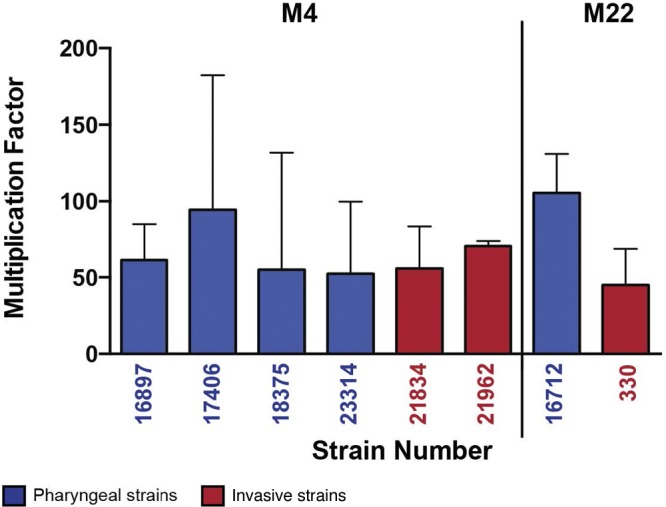
Multiplication of representative serotype M4 and M22 GAS strains in human blood. Human blood was inoculated with randomly selected pharyngeal (blue) and invasive (red) GAS strains as described in the text. Error bars represent standard deviations. Data show multiplication after growth in blood from a single donor, tested in triplicate. Similar results were obtained with an additional donor.
Comment.
The GAS capsule has been the subject of intense investigation since it was discovered by Bordet more than 100 years ago (16). Many lines of evidence have resulted in the prevailing idea that HA capsule is required for colonization of the upper respiratory tract and production of invasive infections in the animal models studied (4–7). Thus, our findings that strains of serotypes M4 and M22 cultured from diseased humans lack HA capsule and biosynthetic genes and proliferate ex vivo in blood were unexpected and run counter to prevailing ideas about GAS pathogenesis. Clearly, HA production is not required for virulence, nor does it contribute to interactions between the pathogen and host in strains of these two M protein serotypes.
Our study raises the question of the molecular mechanism(s) accounting for the absence of the hasABC genes in these strains, and several scenarios are possible. The three genes are contiguous, suggesting that they could be lost en bloc. Genome-wide comparison of M4 and M22 strains indicates that they differ from one another by >10,000 single-nucleotide polymorphisms that are widely dispersed throughout the chromosome. This magnitude of genetic diversity means that they do not share a recent common ancestor. If gene loss is responsible, it could have happened independently on two occasions or on only one occasion long ago, before the subsequent extensive genome-wide divergence of M4 and M22 strains. With the information at hand, we lack the ability to differentiate between these scenarios; that is, both are plausible. The absence of key capsule biosynthesis genes has been observed in Streptococcus pneumoniae (17), group B streptococcus (Streptococcus agalactiae) (18), Haemophilus influenzae (19), and Neisseria meningitidis (20). The data presented here add GAS to the list of bacterial pathogens that have subpopulations that have apparently lost the capacity for capsule synthesis but retained the ability to colonize human mucosal surfaces and cause sterile-site infections.
Serotype M4 and M22 GAS have been recovered from patients in many countries (12). Although it is possible that some strains of these serotypes that we have not studied produce an HA capsule, our data show that GAS strains lacking the capacity to make the HA capsule are geographically widespread. This observation, coupled with epidemiology surveillance reports indicating that serotype M4 and M22 strains can be abundant (12, 15), means that HA-negative organisms have the capacity to successfully spread between humans and persist. Despite the longstanding idea that the HA capsule is critical in pharyngeal infection, our data suggest that serotype M4 and M22 GAS strains may have alternative molecular mechanisms contributing to their success in pathogen-host interaction in the upper respiratory tract and invasive infections. Studies are under way to test this idea.
Bacterial strains and culturing methods.
