ABSTRACT
Due to the limited coding capacity of picornavirus genomic RNAs, host RNA binding proteins play essential roles during viral translation and RNA replication. Here we describe experiments suggesting that AUF1, a host RNA binding protein involved in mRNA decay, plays a role in the infectious cycle of picornaviruses such as poliovirus and human rhinovirus. We observed cleavage of AUF1 during poliovirus or human rhinovirus infection, as well as interaction of this protein with the 5′ noncoding regions of these viral genomes. Additionally, the picornavirus proteinase 3CD, encoded by poliovirus or human rhinovirus genomic RNAs, was shown to cleave all four isoforms of recombinant AUF1 at a specific N-terminal site in vitro. Finally, endogenous AUF1 was found to relocalize from the nucleus to the cytoplasm in poliovirus-infected HeLa cells to sites adjacent to (but distinct from) putative viral RNA replication complexes.
IMPORTANCE
This study derives its significance from reporting how picornaviruses like poliovirus and human rhinovirus proteolytically cleave a key player (AUF1) in host mRNA decay pathways during viral infection. Beyond cleavage of AUF1 by the major viral proteinase encoded in picornavirus genomes, infection by poliovirus results in the relocalization of this host cell RNA binding protein from the nucleus to the cytoplasm. The alteration of both the physical state of AUF1 and its cellular location illuminates how small RNA viruses manipulate the activities of host cell RNA binding proteins to ensure a faithful intracellular replication cycle.
Introduction
Given the relative genetic simplicity of their positive-sense, single-stranded genomic RNAs, picornaviruses have evolved to utilize host proteins during their intracellular replication cycles. Indeed, multiple host factors have been demonstrated to play roles in picornavirus translation and RNA replication (for a review, see references 1 and 2). Among these, members of the heterogeneous ribonucleoprotein (hnRNP) family of proteins, including poly(rC) binding proteins 1 and 2 (PCBP1/2), hnRNP C1/C2, and hnRNP K, have been reported to have essential roles in picornavirus replication (3–16).
In addition to usurping the functions of host proteins during infection, picornaviruses also modify host proteins via viral enzymatic cleavage (Table 1). Notable examples include poliovirus and coxsackievirus proteinase 2A cleavage of eIF4G. Cleavage of eIF4G shuts off host cell cap-dependent translation, thereby eliminating any competition for resources for the synthesis of viral proteins via cap-independent mechanisms (for a review, see reference 17). Poliovirus proteinase 3C/3CD cleavage of PCBP2, a cellular RNA binding protein required for poliovirus translation and RNA replication, is somewhat unique since cleavage of PCBP2 does not simply disrupt cellular processes but may influence template selection for RNA replication versus viral translation (18).
TABLE 1 .
Examples of cellular proteins cleaved during picornavirus infectionsb
| Cellular protein(s) | Picornavirus proteinase(s)a | Effect(s) of cleavage |
|---|---|---|
| PABP | PV 2A, PV 3C, HRV14 3C, CVB 2A | Disrupts interaction with eIF4G, inhibits host cell translation |
| eIF4GI, eIF4GII, eIF4A | PV 2A, CVB 2A, FMDV 3C, L | Disrupts cap-binding complex, shuts off host cell translation |
| PTB | PV 3C | Possible template selection for viral translation versus RNA replication |
| Dystrophin | CVB 2A | Disrupts cytoskeleton |
| MAP-4 | PV 3C | Induces morphological changes in host cells |
| TBP, CREB, TFIIIC, Oct-1 | PV 3C | Inhibits host cell RNA synthesis |
| La autoantigen | PV 3C | Redistributes La to cytoplasm to enhance translation of viral mRNA |
| PCBP1/PCBP2 | PV 3C, 3CD | Regulates template selection for viral translation versus RNA replication |
| Dcp1a, Pan3 | PV 3C, 3CD | Possible role in P body disruption and RNA stability |
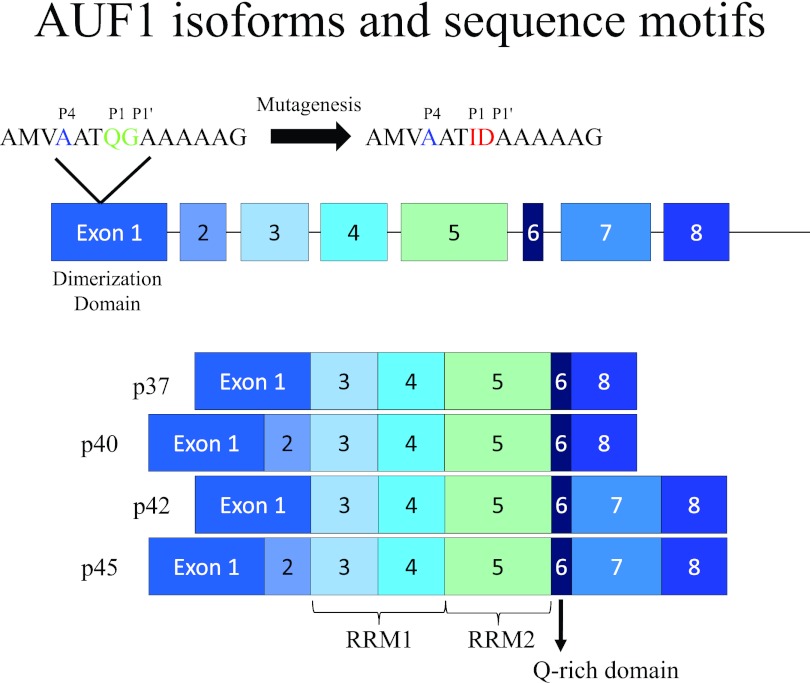
AU-rich binding factor 1 (AUF1), also known as hnRNP D, has characteristics typical of an hnRNP, including nucleocytoplasmic shuttling properties and multiple isoforms (for a review, see references 19 and 20). AUF1 consists of four related isoforms named by their relative molecular masses: p45, p42, p40, and p37 (Fig. 1). These isoforms result from alternative splicing of a single mRNA (20–22). Of the four AUF1 isoforms, p45 is the largest and is the canonical form to which the other isoforms are compared (for a review, see references 19 and 20). All four isoforms have a dimerization domain (encoded in exon 1), as well as two RNA recognition motif (RRM) domains (encoded in exons 3, 4, and 5). AUF1 binds specific proto-oncogene and cytokine mRNAs containing AU-rich element (ARE) sequences, such as c-myc (23), which are then targeted for decay; all four isoforms have been reported to have different roles in this process (22, 24). AUF1 has also been reported to have a role in telomere binding (25) and transcriptional activation (26, 27).
FIG 1.
AUF1 exon structure schematic and proposed 3CD cleavage site. The figure shows the exon structure of the human AUF1 gene and the different protein isoforms (p37, p40, p42, and p45) generated by alternative splicing. The RNA recognition motifs (RRM1 and RRM2), the Q-rich domain, and the dimerization domain are indicated. The top line of the figure shows an expansion of the amino acid sequence in exon 1 starting with amino acid residue 29 and extending through residue 42. This region contains a putative picornavirus 3CD cleavage recognition site with the P1 and P1′ positions Q-G (highlighted in green) preceded by an upstream A in the P4 position (highlighted in blue). For some of the experimental results displayed in Fig. 3, the Q-G pair was mutagenized to I-D (highlighted in red). This amino acid pair is predicted not to be cleaved by the poliovirus or human rhinovirus 3CD proteinase. Adapted with data from the work of Gratacos and Brewer (19).
Much like other hnRNPs, AUF1 has been reported to have a role in the replication cycles of different viruses. It is required for efficient hepatitis C virus translation (28) and has been shown to regulate the C promoter in Epstein-Barr virus (27). For picornaviruses, there is one report demonstrating an increase in AUF1 levels in the cytoplasm of primary cultures of human airway epithelial cells infected with human rhinovirus 16 (HRV16) (29). Although direct evidence for a role for AUF1 in picornavirus infections is currently lacking, this host protein does interact with several proteins that have been reported to function during replication of poliovirus and other picornaviruses. These include poly(A) binding protein (PABP) (30), nucleolin (21), and PCBP1/2 (31). In uninfected cells, these interactions are thought to facilitate a role in regulation of mRNA decay (30), transcriptional activation (21), and the α-globin mRNA stability complex (31), respectively. PABP binds the 3′ poly(A) tract of picornavirus genomes and has been implicated in bridging the 5′ and 3′ ends of the genome via interaction with PCBP (32, 33). Nucleolin interacts with the 3′ noncoding region (NCR) of the poliovirus genome and has been observed to relocalize to the cytoplasm during poliovirus infection; depletion of this protein in cytoplasmic extracts significantly decreases virus production (34). PCBP1/2 proteins interact with the 5′ NCR of poliovirus and are required for viral translation and RNA replication (3–11).
