Abstract
We studied aquaporins in maize (Zea mays), an important crop in which numerous studies on plant water relations have been carried out. A maize cDNA, ZmTIP1, was isolated by reverse transcription-coupled PCR using conserved motifs from plant aquaporins. The derived amino acid sequence of ZmTIP1 shows 76% sequence identity with the tonoplast aquaporin γ-TIP (tonoplast intrinsic protein) from Arabidopsis. Expression of ZmTIP1 in Xenopus laevis oocytes showed that it increased the osmotic water permeability of oocytes 5-fold; this water transport was inhibited by mercuric chloride. A cross-reacting antiserum made against bean α-TIP was used for immunocytochemical localization of ZmTIP1. These results indicate that this and/or other aquaporins is abundantly present in the small vacuoles of meristematic cells. Northern analysis demonstrated that ZmTIP1 is expressed in all plant organs. In situ hybridization showed a high ZmTIP1 expression in meristems and zones of cell enlargement: tips of primary and lateral roots, leaf primordia, and male and female inflorescence meristems. The high ZmTIP1 expression in meristems and expanding cells suggests that ZmTIP1 is needed (a) for vacuole biogenesis and (b) to support the rapid influx of water into vacuoles during cell expansion.
The vacuole is a multifunctional organelle with important roles in space filling, osmotic adjustment, storage, and digestion. In dividing cells the vacuolar compartment is represented by small vacuoles that first increase in number as a result of de novo biogenesis, and then expand and coalesce to form one or several highly lobed structures that occupy most of the cellular volume. Vacuole biogenesis and enlargement require the transport of osmotically active substances across the tonoplast, followed by the rapid influx of water into the vacuole. This influx generates the turgor pressure that drives cell expansion and maintains cell shape. Recent studies (Maurel et al., 1997; Niemietz and Tyerman, 1997) show that the tonoplast is highly permeable to water and that this high permeability is caused by the presence of mercuric-chloride-inhibitable water channels that permit the rapid passage of water with a low energy of activation. Such observations are consistent with the presence of aquaporins in the tonoplast.
Aquaporins form a large family (Weig et al., 1997) of proteins present in the plasma membrane (PIPs) and tonoplast (TIPs) that increase the hydraulic conductivity of the plasma membrane when expressed in Xenopus laevis oocytes (for review, see Maurel, 1997). They are 25- to 29-kD membrane proteins with primary sequences similar to those of the MIP family (Park and Saier, 1996). MIPs have six transmembrane domains with cytosolic amino and carboxy termini and short, conserved amino acid motifs, including the signature sequence SGxHxNPA, which is repeated in the second half of the protein as NPA. Some of these proteins transport small solutes, others transport small solutes and water, and still others transport only water (Park and Saier, 1996).
The expression patterns of specific plant aquaporins are tissue- and cell-type specific. The aquaporin α-TIP from common bean accumulates during seed maturation (Johnson et al., 1989; Melroy and Herman, 1991), and the aquaporins γ-TIP and δ-TIP from Arabidopsis are preferentially expressed in elongating root cells and in the parenchymal cells of vascular tissues, respectively (Ludevid et al., 1992; Daniels et al., 1996). The plasma membrane aquaporin RD28 from Arabidopsis is found in all plant organs, but is absent from seeds (Daniels et al., 1994). Several other studies have revealed the organ- and cell-type-specific expression patterns of TIP and PIP aquaporins (Yamamoto et al., 1991; Kammerloher et al., 1994; Opperman et al., 1994; Kaldenhoff et al., 1995; Yamada et al., 1995). The variety of the expression patterns suggests that aquaporins may function in long-distance transport (xylem and phloem loading and unloading), in short-distance transcellular water flow, and in intracellular osmotic adjustment.
Maize (Zea mays) has been used extensively to study water transport and its regulation by environmental parameters (Westgate and Boyer, 1985; Sharp et al., 1988; Zhu and Steudle, 1991), and for this reason we decided to characterize its aquaporins and study their expression patterns. The presence of conserved sequence motifs in plant MIPs allows their cDNAs to be isolated by RT-PCR. In this paper we report the isolation and properties of a highly expressed maize tonoplast aquaporin cDNA, ZmTIP1, and document its expression in tissues of maize that are actively dividing and beginning to elongate: meristems of primary roots and lateral roots, leaf primordia, and male and female inflorescence meristems. We interpret our results to indicate that TIPs may be needed for vacuole biogenesis and enlargement in cells that are still dividing and beginning to expand.