Strains used in this study are listed in Tables S1 and S2 in the supplemental material. Serotype M4 and M22 strains used for whole-genome sequencing were cultured from subjects enrolled in a study to examine the human immune response to GAS (21). Pharyngitis strains were identified in an epidemiological study conducted in Ontario, Canada (13), or were isolated in a study of pediatric pharyngitis in Houston, TX (22). Invasive serotype M4 GAS isolates were identified during outbreak surveillance in Montana. Invasive serotype M22 GAS strains were identified in a previous study (23). The reference M4 GAS strain (MGAS10750) was isolated from a patient with pharyngitis in Florida in 2001 (24). The serotype M3 GAS strain MGAS315 was cultured from a patient with a toxic shock-like syndrome in Texas in 1991 (23). An HA capsule-deficient isogenic mutant strain was generated from wild-type strain MGAS315 by insertional inactivation of hasA with a spectinomycin resistance cassette as previously described (25). Strains used for HA and bactericidal assays are shown in Table 1. GAS strains were grown in Todd-Hewitt broth supplemented with 2% yeast extract (THY) or on Trypticase soy agar supplemented with 5% sheep blood (SBA) at 37°C with 5% CO2. When needed, spectinomycin was added at 150 µg/ml.
Genome sequencing of sequential pharyngeal strains.
Methods for genome sequencing and data processing and analysis have been described (8).
Targeted PCR of the hasABC operon.
Amplification of target genes was carried out as previously described (13) with the following modifications. Phusion DNA polymerase (New England Biolabs, Ipswich, MA) was used to amplify the hasABC operon or hasA in serotype M4 and M22 GAS or serotype M3 GAS using primers 1 (5′-TTTGCAATTAGTTCTGGGCT-3′) and 2 (5′-CAATTCTTTATCAGCCCTCG-3′) or 3 (5′-TAATCTATTAACGCGACTTA-3′) and 4 (5′-TGATGGATAAAAAGGCAAAAAG-3′), respectively. Primers 1 and 2 reside in the regions flanking the hasABC operon and amplify an approximately 5,000-bp fragment in serotype M3 GAS. These two primer sites are conserved in serotype M4 and M22 strains but amplify a 1,500-bp fragment due to the absence of the hasABC genes (Fig. 1A).
Hyaluronic acid assays.
GAS strains were grown to mid-exponential phase in 10 ml of THY, harvested by centrifugation, washed twice with sterile water, and suspended in 500 µl of sterile water. The cells were lysed with chloroform, and the aqueous phase was assayed for HA using an enzyme-linked binding-protein assay according to the manufacturer’s instructions (hyaluronic acid test kit; Corgenix, Broomfield, CO). As a positive control, we used the HA capsule-producing serotype M3 strain MGAS315. A capsule-negative strain of MGAS315 made by insertional inactivation of hasA (MGAS315ΔhasA) was used as a negative control.
Bactericidal assays.
The bactericidal assays were conducted as described by Lancefield (26). At least two healthy nonimmune adult donors were used for each experiment. Bacteria were grown to mid-exponential phase, pelleted, and suspended in an equal volume of phosphate-buffered saline (PBS). Each strain was subsequently diluted to approximately 1 × 103 CFU, from which 10 to 100 CFU of GAS was used to inoculate 300 µl of fresh human blood. Samples were incubated at 37°C with 5% CO2 with gentle rotation for 3 h, serially diluted in PBS, and immediately plated on SBA. The growth multiplication factor was calculated by dividing the number of CFU/ml after 3 h of incubation by the number in the starting inoculum. Experiments assessing the ability of GAS to grow in human blood were conducted under a human subject protocol approved by The Methodist Hospital Research Institute Institutional Review Board.