In this paper, we describe data that suggest that AUF1 is a player in picornavirus infection. We report modification of this protein during poliovirus or human rhinovirus infection and have observed that all four isoforms of AUF1 are cleaved during infection with poliovirus, HRV14, or HRV16 in HeLa cells. Using in vitro proteinase assays with recombinant enzymes and substrates, we present evidence identifying the primary cleavage site in all four isoforms that is cleaved by viral proteinase 3CD. We have also determined that full-length and truncated forms of AUF1 will interact with the 5′ NCR of poliovirus or human rhinovirus 16 genomes in vitro. In addition, AUF1 was observed to relocalize from the nucleus to the cytoplasm and to localize in sites adjacent to (but distinct from) putative viral RNA replication complexes during poliovirus infection. Taken together, these data strongly support a role for AUF1 during poliovirus or human rhinovirus infections.
RESULTS
Modification of AUF1 during poliovirus or human rhinovirus infection.
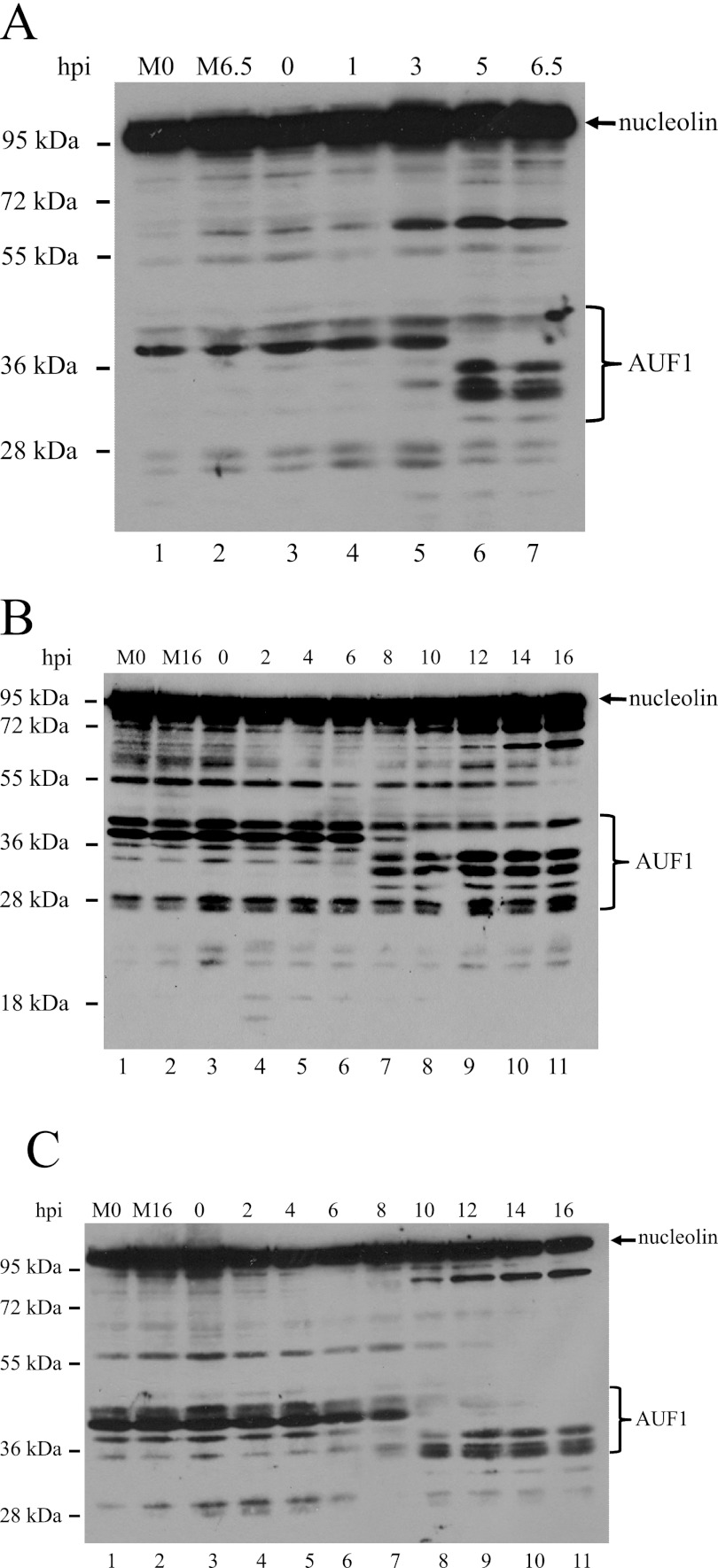
Given the roles that different classes of RNA binding proteins play in infections by different picornaviruses, we initially examined possible alterations of proteins involved in cellular mRNA decay during picornavirus infection. Cytoplasmic S10 lysates were generated from poliovirus-infected HeLa cells and examined for host protein modifications. Although AUF1, HuR, Xrn1, Dcp2, and nucleophosmin were explored via Western blot analysis, AUF1 was the only protein observed to display an altered gel migration rate (data not shown). To extend this finding to other picornaviruses, we subjected extracts from poliovirus- or human rhinovirus-infected cells to SDS-PAGE, electroblotted the proteins to a polyvinylidene difluoride (PVDF) membrane, and carried out a Western blot analysis using anti-AUF1 and antinucleolin (as a loading control) antibodies to visualize specific protein species (Fig. 2). The results confirmed our initial data for poliovirus (Fig. 2A, lanes 5 to 7). In comparing the AUF1 protein profile observed at 0 h (Fig. 2A, lane 3) to those observed 5 h postinfection (hpi) and 6.5 hpi (Fig. 2A, lanes 6 and 7), a very different protein profile emerges during the course of poliovirus infection. The data show the disappearance of AUF1 protein species in the 36- to 45-kDa range with the concomitant appearance of lower-molecular-mass species in the 29- to 36-kDa range. It appears that several (or all) of the four isoforms of AUF1 are cleaved during poliovirus infection, with incomplete cleavage of some isoforms.
FIG 2.
Cleavage of AUF1 during poliovirus, human rhinovirus 14, or human rhinovirus 16 infection. HeLa cells were infected with poliovirus (A) (lanes 3 to 7), human rhinovirus 14 (B) (lanes 3 to 11), or human rhinovirus 16 (C) (lanes 3 to 11), and NP-40 lysates were generated at different times after infection. Lysates from mock-infected cells (M0 and M6.5 or M16, lanes 1 and 2) were generated at 0 hpi or at 6.5 hpi (for poliovirus) or 16 hpi (for human rhinovirus). Lysates were subjected to SDS-PAGE and Western blot analysis using anti-AUF1 rabbit polyclonal antibody and antinucleolin rabbit polyclonal antibody (as a loading control) to visualize proteins. For size comparison, a molecular mass marker was included (left side of the autoradiograph of the gel). The electrophoretic mobilities of endogenous AUF1, cleaved and uncleaved, and nucleolin are indicated by the bracket and arrow, respectively, on the right.
To determine if AUF1 is cleaved in cells infected by other picornaviruses, or if these events are restricted to cells infected with poliovirus, we analyzed extracts from HRV14 (Fig. 2B)- or HRV16 (Fig. 2C)-infected HeLa cells and observed that AUF1 is cleaved during infection by both of these viruses. Interestingly, the kinetics of AUF1 cleavage patterns differ for all three viruses. Compared to poliovirus infection (Fig. 2A, lanes 5, 6, and 7), in which the cleavage event is detectable at 3 hpi, AUF1 was not cleaved in HRV14-infected cells until 8 hpi (Fig. 2B, lane 7). In HRV16-infected cells, readily detectable cleavage products of AUF1 were not observed until 10 hpi (Fig. 2C, lane 8) although some cleavage products were barely detected at 8 hpi. This delay in AUF1 cleavage for both human rhinoviruses may be attributed to the longer replication cycle and lower incubation temperature than those of poliovirus infection in HeLa cells. Additionally, it appears that not all endogenous AUF1 isoforms are completely cleaved during HRV14 infection (Fig. 2B), an observation that is in contrast to infection with HRV16 but similar to the data from poliovirus-infected cells. Overall, we conclude that all four isoforms of AUF1 appear to be cleaved during infection of HeLa cells by poliovirus or human rhinovirus.