MATERIALS AND METHODS
Plant Growth Conditions
Maize (Zea mays, Oh43 line) was grown in a greenhouse under a 16-h light/8-h dark photoperiod. Seedlings were grown on moistened filter paper at 30°C in the dark.
RNA Extraction
Total RNA was obtained from seeds, embryos, and endosperm at 19 d after pollination from shoots and roots of germinating seedlings, from leaves of 1- to 2-week-old plants, and from developing ears and tassels approximately 2 cm in size. Endosperm samples were isolated from developing seeds by cutting off the top of the seed coat, extracting the seed contents with a small spatula, and removing the embryo from the endosperm. All tissue samples were frozen in liquid N2 after isolation and stored at −70°C. Total RNA was extracted as previously described (Cone et al., 1986). Poly(A+) RNA was isolated from total RNA using the Poly(A+) Tract Kit (Promega) following the instructions of the manufacturer.
Identification of MIP cDNAs by RT-PCR
cDNA was synthesized from 0.5 μg of seed mRNA using oligo(dT)12–18 as a primer and Moloney murine leukemia virus RT (GIBCO-BRL). Partial ZmTIP1 cDNA was amplified by PCR using degenerate TIP2 and TIP4 primers (Weig et al., 1997), and the reaction products were separated and cloned as described previously (Weig et al., 1997).
ZmTIP1 cDNA Cloning
Full-length ZmTIP1 cDNA was obtained using the 5′/3′ RACE kit (Boehringer Mannheim) following the instructions of the manufacturer. For the ZmTIP1 5′/3′ RACE, three antisense- and one sense-specific primers (MRACE3, 5′-GCGATGGTGCCCAGGCTGCC-3′; MRACE7, 5′-GGTCCACCGCCGTGGCGTAC-3′; MRACE10, 5′-CAGCACGTGCGCCACCCAGTA-3′; and MRACE5, 5′-GCAGGCCACGGGCACCTTCG-3′) were used. The PCR products were cloned into pCRII (TA cloning kit, Invitrogen) and sequenced. The full-length ZmTIP1 cDNA was amplified using Pfu polymerase (Stratagene) with proofreading activity and specific primers to the 5′- and 3′-noncoding regions (ZMTIP1–1, 5′-CGGAATTCTCCAGCTCCAATCACAGTC-3′; and ZMTIP1–2, 5′-CGGAATTCACGGTTACAAGCAG-3′) incorporating EcoRI sites on both ends, and subcloned into EcoRI site of Bluescript II SK+ (Stratagene).
Plasmid Constructions and in Vitro RNA Synthesis
cDNA encoding ZMTIP1 was amplified by PCR with specific primers (ZMTIP1–3, 5′-GGCGGATCCTACCATGCCGATCAATAGGAT-3′; and ZMTIP1–4, 5′-CGATGGATCCACGTGCACGAG-3′) incorporating BamHI sites on both ends, and subcloned into the BglII site of a pSP64T-derived Bluescript vector carrying 5′- and 3′-untranslated sequences of a β-globin gene from Xenopus laevis (Preston et al., 1992). The orientation of the insert was determined by restriction mapping and sequencing. Capped complementary RNA encoding ZMTIP1 was synthesized in vitro using T3 RNA polymerase, and was purified as described by Preston et al. (1992).
The 3′-untranslated region of ZmTIP1 was amplified by PCR with specific primers (ZMTIP1–5, 5′-CACCGGATCCTAAAAGCCGAAG-3′; and ZMTIP1–2) incorporating BamHI and EcoRI sites on the ends, and subcloned into the corresponding sites of pBluescript II SK+ (Stratagene) (pBS3′-ZmTIP1).
Part of ZmTIP1 cDNA encoding the carboxy-terminal 62 amino acid residues of ZMTIP1 was amplified by PCR with T7 and ZMTIP1–7 (5′-GGCGGCGAATTCGACGGCGC-3′) primers. The PCR product was digested with EcoRI and SalI and subcloned in the corresponding sites of pGEX-4T-1 (Pharmacia) (pGEX-C-Zmtip1).
Osmotic Water-Permeability Assay
X. laevis oocytes were prepared and injected as previously described (Daniels et al., 1996), and the osmotic water permeability of the plasma membrane was determined (Weig et al., 1997).