SUPPLEMENTAL MATERIAL
The hasABC operon of GAS is highly conserved between sequenced serotypes. (A) Unrooted neighbor-joining phylogenetic tree assembled from single nucleotide polymorphisms (SNPs) in the hasABC operon of GAS serotypes for which whole genome sequence is available. (B) Matrix displaying the number of differences (SNPs) in the hasABC operon between GAS serotypes, as in panel A. All serotypes have more than 99% identity in hasABC. The hasABC genes were extracted from genome sequences of the following GAS strains (serotype; GenBank accession number): M1476 (M1; AP012491), MGAS5005 (M1; NC_007297), SF370 (M1; NC_002737), MGAS10270 (M2; NC_008022), MGAS315 (M3; NC_004070), Manfredo (M5; NC_009332), MGAS10394 (M6; NC_006086), MGAS2096 (M12; NC_008023), MGAS9429 (M12; NC_008021), MGAS8232 (M18; NC_003485), MGAS6180 (M28; NC_007296), NZ131 (M49; CP000829), Alab49 (M53; NC_017596), MGAS1882 (M59; NC_017053), MGAS15252 (M59; NC_017040), MGAS11027 (M89; unpublished data), and MGAS11610 (M89; unpublished data). Sequence alignments and phylogenetic trees were generated with Geneious v5.6 (Biomatters Ltd., Auckland, New Zealand). Download Figure S1, DOCX file, 0.1 MB.
Human disease isolates of group A streptococcus serotype M4.
Human disease isolates of group A streptococcus serotype M22.
ACKNOWLEDGMENTS
A.R.F. was supported by the Pediatric Infectious Disease Society—St. Jude Children’s Research Hospital Award in Basic Research and the Robert Wood Johnson Foundation—Harold Amos Medical Faculty Development Program Award.
Some strains were supplied by D. R. Johnson and E. L. Kaplan for a fee.
Footnotes
Citation Flores AR, Jewell BE, Fittipaldi N, Beres SB, and Musser JM. 2012. Human disease isolates of serotype M4 and M22 group A streptococcus lack genes required for hyaluronic acid capsule biosynthesis. mBio 3(6):e00413-12. doi:10.1128/mBio.00413-12.
REFERENCES
- 1. Stollerman GH, Dale JB. 2008. The importance of the group a Streptococcus capsule in the pathogenesis of human infections: a historical perspective. Clin. Infect. Dis. 46:1038–1045 [DOI] [PubMed] [Google Scholar]
- 2. Foley MJ, Wood WB., Jr 1959. Studies on the pathogenicity of group A Streptococci. II. The antiphagocytic effects of the M protein and the capsular gel. J. Exp. Med. 110:617–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kass EH, Seastone CV. 1944. The role of the mucoid polysaccharide (hyaluronic acid) in the virulence of group a hemolytic Streptococci. J. Exp. Med. 79:319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wessels MR, Moses AE, Goldberg JB, DiCesare TJ. 1991. Hyaluronic acid capsule is a virulence factor for mucoid group A Streptococci. Proc. Natl. Acad. Sci. U. S. A. 88:8317–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Husmann LK, Yung DL, Hollingshead SK, Scott JR. 1997. Role of putative virulence factors of Streptococcus pyogenes in mouse models of long-term throat colonization and pneumonia. Infect. Immun. 65:1422–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ashbaugh CD, Warren HB, Carey VJ, Wessels MR. 1998. Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J. Clin. Invest. 102:550–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ashbaugh CD, et al. 2000. Bacterial determinants of persistent throat colonization and the associated immune response in a primate model of human group A Streptococcal pharyngeal infection. Cell. Microbiol. 2:283–292 [DOI] [PubMed] [Google Scholar]
- 8. Shea PR, et al. 2011. Distinct signatures of diversifying selection revealed by genome analysis of respiratory tract and invasive bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 108:5039–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hernandez D, François P, Farinelli L, Osterås M, Schrenzel J. 2008. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 18:802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yother J. 2011. Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Annu. Rev. Microbiol. 65:563–581 [DOI] [PubMed] [Google Scholar]
- 11. Cieslewicz MJ, et al. 2005. Structural and genetic diversity of group B Streptococcus capsular polysaccharides. Infect. Immun. 73:3096–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. 2009. Global emm type distribution of group A Streptococci: systematic review and implications for vaccine development. Lancet Infect. Dis. 9:611–616 [DOI] [PubMed] [Google Scholar]