Picornavirus proteinase 3CD catalyzes the cleavage of AUF1.
It is possible that cleavage of AUF1 during a poliovirus or human rhinovirus infection was an indirect effect of the infection. However, since these viruses encode two proteinases (2A and 3C/3CD), both of which have been shown to specifically cleave several host proteins, it is likely that cleavage was mediated directly by a viral enzyme. To determine which viral proteinase, 2A or 3CD, might be responsible for cleavage of AUF1, we first analyzed sequences of all four AUF1 isoforms for potential proteinase cleavage sites (Fig. 1). We predicted that protein 2A was not responsible for cleavage because although a recognized cleavage site, tyrosine-glycine (35), is found 16 amino acids from the C terminus of AUF1 in all four isoforms, the predicted cleavage products did not match the estimated size of the products observed in Fig. 2. If 2A was the only viral proteinase responsible for cleaving AUF1, we would expect to observe protein products larger than those that are observed since the consensus 2A cleavage site for all isoforms would generate truncated proteins only 2 kDa less than their original size. However, protein products were observed with electrophoretic mobilities during SDS-PAGE corresponding to molecular masses approximately 5 kDa less than those of the uncleaved forms. Additionally, AUF1 isoforms p42 and p45 both have an amino acid sequence insertion (Fig. 1, exon 7) containing multiple predicted 2A cleavage sites, which also did not correlate with cleavage products observed in Fig. 2.
Located in all four isoforms, 35 amino acids from the N terminus, is an optimal 3C/3CD proteinase cleavage site: in the P1-P1' position is a glutamine-glycine pair (36–39), and an alanine is located 4 amino acids upstream of the putative cleavage site, an optimal P4 residue (Fig. 1). Predicted 3C/3CD cleavage at this site would disrupt a region reported to enhance binding to AU-rich elements and mediate AUF1 dimerization (40). Isoforms p42 and p45 contain additional 3C/3CD cleavage sites in their inserts; however, these are not considered optimal 3CD cleavage sites due to the lack of an alanine in the P4 position (41).
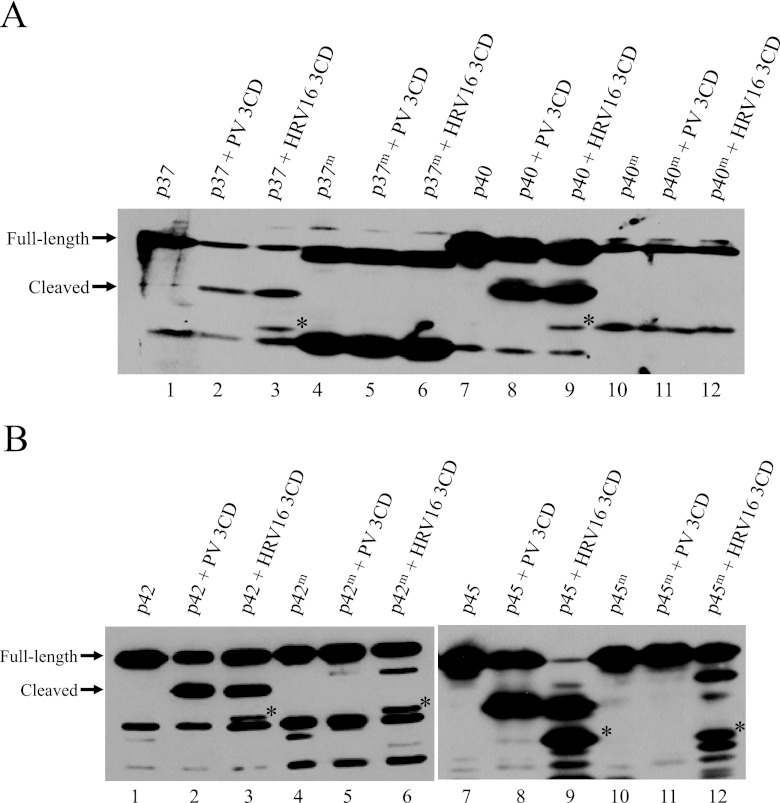
We hypothesized that 3C/3CD proteinase could catalyze the cleavage of AUF1 during infection because the putative N-terminal cleavage site, located in every isoform, would generate cleavage products closer in size to what was observed in Fig. 2. To determine if 3CD was responsible for AUF1 cleavage, recombinant wild-type AUF1 isoforms (Fig. 3A and 3B, lanes 1 and 7) or AUF1 proteins containing the optimal 3CD cleavage site the Q-G pair mutated to an uncleavable I-D pair (lanes 4 and 10 in Fig. 3A and 3B) were incubated with recombinant poliovirus 3CD proteinase (Fig. 3A and 3B, lanes 2, 5, 8, and 11) or HRV16 3CD proteinase (Fig. 3A and 3B, lanes 3, 6, 9, and 12). Both viral proteinases were able to cleave recombinant AUF1, but cleavage of the mutated AUF1 proteins was substantially—if not completely—abrogated. Interestingly, when wild-type AUF1 isoforms were incubated with HRV16 3CD, an additional cleavage product was detected (denoted by an asterisk in Fig. 3), suggesting that the rhinovirus proteinase recognizes additional cleavage sites in AUF1. This finding is consistent with data suggesting that HRV 3C/3CD exhibits less stringency for cleavage sites than do other picornavirus 3C/3CD proteinases (41). Our data suggest that 3CD proteinase is responsible for cleavage of AUF1 during picornavirus infection.
FIG 3.
In vitro cleavage of all isoforms of AUF1 by picornavirus 3CD is abrogated by a mutation within the AUF1 amino-terminal dimerization domain. Purified recombinant wild-type AUF1 isoforms p37 and p40 (A) (lanes 1 and 7) or p42 and p45 (B) (lanes 1 and 7) were incubated with active, recombinant 3CD from poliovirus (lanes 2 and 8) or human rhinovirus 16 (lanes 3 and 9). Recombinant 3CD proteins were rendered uncleavable by an insertion at the P3 position of the 3C-3D junction (PV 3CD [as described in reference 64] and HRV16 3CD [A. J. Chase and B. L. Semler, unpublished data]). The putative 3CD cleavage site (Fig. 1) was mutated in all four isoforms of AUF1 from a Q-G to an I-D amino acid pair, as denoted by a superscript “m.” Recombinant AUF1 proteins containing mutations at this putative 3CD cleavage site (p37m, p40m, p42m, and p45m) were incubated with active, recombinant 3CD from poliovirus (lanes 5 and 11) or human rhinovirus 16 (lanes 6 and 12). Reaction mixtures were incubated for 3 h at 30°C and subjected to SDS-PAGE and Western blot analysis using anti-AUF1 antibodies to visualize proteins. Asterisks denote additional cleavage products apparent when specific isoforms of AUF1 are incubated with HRV16 3CD (A, lanes 3 and 9; B, lanes 3, 6, 9, and 12).
AUF1 interacts with the 5′NCR of poliovirus and human rhinovirus genomic RNAs.
Initial experiments were carried out to determine the array of host proteins that bind to the 5′ NCR of the poliovirus genome, a region that encompasses intricate stem-loop secondary structures required for viral translation and RNA replication. RNA affinity assays using the 5′ NCR of the poliovirus genome were carried out to pull down proteins from uninfected HeLa cell ribosomal salt wash (RSW); isolated fractions were subjected to mass spectrometry analysis, which revealed AUF1, among others, to be a binding partner (data not shown). Further RNA affinity assays using the 5′ NCR of poliovirus type 1 or human rhinovirus 16 RNA were carried out to pull down interacting proteins from lysates from uninfected, poliovirus-infected, or HRV16-infected HeLa cells (Fig. 4). Using S10 extracts from mock-infected or poliovirus-infected HeLa cells, we determined that full-length (Fig. 4A, lane 5) and cleaved (Fig. 4A, lane 6) AUF1 products interacted with the 5′ NCR of the poliovirus genome. Although there is a high background of AUF1 isoforms from S10 extracts derived from poliovirus-infected HeLa cells binding to resin alone (Fig. 4A, lane 4), there is a marked increase above this background when the poliovirus 5′ NCR is linked to the resin (Fig. 4A, lane 6). We obtained similar results when proteins were pulled down using NP-40 lysates from mock-infected or human rhinovirus 16-infected cells and the HRV16 5′ NCR (Fig. 4B, lanes 5 and 6). Interestingly, we also observed that nucleolin, our loading control, interacted with the HRV16 5′ NCR in lysates from mock-infected cells (Fig. 4B, lane 5); however, this interaction was not detectable when lysates from HRV16-infected cells were analyzed (Fig. 4B, lane 6). Since it has been reported elsewhere that AUF1 and nucleolin are binding partners in uninfected cells (21), the interaction of nucleolin with the human rhinovirus 5′ NCR may be the result of an indirect binding event mediated by AUF1. It is possible that cleavage of AUF1 during HRV16 infection limits the ability of these two host proteins to interact with one another. Overall, full-length and truncated AUF1 both are capable of binding to the 5′ NCR of poliovirus and human rhinovirus 16 under the conditions employed in our assays.