DNA Gel-Blot Analysis
Total DNA was extracted from leaf tissue as described previously (Schmidt et al., 1987). DNA blots and hybridizations were as described previously (Evola et al., 1986). For probe synthesis, the 3′-untranslated region of ZmTIP1 cDNA was gel purified and radiolabeled using a kit (Rediprime, Amersham) following the instructions of the manufacturer. Hybridizations were performed at 42°C in 50% formamide. Washes were performed four times for 15 min each in 0.1× SSC (1× SSC is 150 mm NaCl and 15 mm Na3C6H5O7) and 0.1% SDS at 60°C.
RNA Gel-Blot Analysis
Total RNA samples (20 μg each) were fractionated by electrophoresis on a Hepes-formaldehyde 1.5% agarose gel following the protocol of Tsang et al. (1993), and were transferred to Hybond-N nylon membranes (Amersham) using standard blotting techniques (Sambrook et al., 1989). Ethidium-bromide-stained rRNAs were used as the internal loading control. The RNA was bound to the membrane with UV illumination and baking at 80°C for 1 to 2 h. The prehybridization and hybridization were performed at 42°C in 50% formamide, 5× SSPE (1× SSPE is 180 mm NaCl, 1 mm EDTA, and 10 mm Na2HPO4, pH 7.7), 5× Denhardt's solution (1× Denhardt's solution is 0.02% [w/v] BSA, 0.02% [w/v] Ficoll, 0.02% [w/v] PVP), 0.5% SDS, and 100 μg mL−1 of yeast tRNA. The random-primer-labeled probes were generated using the Rediprime kit following the instructions of the manufacturer. Hybridized membranes were washed under high-stringency conditions (0.2× SSPE, 0.2 SDS at 65°C for 20 min), and then exposed to radiographic film with intensifying screens at −70°C.
GST-C-ZMTIP1 Expression in Escherichia coli and Immunodetection
pGEX-C-ZmTIP1 plasmid was introduced in the M15 bacterial strain (Qiagen, Santa Clarita, CA), and GST-C-ZMTIP1 expression was induced by 2 mm isopropyl β-d-thiogalactopyranoside for 2 h. Appropriate quantities of total protein extract were fractionated by 12.5% SDS-PAGE, transferred to nitrocellulose, and the proteins detected using the rabbit antisera raised against Arabidopsis γ-TIP (Höfte et al., 1992) and bean α-TIP (Johnson et al., 1989). Goat anti-rabbit IgG coupled to horseradish peroxidase (Bio-Rad) was used as the secondary antibody.
Immunocytochemical Localization
The immunocytochemical localization of ZmTIP1 in the embryo after 1 d of germination was performed with antiserum raised against bean seed α-TIP, as described previously (Melroy and Herman, 1991).
RNA in Situ Hybridization
Maize tissues were fixed in 50% (v/v) ethanol, 5% (v/v) acetic acid, and 3.7% (v/v) formaldehyde at room temperature for 4 h with occasional degassing under a vacuum for 15 min. After fixation, the tissues were dehydrated through an alcohol series and embedded in Paraplast Plus (Oxford Labware, St. Louis, MO). The tissues were sectioned into 8- to 10-μm slices, dewaxed with Histoclear (National Diagnostics, Atlanta, GA), and hydrated by passing through an alcohol series to water. Sections were prepared for in situ hybridization as described previously (Marrison and Leech, 1994).
The in situ hybridizations were performed as described previously (Marrison and Leech, 1994) with some modifications. ZmTIP1 sense- and antisense-labeled probes were generated using pBS3′-ZmTIP1 linearized with EcoRI or BamHI and transcribed using a digoxigenin RNA-labeling mixture (Boehringer Mannheim) with either T3 or T7 RNA polymerase (Promega), respectively. The probes were hybridized to the tissue sections overnight at 50°C at a concentration of 200 to 400 ng mL−1 in 40 μL of hybridization buffer (6× SSC, 3% [w/v] SDS, 50% [v/v] formamide, and 100 μg mL−1 tRNA). After hybridization, the sections were incubated twice in wash buffer (2× SSC and 50% [v/v] formamide) at 50°C for 90 min; treated with RNase A (10 μg mL−1 in 2× SSC) at 37°C for 30 min; and washed at 50°C for 1 h in wash buffer. The sections were incubated in a blocking solution (Boehringer Mannheim, 0.5% in TBS) for 1 h; in 1% (w/v) BSA and 0.3% (v/v) Triton X-100 in TBS for 30 min; and in the same solution containing alkaline phosphatase-conjugated antibodies (Boehringer Mannheim) at a 1/1000 dilution for 90 min. Unbound antibody conjugate was removed and ZmTIP1 transcripts were detected according to the method of Marrison and Leech (1994).