- 13. Shea PR, et al. 2011. Group A Streptococcus emm gene types in pharyngeal isolates, Ontario, Canada, 2002–2010. Emerg. Infect. Dis. 17:2010–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ashbaugh CD, Albertí S, Wessels MR. 1998. Molecular analysis of the capsule gene region of group A Streptococcus: the hasAB genes are sufficient for capsule expression. J. Bacteriol. 180:4955–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O’Loughlin RE, et al. 2007. The epidemiology of invasive group A Streptococcal infection and potential vaccine implications: United States, 2000–2004. Clin. Infect. Dis. 45:853–862 [DOI] [PubMed] [Google Scholar]
- 16. Bordet J. 1907. A contribution to the study of antistreptococcal serum. John Wiley and Sons, New York, NY. [Google Scholar]
- 17. Hanage WP, Kaijalainen T, Saukkoriipi A, Rickcord JL, Spratt BG. 2006. A successful, diverse disease-associated lineage of nontypeable pneumococci that has lost the capsular biosynthesis locus. J. Clin. Microbiol. 44:743–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Creti R, et al. 2012. Identification and molecular characterization of a S. agalactiae strain lacking the capsular locus. Eur. J. Clin. Microbiol. Infect. Dis. 31:233–235 [DOI] [PubMed] [Google Scholar]
- 19. Moxon ER, Deich RA, Connelly C. 1984. Cloning of chromosomal DNA from Haemophilus influenzae. Its use for studying the expression of type b capsule and virulence. J. Clin. Invest. 73:298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Claus H, Maiden MC, Maag R, Frosch M, Vogel U. 2002. Many carried Meningococci lack the genes required for capsule synthesis and transport. Microbiology 148:1813–1819 [DOI] [PubMed] [Google Scholar]
- 21. Johnson DR, Kurlan R, Leckman J, Kaplan EL. 2010. The human immune response to streptococcal extracellular antigens: clinical, diagnostic, and potential pathogenetic implications. Clin. Infect. Dis. 50:481–490 [DOI] [PubMed] [Google Scholar]
- 22. Green NM, et al. 2005. Genetic diversity among type emm28 group A Streptococcus strains causing invasive infections and pharyngitis. J. Clin. Microbiol. 43:4083–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Musser JM, et al. 1991. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc. Natl. Acad. Sci. U. S. A. 88:2668–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beres SB, Musser JM. 2007. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS One 2:e800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lukomski S, et al. 2000. Nonpolar inactivation of the hypervariable Streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect. Immun. 68:535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lancefield RC. 1957. Differentiation of group A Streptococci with a common R antigen into three serological types, with special reference to the bactericidal test. J. Exp. Med. 106:525–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The hasABC operon of GAS is highly conserved between sequenced serotypes. (A) Unrooted neighbor-joining phylogenetic tree assembled from single nucleotide polymorphisms (SNPs) in the hasABC operon of GAS serotypes for which whole genome sequence is available. (B) Matrix displaying the number of differences (SNPs) in the hasABC operon between GAS serotypes, as in panel A. All serotypes have more than 99% identity in hasABC. The hasABC genes were extracted from genome sequences of the following GAS strains (serotype; GenBank accession number): M1476 (M1; AP012491), MGAS5005 (M1; NC_007297), SF370 (M1; NC_002737), MGAS10270 (M2; NC_008022), MGAS315 (M3; NC_004070), Manfredo (M5; NC_009332), MGAS10394 (M6; NC_006086), MGAS2096 (M12; NC_008023), MGAS9429 (M12; NC_008021), MGAS8232 (M18; NC_003485), MGAS6180 (M28; NC_007296), NZ131 (M49; CP000829), Alab49 (M53; NC_017596), MGAS1882 (M59; NC_017053), MGAS15252 (M59; NC_017040), MGAS11027 (M89; unpublished data), and MGAS11610 (M89; unpublished data). Sequence alignments and phylogenetic trees were generated with Geneious v5.6 (Biomatters Ltd., Auckland, New Zealand). Download Figure S1, DOCX file, 0.1 MB.
Human disease isolates of group A streptococcus serotype M4.
Human disease isolates of group A streptococcus serotype M22.