FIG 4.
Full-length and virally truncated forms of AUF1 interact with the 5′ NCR of poliovirus and human rhinovirus 16. Transcripts of the poliovirus 5′ NCR (A) or human rhinovirus 16 5′ NCR (B) were biotinylated with biotin-CTP. Streptavidin agarose resin was first incubated with either tRNA alone (lanes 3 and 4) or biotinylated 5′-NCR transcripts (lanes 5 and 6). All experiments were carried out in the presence of a molar excess of tRNA. Lysates from mock-infected or poliovirus-infected (A) or human rhinovirus 16-infected (B) HeLa cells were incubated with streptavidin-bound RNA in RNA affinity assays. Bound complexes were analyzed by SDS-PAGE and Western blot analysis using anti-AUF1 antibody (A) or anti-AUF1 and antinucleolin (as a loading control) antibodies (B). In lanes 1 and 2, 20% of the experimental input sample was loaded from mock or infected lysates, respectively. Electrophoretic mobilities of AUF1 and nucleolin are indicated by a bracket or arrow, respectively.
Relocalization of AUF1 during poliovirus infection.
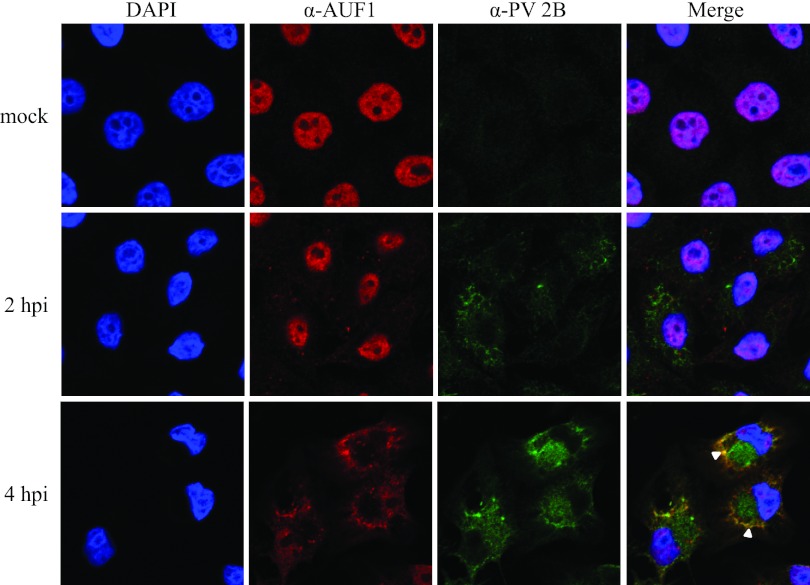
While there are detectable isoforms of AUF1 in extracts derived from the cytoplasm of uninfected HeLa cells (Fig. 2), it has been reported that this host protein is primarily localized to the nucleus (42, 43). Indeed, this was our observation when we carried out immunofluorescence assays to determine if AUF1 localization was altered during poliovirus infection (Fig. 5). In mock-infected HeLa cells, AUF1 was detected primarily in the nucleus; however, by 4 h postinfection with poliovirus, a dramatic relocalization of AUF1 from the nucleus to the cytoplasm was observed. In the cytoplasm of infected cells, AUF1 displayed a distinct localization pattern, partly around the periphery of the nucleus and a region of the cytoplasm devoid of AUF1 (Fig. 5, α-AUF1 panel at 4 hpi). To determine if the above-noted cytoplasmic region might include poliovirus RNA replication complexes, we costained cells with anti-2B monoclonal antibodies (Fig. 5, α-PV 2B panel at 4 hpi). Protein 2B is a nonstructural viral polypeptide with proposed functions associated with rearrangement of cytoplasmic membranes (in its precursor form, 2BC) and formation of membrane pores (for a review, see reference 44), functions that alter the host cell cytoplasm as a prerequisite to the formation of viral RNA replication complexes. The results shown in Fig. 5 reveal the partial colocalization of AUF1 and poliovirus protein 2B (white arrowheads in the 4-hpi merged image) but a lack of colocalization in the 2B-containing cytoplasmic region (green stain in the 4-hpi merged image) adjacent to the nucleus.
FIG 5.
Relocalization of AUF1 and partial colocalization with viral protein 2B during poliovirus infection. HeLa cells seeded on coverslips were either mock infected or infected with poliovirus and fixed at 2 or 4 hpi. Cells were immunostained with anti-AUF1 antibody (shown in red) or anti-poliovirus 2B antibody (shown in green) and stained with DAPI to visualize the nucleus (shown in blue). Colocalization of AUF1 and 2B, indicated in the merged image by white arrowheads, was examined by confocal microscopy.
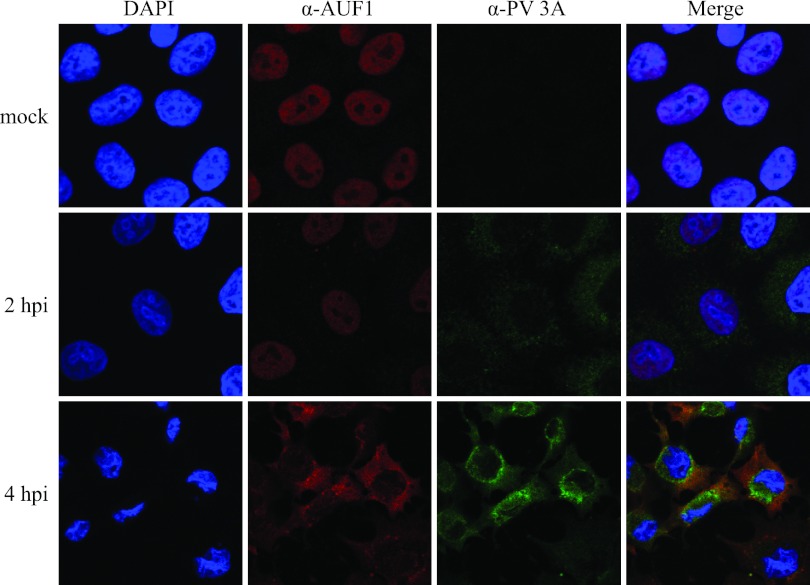
To determine if relocalized AUF1 is associated with the viral RNA synthesizing machinery in the cytoplasm of poliovirus-infected HeLa cells, we analyzed possible colocalization of AUF1 with poliovirus protein 3A. This protein is known to interact with cytoplasmic membranes via its hydrophobic domain; with the viral RNA-dependent RNA polymerase, 3Dpol (and its precursor, 3CD); and with a guanine nucleotide exchange factor (GEF) to recruit this protein as well as its substrate (Arf1) to sites of RNA replication in poliovirus- or coxsackievirus-infected cells (for a review, see reference 44). Antibodies to protein 3A will also interact with its precursor polypeptide 3AB, which has been shown to interact with RNA sequences in stem-loop I within the 5′ NCR and with sequences in the 3′ NCR of the poliovirus genome (45–48). This interaction may be necessary to anchor the viral RNA replication complex to cytoplasmic membranes via a hydrophobic domain located in the C terminus of 3A, thus making 3A a useful marker for viral RNA replication complexes (49–52). As demonstrated in Fig. 6, antibodies to poliovirus 3A protein detect significant levels of protein in the cytoplasm of poliovirus-infected cells at 4 hpi. As previously demonstrated, AUF1 was relocalized from the nucleus to the cytoplasm by 4 hpi. However, this cytoplasmic location appeared to be distinct from that of protein 3A, as shown in the merged image at 4 hpi. These experiments reveal that AUF1 not only relocalizes from the nucleus to the cytoplasm of HeLa cells during poliovirus infection but also partially colocalizes with the poliovirus protein 2B and appears to be excluded from cytoplasmic domains containing viral protein 3A.
FIG 6.
AUF1 is excluded from 3A-containing cytoplasmic regions in poliovirus-infected cells. HeLa cells seeded on coverslips were infected with poliovirus, fixed at 2 or 4 h postinfection, and immunostained with anti-AUF1 (shown in red) and anti-3A (shown in green) antibodies; nuclei were identified by DAPI staining (shown in blue). Proteins were visualized using confocal microscopy.
Amplification of viral genomic RNAs is required for relocalization and cleavage of AUF1 in poliovirus-infected HeLa cells.
We next determined if AUF1 was cleaved during the initial viral translation of genomic RNAs released from uncoated poliovirus particles, or if cleavage occurred only after an increase in viral gene products following viral RNA replication. The latter possibility is consistent with the timing of AUF1 cleavage (Fig. 2A) and relocalization from the nucleus to the cytoplasm (Fig. 5) in poliovirus-infected HeLa cells between 3 hpi and 5 hpi. To differentiate between these possibilities, a poliovirus infection was carried out in HeLa cells that were mock treated or pretreated with 2 mM guanidine HCl (GuaHCl) (Fig. 7). Guanidine HCl does not affect viral translation, but it inhibits viral RNA synthesis via inactivation of the viral nonstructural protein 2C (53, 54). Lanes 1 to 6 in Fig. 7A demonstrate the cleavage of AUF1 observed during a typical poliovirus infection; accumulation of lower-molecular-weight products was apparent by 4 hpi. However, when cells were pretreated with 2 mM GuaHCl (Fig. 7A, lanes 7 to 12), no difference in the AUF1 protein migration pattern was observed during the course of poliovirus infection, indicating a lack of AUF1 cleavage. When GuaHCl-treated HeLa cells were infected with poliovirus and analyzed by immunofluorescence, AUF1 did not relocalize from the nucleus to the cytoplasm, displaying the same pattern as that detected in uninfected cells (Fig. 7B, compare “mock” panel to 5-hpi panel). It should be noted that poliovirus protein 3A was not detectable at 5 hpi in GuaHCl-treated cells, although it was readily detected by 4 hpi in untreated cells (Fig. 6). These data suggest that although GuaHCl does not inhibit viral translation, viral proteins could not accumulate to levels normally observed during infection due to the inhibition of RNA synthesis and, subsequently, the production of newly synthesized, positive-strand RNA templates for translation. Thus, it is likely that relocalization and cleavage of AUF1 require higher levels of viral protein products, including those involved in protein processing or viral RNA synthesis (e.g., 3CD and 3A), than those that are produced during the initial stages of a poliovirus infection.
FIG 7.
Guanidine HCl prevents cleavage and relocalization of AUF1 during poliovirus infection. (A) HeLa cells were mock treated (lanes 1 to 6) or treated with 2 mM guanidine HCl (GuaHCl; lanes 7 to 12) during mock infection (lanes 1 and 7) or poliovirus infection (lanes 2 to 6 and lanes 8 to 12). Lysates were collected at indicated hours postinfection (hpi) and subjected to SDS-PAGE and Western blot analysis using anti-AUF1 and antinucleolin (as a loading control) antibodies to visualize proteins. Electrophoretic mobilities of nucleolin and AUF1 are indicated to the right by an arrow and a bracket, respectively. (B) HeLa cells seeded on coverslips were mock infected or infected with poliovirus and treated with 2 mM guanidine HCl. Cells were fixed at either 0 or 5 hpi, immunostained with anti-AUF1 antibody (in red) and anti-3A antibody (in green), and stained with DAPI (blue) to visualize the nucleus. Proteins were visualized using a Zeiss Axiovert 200 M inverted microscope.
DISCUSSION
In this work, we have presented evidence for the inclusion of AUF1 in the cadre of established host RNA binding proteins that are cleaved during picornavirus infection. AUF1 is cleaved during poliovirus or human rhinovirus infection, and this cleavage event is dependent upon viral genome RNA amplification. In vitro, AUF1 binds to the 5′ NCR of the poliovirus or human rhinovirus genome and is cleaved by recombinant viral 3CD proteinase derived from either poliovirus or human rhinovirus. Additionally, AUF1 relocalizes from the nucleus to the cytoplasm during poliovirus infection and partially colocalizes with poliovirus protein 2B but not viral protein 3A. These latter observations suggest that poliovirus may redirect AUF1 to cytoplasmic sites of membrane rearrangement or pore formation (which result, in part, from the functions of protein 2B) that are distinct from (or perhaps excluded from) sites that are actively involved in viral RNA replication, where viral protein 3A would be expected to localize. While the mechanism of relocalization of AUF1 has not yet been determined, we predict that at least one of the poliovirus or human rhinovirus proteinases is responsible. Under normal cell growth conditions, the localization of AUF1 is complex and not completely understood since each isoform contains multiple localization signals (for a review, see reference 19). It has been suggested that the hetero-oligomerization of AUF1 isoforms could contribute to their shuttling properties; as such, it is possible that the release of the dimerization domain following 3CD cleavage during infection could lead to a redistribution of AUF1 (55). However, since all four isoforms contain a transportin 1 binding site in exon 8 (56) (Fig. 1), it is equally likely that the disruption of this pathway by the 2A proteinase leads to cytoplasmic accumulation of AUF1 during infection (57). Additional colocalization studies with other viral proteins expressed alone or in the context of an infection, together with functional studies, should reveal putative steps in the intracellular replication cycle of poliovirus affected by the relocalization of AUF1.
It is well documented that host RNA binding proteins are required for picornavirus replication (2), but to our knowledge, this is the first report of a host mRNA decay-associated protein identified to interact with picornavirus RNA and possibly act as a player in picornavirus replication. Other mRNA decay proteins, including the decapping enzyme Dcp1a and the deadenylase complex component Pan3, are degraded in poliovirus-infected cells; however, a functional role for this degradation has not been established (58). It is possible that cleavage of mRNA decay components during infection is a viral defense against the host mRNA decay machinery; as such, our early studies focused on elucidating an antiviral role for AUF1 during picornavirus infection. To date, data from our unpublished in vitro and cell culture analyses are equivocal on this putative role. Knockdown experiments with small interfering RNA (siRNA) were carried out in which all four isoforms of AUF1 were reduced to ~25% of their normal cytoplasmic levels. Transfected cells were subsequently infected with poliovirus or human rhinovirus 16, but a significant increase or decrease in viral titers was not observed (data not shown). However, we did not subject extracts from these transfected cells after infection to Western blot analysis to determine if cytoplasmic levels of AUF1 were altered from baseline expression. Based on our subsequent immunofluorescence data, it is possible that relocalization of AUF1 from the nucleus to the cytoplasm during infection provided sufficient AUF1 to fulfill its putative antiviral role or its putative function(s) in the poliovirus replication cycle. Ongoing studies with cell lines in which the AUF1 locus has been knocked out should assess how the constitutive absence of AUF1 affects poliovirus replication.
Deducing the functional role of AUF1 during picornavirus infection may prove challenging because, much as its role in mRNA decay is indirect, it may serve a similar indirect, perhaps redundant, role in picornavirus replication. Due to the dependence that picornaviruses have on host proteins, it would be advantageous for the virus to utilize more than one protein for the same function during replication. AUF1 isoforms have been reported to interact with PCBP1 and PCBP2 (31), nucleolin (21), and PABP (30), which have been demonstrated to interact with the 5′ NCR, the 3′ NCR, and the 3′ poly(A) tract of poliovirus RNA, respectively (3, 10, 32–34, 59). Our data in this study demonstrate that AUF1 interacts with the 5′ NCR of the genomes of poliovirus and human rhinovirus and that, in extracts from uninfected cells, nucleolin interacts with AUF1 bound to the human rhinovirus 5′ NCR (Fig. 4B). Given the interactions known to occur between AUF1 and these proteins in uninfected cells, it is likely that AUF1 interacts with these same proteins during a picornavirus infection, perhaps to carry out functions that impact viral replication. This model may also explain why PABP depletion does not have a strongly deleterious effect on poliovirus replication (33), i.e., the presence of AUF1 may be sufficient to bridge the 5′ and 3′ ends of the viral genome for replication complex formation and subsequent translocation of the RNA-dependent RNA polymerase 3Dpol to the 3′ end of template RNAs (32). It has been reported previously that nucleolin has a role in poliovirus infection (60), so it is also possible that the reduction in virus titers in nucleolin-depleted extracts is due to failure of AUF1 to bridge the interaction between the two ends of the viral genome. To test these and other possibilities, it will be necessary to reduce the endogenous levels of nucleolin or PABP in tandem with AUF1 depletion and assay for the effect on poliovirus titers in HeLa cells. In addition, it will be important to determine the specific binding site(s) for AUF1 in the 5′ NCR and to subsequently mutate these sites to determine if any resulting viruses have a reduced growth phenotype in HeLa cells.
An alternative to a direct role for AUF1 in picornavirus replication is an indirect role via RNP complexes that maintain the stability of the viral genome, as has been suggested for PCBP2 (60). Although AUF1 has primarily been reported to contribute to mRNA destabilization, it has also been reported to be a component of the α-globin mRNA stability complex via interaction with PCBPs (31). In addition, binding of PCBPs to poliovirus RNA has been suggested to protect it from exonucleolytic degradation, resulting in increased genome stability (60). Although our ongoing analyses have not revealed a role for AUF1 in the stabilization or destabilization of poliovirus RNA during the early stages of infection (data not shown), it is possible that AUF1 has a more important role in stability at later times during infection, when newly synthesized RNAs and viral RNAs destined for packaging in progeny virions must be protected from cellular RNases.
Finally, when considering the location of the 3CD cleavage site near the N terminus of AUF1, it is likely that this cleavage disrupts its high-affinity AU-rich element binding and prohibits dimerization/multimerization of AUF1 proteins (40). Such disruptions would be predicted to impair the ability of AUF1 to nucleate assembly of mRNA degradation complexes on target mRNAs (19). Thus, picornaviruses like poliovirus and human rhinovirus may be using a two-pronged strategy by disrupting some of the normal cellular functions of AUF1 (including suppression of an RNA degradation pathway) as well as modifying its structure and cellular location for viral replication activities.
MATERIALS AND METHODS
Preparation of extracts from uninfected and infected HeLa cells.
HeLa cells were grown in suspension culture in Spinner minimal essential medium (S-MEM) supplemented with 8% newborn calf serum (NCS). To generate cytoplasmic extracts from poliovirus-infected cells, cells were pelleted and washed twice with 1× phosphate-buffered saline (PBS). Pelleted cells were resuspended in half of the required total volume of serum-free S-MEM and infected with wild-type poliovirus at a multiplicity of infection (MOI) of 20; adsorption took place at room temperature for 30 min. After adsorption, the remaining medium (supplemented with 8% newborn calf serum) and HEPES, pH 7.4 (20 mM final concentration), were added; the infection was carried out at 37°C. S10 cytoplasmic extracts generated from poliovirus-infected HeLa cells at 0, 2, 4, and 6 h postinfection, as well as 0- and 6-h time points from mock-infected HeLa cells, were prepared as described elsewhere (61), with the exception that they were not subjected to treatment with micrococcal nuclease.
To prepare ribosomal salt wash (RSW) fractions, S10 cytoplasmic extract from uninfected HeLa cells was centrifuged at a maximum relative centrifugal force (RCF) of 370,500 for 1 h at 4°C. The supernatant was discarded, and the pellet was resuspended in 1× hypotonic buffer (20 mM HEPES, pH 7.4, 10 mM magnesium acetate [MgOAc], 1 mM dithiothreitol [DTT]) and adjusted to an optical density at 260 nm (OD260) of 240 to 480 units/ml (1 µl of sample resuspended in 400 µl of 1× hypotonic buffer equals OD260 units/ml). KCl was added to a final concentration of 0.5 M, and the mixture was incubated at 4°C for 15 min. The resuspended pellet was then centrifuged again at a maximum RCF of 370,500 for 1 h at 4°C. The supernatant was dialyzed into unlinkase buffer (20 mM Tris-HCl, pH 7.5, 1 mM DTT, 5% glycerol). Dialyzed fractions were aliquoted and stored at −70°C.
NP-40 lysates from mock-, poliovirus type 1-, human rhinovirus 14-, and human rhinovirus 16-infected HeLa cells were generated from 150-mm plates. Cells were washed with 1× PBS and infected with poliovirus at an MOI of 20 or HRV at an MOI of 10. Adsorption was carried out at room temperature for 30 min (for poliovirus) or 60 min (for HRV) in 1× PBS, and then Dulbecco modified Eagle medium (DMEM) (supplemented with 8% NCS) was added; infection was carried out at 37°C for poliovirus or 34°C for HRV. Cells were washed with 1× PBS and harvested every 2 h over a 6.5 (poliovirus)- or 16 (HRV)-hour time course and resuspended in NP-40 lysis buffer (50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 150 mM NaCl, 1% NP-40) for 30 min on ice. After incubation on ice, cell debris was pelleted, supernatant was collected, and total protein concentration was measured via the Bradford assay. Cellular extracts were subjected to SDS-PAGE, and proteins were electroblotted to a PVDF membrane that was then probed using anti-AUF1 (Millipore) and antinucleolin (Abcam) antibodies (with expression levels of nucleolin serving as a loading control). Protein bands were visualized using chemiluminescence.
For analysis of poliovirus-infected cells treated with or without guanidine HCl, infection was carried out as described above for generation of cytoplasmic extracts using NP-40 lysis buffer, with the exception of the addition of 2 mM guanidine HCl after adsorption.
RNA affinity assays and mass spectrometry analysis.
A plasmid encoding the poliovirus 5′ NCR (62) or the human rhinovirus 16 5′ NCR was linearized with EcoRI and EcoICRI (respectively) and transcribed in the presence of biotinylated CTP using the MEGAscript T7 transcription kit (Ambion). Transcription reaction mixtures were treated with DNase I, quenched with 700 mM ammonium acetate, phenol-chloroform extracted, and ethanol precipitated. Biotinylated transcript was bound to streptavidin agarose in 50 mM KCl buffer (50 mM KCl, 5% glycerol, 1 mM DTT, 0.5 mM EDTA, 25 µg/ml tRNA) for 1 h at 4°C. Precleared HeLa S10 (from mock- or poliovirus-infected cells) or NP-40-derived cytoplasmic lysate (from mock- or HRV16-infected HeLa cells) was incubated with the RNA-bound resin in 50 mM KCl buffer for 2 h at 4°C, and unbound complexes were washed from the resin using 100 mM KCl buffer (100 mM KCl, 5% glycerol, 1 mM DTT, 0.5 mM EDTA, 25 µg/ml tRNA). Complexes associated with the resin were resuspended in 2× Laemmli sample buffer (LSB) and subjected to SDS-PAGE. Proteins were electroblotted to a PVDF membrane and subjected to Western blot analysis using antibodies targeting a specific protein of interest; protein bands were visualized using chemiluminescence. To identify proteins bound to the 5′ NCR of poliovirus, fractions eluted from RNA affinity chromatography were digested with trypsin and subjected to nano-liquid chromatography–tandem mass spectrometry (nanoLC-MS/MS) analysis.
Preparation and purification of recombinant proteins; cleavage assays.
pET23b-AUF1 expression constructs for isoforms p37, p40, p42, and p45 were generously provided by Robert J. Schneider (New York University School of Medicine). The expression constructs were transformed into freshly competent BL21 cells; a 5-ml overnight culture of transformed cells was used to inoculate 1 liter of LB at 37°C and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when the OD600 was 0.2 to 0.4. Cells were pelleted, and purification of recombinant His-tagged AUF1 proteins was carried out as described elsewhere (63).
The pET15b expression constructs for poliovirus and human rhinovirus 16 3CD were transformed into freshly competent BL21 cells; a 5-ml overnight culture of transformed cells was used to inoculate 1 liter of LB at 37°C and induced with 1 mM IPTG when the OD600 was 0.2 to 0.4. Transformed BL21 cells were induced for 2 to 4 h at 25°C, pelleted, resuspended in 30 ml of buffer A (20 mM Tris-HCl, pH 7.5, 25 mM NaCl, 1 mM DTT, 5% glycerol), and lysed using a French pressure cell, and the cell suspension was centrifuged at 10,000 rpm for 30 min to pellet insoluble proteins. The supernatant was decanted, the pellet was washed three times with buffer A, and a final wash was carried out using buffer A lacking DTT. The pellet was resuspended in 20 ml of high-salt I60 buffer (20 mM Tris-HCl, pH 7.9, 1 M NaCl, 60 mM imidazole, 0.5% 1-O-n-octyl-beta-d-glucopyranoside, 10% glycerol) and incubated on ice for 30 min. Following a 30-min spin at 10,000 rpm, the pellet was resuspended in the same buffer and incubated on ice for an additional 30 min. The recombinant 3CD was batch-bound to nickel resin, washed with I60 buffer, and eluted with high-salt I200 buffer (20 mM Tris-HCl, pH 7.9, 1 M NaCl, 200 mM imidazole, 10% glycerol). Fractions found to have the highest protein concentration, determined by the Bradford assay, were stored at −70°C.
Cleavage assays were carried out using recombinant 3CD and AUF1. Fifty picomoles of each AUF1 isoform was incubated with 50 pmol of 3CD in 1× hypotonic buffer with 150 mM KCl (20 mM HEPES, pH 7.4, 10 mM MgOAc, 1 mM DTT, 150 mM KCl) for 3 h at 30°C. Reactions were stopped using 2× Laemmli sample buffer (LSB), reaction mixtures were subjected to SDS-PAGE, and proteins were electroblotted to a PVDF membrane. The membrane was probed using anti-AUF1 antibodies (Millipore), and protein bands were visualized using chemiluminescence.
Immunofluorescence.
HeLa cells were seeded on coverslips and either mock infected or infected with poliovirus at an MOI of 20 as described above. At specific times postinfection, cells were washed with 1× PBS and fixed with 3.7% paraformaldehyde at room temperature for 15 min. After two washes with 1× PBS, cells were permeabilized with 0.5% NP-40 in PBS, washed with 1% NCS-PBS, and blocked in 3% milk in 1× PBS. Cells were washed with 1% NCS-PBS and incubated with rabbit anti-AUF1 antibodies from Millipore (1:100) and either mouse anti-poliovirus 3A or mouse anti-poliovirus 2B antibodies (1:500) diluted in 3% milk-PBS. Poliovirus 3A and 2B antibodies were generously provided by George Belov (University of Maryland). After incubation with primary antibodies, cells were washed with 1% NCS-PBS and incubated with Alexa Fluor 594 goat anti-rabbit IgG and either Alexa Fluor 488 goat anti-mouse IgG (1:1,000) (Molecular Probes) or Alexa Fluor 488 donkey anti-mouse IgG (1:200) (Jackson ImmunoResearch). Cells were washed in 1% NCS-PBS and incubated with 4′,6-diamidino-2-phenylindole (DAPI). Coverslips were mounted onto slides using mounting medium and dried overnight. Cells were visualized using either a Zeiss Axiovert 200 M inverted microscope or a Zeiss LSM700 laser scanning confocal microscope. Images were processed with either LSM510 or Zen software.
ACKNOWLEDGMENTS
We are grateful to Kerry Fitzgerald for critical comments on the manuscript. We thank George Belov for providing monoclonal antibodies to poliovirus proteins 2B and 3A and Robert Schneider for AUF1 expression constructs. We also thank Hung Nguyen and MyPhuong Tran for their expert technical assistance. We thank Adeela Syed for confocal software training.
Confocal microscopy images were generated at the UCI Optical Biology Core facility, which is supported by Comprehensive Cancer Center award P30CA062203 from the National Cancer Institute. This research was supported by National Institutes of Health grant AI065357 associated with the Rocky Mountain Regional Center of Excellence in Biodefense (J.W.), by Public Health Service grant AI026765 from the National Institutes of Health (B.L.S.), by the California Center for Antiviral Drug Discovery (a Multicampus Research Program Initiative from the University of California), and by a Senior Investigator Award from the American Asthma Foundation (B.L.S.). J.M.R. was supported by a postdoctoral fellowship from the George E. Hewitt Foundation for Medical Research.
Footnotes
Citation Rozovics JM, et al. 2012. Picornavirus modification of a host mRNA decay protein. mBio 3(6):e00431-12. doi:10.1128/mBio.00431-12.
REFERENCES
- 1. Rozovics JM, Semler BL. 2010. Genome replication I: the players, p 107–125 In Ehrenfield E, Domingo E, Roos RP, The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 2. Daijogo S, Semler BL. 2011. Mechanistic intersections between picornavirus translation and RNA replication. Adv. Virus Res. 80:1–24 [DOI] [PubMed] [Google Scholar]
- 3. Gamarnik AV, Andino R. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3:882–892 [PMC free article] [PubMed] [Google Scholar]
- 4. Blyn LB, Towner JS, Semler BL, Ehrenfeld E. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andino R, Rieckhof GE, Baltimore D. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369–380 [DOI] [PubMed] [Google Scholar]
- 6. Toyoda H, Franco D, Fujita K, Paul AV, Wimmer E. 2007. Replication of poliovirus requires binding of the poly(rC) binding protein to the cloverleaf as well as to the adjacent C-rich spacer sequence between the cloverleaf and the internal ribosomal entry site. J. Virol. 81:10017–10028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sean P, Nguyen JH, Semler BL. 2008. The linker domain of poly(rC) binding protein 2 is a major determinant in poliovirus cap-independent translation. Virology 378:243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barton DJ, O’Donnell BJ, Flanegan JB. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Andino R, Rieckhof GE, Achacoso PL, Baltimore D. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blyn LB, et al. 1996. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 93:11115–11120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walter BL, Nguyen JH, Ehrenfeld E, Semler BL. 1999. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA 5:1570–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brunner JE, et al. 2005. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J. Virol. 79:3254–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roehl HH, Semler BL. 1995. Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3′ end of viral negative-strand RNA. J. Virol. 69:2954–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin JY, et al. 2008. Heterogeneous nuclear ribonuclear protein K interacts with the enterovirus 71 5′ untranslated region and participates in virus replication. J. Gen. Virol. 89:2540–2549 [DOI] [PubMed] [Google Scholar]
- 15. Zell R, Sidigi K, Bucci E, Stelzner A, Görlach M. 2002. Determinants of the recognition of enteroviral cloverleaf RNA by coxsackievirus B3 proteinase 3C. RNA 8:188–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ertel KJ, Brunner JE, Semler BL. 2010. Mechanistic consequences of hnRNP C binding to both RNA termini of poliovirus negative-strand RNA intermediates. J. Virol. 84:4229–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fitzgerald KD, Semler BL. 2009. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim. Biophys. Acta 1789:518–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perera R, Daijogo S, Walter BL, Nguyen JH, Semler BL. 2007. Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein. J. Virol. 81:8919–8932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gratacós FM, Brewer G. 2010. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip. Rev. RNA 1:457–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guhaniyogi J, Brewer G. 2001. Regulation of mRNA stability in mammalian cells. Gene 265:11–23 [DOI] [PubMed] [Google Scholar]
- 21. Dempsey LA, Hanakahi LA, Maizels N. 1998. A specific isoform of hnRNP D interacts with DNA in the LR1 heterodimer: canonical RNA binding motifs in a sequence-specific duplex DNA binding protein. J. Biol. Chem. 273:29224–29229 [DOI] [PubMed] [Google Scholar]
- 22. Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. 1998. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics 48:195–202 [DOI] [PubMed] [Google Scholar]
- 23. Brewer G. 1991. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol. 11:2460–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Laroia G, Sarkar B, Schneider RJ. 2002. Ubiquitin-dependent mechanism regulates rapid turnover of AU-rich cytokine mRNAs. Proc. Natl. Acad. Sci. U. S. A. 99:1842–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eversole A, Maizels N. 2000. In vitro properties of the conserved mammalian protein hnRNP D suggest a role in telomere maintenance. Mol. Cell. Biol. 20:5425–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brys A, Maizels N. 1994. LR1 regulates c-myc transcription in B-cell lymphomas. Proc. Natl. Acad. Sci. U. S. A. 91:4915–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fuentes-Pananá EM, Peng R, Brewer G, Tan J, Ling PD. 2000. Regulation of the Epstein-Barr virus C promoter by AUF1 and the cyclic AMP/protein kinase A signaling pathway. J. Virol. 74:8166–8175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paek KY, Kim CS, Park SM, Kim JH, Jang SK. 2008. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J. Virol. 82:12082–12093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Spurrell JC, Wiehler S, Zaheer RS, Sanders SP, Proud D. 2005. Human airway epithelial cells produce IP-10 (CXCL10) in vitro and in vivo upon rhinovirus infection. Am. J. Physiol. Lung Cell. Mol. Physiol. 289:L85–L95 [DOI] [PubMed] [Google Scholar]
- 30. Lu JY, Bergman N, Sadri N, Schneider RJ. 2006. Assembly of AUF1 with eIF4G-poly(A) binding protein complex suggests a translation function in AU-rich mRNA decay. RNA 12:883–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kiledjian M, DeMaria CT, Brewer G, Novick K. 1997. Identification of AUF1 (heterogeneous nuclear ribonucleoprotein D) as a component of the alpha-globin mRNA stability complex. Mol. Cell. Biol. 17:4870–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Herold J, Andino R. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Svitkin YV, Costa-Mattioli M, Herdy B, Perreault S, Sonenberg N. 2007. Stimulation of picornavirus replication by the poly(A) tail in a cell-free extract is largely independent of the poly(A) binding protein (PABP). RNA 13:2330–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Waggoner S, Sarnow P. 1998. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J. Virol. 72:6699–6709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Toyoda H, et al. 1986. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell 45:761–770 [DOI] [PubMed] [Google Scholar]
- 36. Hanecak R, Semler BL, Anderson CW, Wimmer E. 1982. Proteolytic processing of poliovirus polypeptides: antibodies to polypeptide P3-7c inhibit cleavage at glutamine-glycine pairs. Proc. Natl. Acad. Sci. U. S. A. 79:3973–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emini EA, Elzinga M, Wimmer E. 1982. Carboxy-terminal analysis of poliovirus proteins: termination of poliovirus RNA translation and location of unique poliovirus polyprotein cleavage sites. J. Virol. 42:194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lawson MA, Dasmahapatra B, Semler BL. 1990. Species-specific substrate interaction of picornavirus 3C proteinase suballelic exchange mutants. J. Biol. Chem. 265:15920–15931 [PubMed] [Google Scholar]
- 39. Semler BL, Hanecak R, Anderson CW, Wimmer E. 1981. Cleavage sites in the polypeptide precursors of poliovirus protein P2-X. Virology 114:589–594 [DOI] [PubMed] [Google Scholar]
- 40. DeMaria CT, Sun Y, Long L, Wagner BJ, Brewer G. 1997. Structural determinants in AUF1 required for high affinity binding to A + U-rich elements. J. Biol. Chem. 272:27635–27643 [DOI] [PubMed] [Google Scholar]
- 41. Palmenberg AC. 1990. Proteolytic processing of picornaviral polyprotein. Annu. Rev. Microbiol. 44:603–623 [DOI] [PubMed] [Google Scholar]
- 42. Zhang W, et al. 1993. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 13:7652–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Laroia G, Cuesta R, Brewer G, Schneider RJ. 1999. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284:499–502 [DOI] [PubMed] [Google Scholar]
- 44. van Kuppeveld F, Belov G, Ehrenfeld E. 2010. Remodeling cellular membranes, p 181–193 In Ehrenfeld E, Domingo E, Roos RP, The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 45. Xiang W, Harris KS, Alexander L, Wimmer E. 1995. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 69:3658–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harris KS, et al. 1994. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 269:27004–27014 [PubMed] [Google Scholar]
- 47. Paul AV, Cao X, Harris KS, Lama J, Wimmer E. 1994. Studies with poliovirus polymerase 3Dpol. Stimulation of poly(U) synthesis in vitro by purified poliovirus protein 3AB. J. Biol. Chem. 269:29173–29181 [PubMed] [Google Scholar]
- 48. Xiang W, Cuconati A, Paul AV, Cao X, Wimmer E. 1995. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA 1:892–904 [PMC free article] [PubMed] [Google Scholar]
- 49. Giachetti C, Hwang SS, Semler BL. 1992. cis-acting lesions targeted to the hydrophobic domain of a poliovirus membrane protein involved in RNA replication. J. Virol. 66:6045–6057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Semler BL, Anderson CW, Hanecak R, Dorner LF, Wimmer E. 1982. A membrane-associated precursor to poliovirus VPg identified by immunoprecipitation with antibodies directed against a synthetic heptapeptide. Cell 28:405–412 [DOI] [PubMed] [Google Scholar]
- 51. Towner JS, Ho TV, Semler BL. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271:26810–26818 [DOI] [PubMed] [Google Scholar]
- 52. Xiang W, Cuconati A, Hope D, Kirkegaard K, Wimmer E. 1998. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J. Virol. 72:6732–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Crowther D, Melnick JL. 1961. Studies of the inhibitory action of guanidine on poliovirus multiplication in cell cultures. Virology 15:65–74 [DOI] [PubMed] [Google Scholar]
- 54. Pfister T, Wimmer E. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274:6992–7001 [DOI] [PubMed] [Google Scholar]
- 55. Sarkar B, Lu JY, Schneider RJ. 2003. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J. Biol. Chem. 278:20700–20707 [DOI] [PubMed] [Google Scholar]
- 56. Suzuki M, et al. 2005. Two separate regions essential for nuclear import of the hnRNP D nucleocytoplasmic shuttling sequence. FEBS J. 272:3975–3987 [DOI] [PubMed] [Google Scholar]
- 57. Gustin KE, Sarnow P. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 20:240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dougherty JD, White JP, Lloyd RE. 2011. Poliovirus-mediated disruption of cytoplasmic processing bodies. J. Virol. 85:64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Parsley TB, Towner JS, Blyn LB, Ehrenfeld E, Semler BL. 1997. Poly(rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124–1134 [PMC free article] [PubMed] [Google Scholar]
- 60. Murray KE, Roberts AW, Barton DJ. 2001. Poly(rC) binding proteins mediate poliovirus mRNA stability. RNA 7:1126–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Walter BL, Parsley TB, Ehrenfeld E, Semler BL. 2002. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 76:12008–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dildine SL, Semler BL. 1992. Conservation of RNA-protein interactions among picornaviruses. J. Virol. 66:4364–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wilson GM, Brewer G. 1999. Identification and characterization of proteins binding A + U-rich elements. Methods 17:74–83 [DOI] [PubMed] [Google Scholar]
- 64. Semler BL, Johnson VH, Dewalt PG, Ypma-Wong MF. 1987. Site-specific mutagenesis of cDNA clones expressing a poliovirus proteinase. J. Cell. Biochem. 33:39–51 [DOI] [PubMed] [Google Scholar]
- 65. Badorff C, et al. 2000. Enteroviral protease 2A directly cleaves dystrophin and is inhibited by a dystrophin-based substrate analogue. J. Biol. Chem. 275:11191–11197 [DOI] [PubMed] [Google Scholar]
- 66. Clark ME, Hämmerle T, Wimmer E, Dasgupta A. 1991. Poliovirus proteinase 3C converts an active form of transcription factor IIIC to an inactive form: a mechanism for inhibition of host cell polymerase III transcription by poliovirus. EMBO J. 10:2941–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Clark ME, Lieberman PM, Berk AJ, Dasgupta A. 1993. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol. Cell. Biol. 13:1232–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Das S, Dasgupta A. 1993. Identification of the cleavage site and determinants required for poliovirus 3CPro-catalyzed cleavage of human TATA-binding transcription factor TBP. J. Virol. 67:3326–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Devaney MA, Vakharia VN, Lloyd RE, Ehrenfeld E, Grubman MJ. 1988. Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap-binding protein complex. J. Virol. 62:4407–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW. 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 257:14806–14810 [PubMed] [Google Scholar]
- 71. Joachims M, Etchison D. 1992. Poliovirus infection results in structural alteration of a microtubule-associated protein. J. Virol. 66:5797–5804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Joachims M, Harris KS, Etchison D. 1995. Poliovirus protease 3C mediates cleavage of microtubule-associated protein 4. Virology 211:451–461 [DOI] [PubMed] [Google Scholar]
- 73. Joachims M, Van Breugel PC, Lloyd RE. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73:718–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kuyumcu-Martinez NM, Joachims M, Lloyd RE. 2002. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J. Virol. 76:2062–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liebig HD, et al. 1993. Purification of two picornaviral 2A proteinases: interaction with eIF-4 gamma and influence on in vitro translation. Biochemistry 32:7581–7588 [DOI] [PubMed] [Google Scholar]
- 76. Shiroki K, et al. 1999. Intracellular redistribution of truncated la protein produced by poliovirus 3Cpro-mediated cleavage. J. Virol. 73:2193–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yalamanchili P, Datta U, Dasgupta A. 1997. Inhibition of host cell transcription by poliovirus: cleavage of transcription factor CREB by poliovirus-encoded protease 3Cpro. J. Virol. 71:1220–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yalamanchili P, Harris K, Wimmer E, Dasgupta A. 1996. Inhibition of basal transcription by poliovirus: a virus-encoded protease (3Cpro) inhibits formation of TBP-TATA box complex in vitro. J. Virol. 70:2922–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yalamanchili P, Weidman K, Dasgupta A. 1997. Cleavage of transcriptional activator oct-1 by poliovirus encoded protease 3Cpro. Virology 239:176–185 [DOI] [PubMed] [Google Scholar]