Photographs were made using a light microscope (Optiphot-2, Nikon). The slides were digitized using a slide scanner (CoolScan, Nikon). Brightness and contrast were adjusted using Photoshop 3.0 (Adobe Systems, Mountain View, CA). Composite figures were prepared in Canvas 3.5 (Deneba Software, Miami, FL) and printed using a dye-sublimation color printer (Phaser IIsdx, Tektronix, Wilsonville, OR).
RESULTS
Isolation of ZmTIP1 cDNA
A comparison of plant aquaporin amino acid sequences showed the presence of several conserved regions. Two of them, HI/VNPAVT and WI/VF/YWVGP, were used to design degenerate oligonucleotide primers for RT-PCR (Weig et al., 1997). Using these primers with cDNAs prepared from maize seeds 19 d after pollination, we obtained a PCR-amplified fragment (0.42 kb) containing a sequence homologous to plant TIP aquaporins. The corresponding full-length cDNA was recovered by 5′/3′ RACE with RNA from maize seeds and roots and named ZmTIP1 (accession no. AF037061).
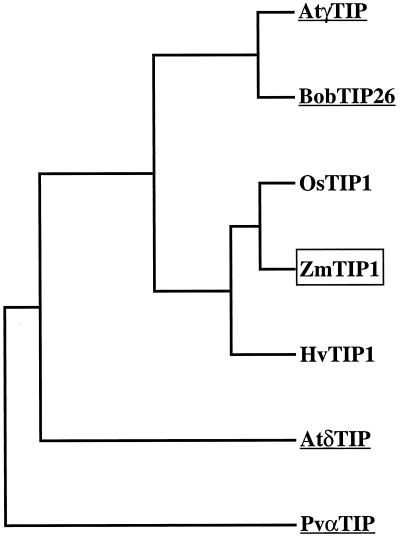
The ZmTIP1 cDNA consists of 1097 bp upstream of the poly(A+) tail, which includes a 93-bp leader sequence, followed by 753 bp of open reading frame encoding 250 amino acids, and, finally, a 251-bp 3′ noncoding region. ZmTIP1 has a calculated Mr of 25,820 and contains the MIP family signature sequence SGxHxNPAVT, which is repeated in the second half of the protein as NPA. A comparison of the amino acid sequences of six other TIPs is shown in Figure 1. ZmTIP1 has the highest sequence identity at the amino acid level with two other monocot TIPs from rice (Liu et al., 1994) and barley (Schünmann and Ougham, 1996) (95.2% and 90.4%, respectively). ZmTIP1 is also related to the known vacuolar aquaporins BobTIP26 from cauliflower (Barrieu et al., 1998; F. Barrieu, D. Marty-Mazars, F. Chaumont, M. Chrispeels, and F. Marty, unpublished data) and γ-TIP from Arabidopsis (Maurel et al., 1993) (77.3% and 76.3% identity, respectively). These proteins cluster together on a dendogram, whereas other TIP aquaporins such as δ-TIP from Arabidopsis (Daniels et al., 1996) and seed α-TIP from bean (Johnson et al., 1990) are more distant (61.1% and 52.7% identity with ZmTIP1, respectively) (Fig. 2).
Figure 1.
Comparison of ZmTIP1 sequence with other plant TIPs. Amino acid sequences were compared with the Clustal W multiple alignment program (Thompson et al., 1994). The amino acid sequences were obtained from the following sources: ZmTIP1 (this work); OsTIP1 (Liu et al., 1994); HvTIP1 (Schünmann and Ougham, 1996); BobTIP26 (Barrieu et al., 1998); Atγ-TIP (Hofte et al., 1992); Atδ-TIP (Daniels et al., 1996); and Pvα-TIP (Johnson et al., 1990). Identical amino acid residues common to at least three sequences are shaded. Numbering refers to the respective amino acid sequence. The position of the Cys residue responsible for the mercury sensitivity of Atγ-TIP and Atδ-TIP is noted by an arrowhead in the consensus line.
Figure 2.
Dendogram of the comparison between ZmTIP1 and other plant TIPs. Amino acid sequences from Figure 1 were compared using the program PILEUP (Genetics Computer Group, Madison, WI). Underlined sequences have been identified as aquaporins.
ZmTIP1 Forms Water Channels in X. laevis Oocytes
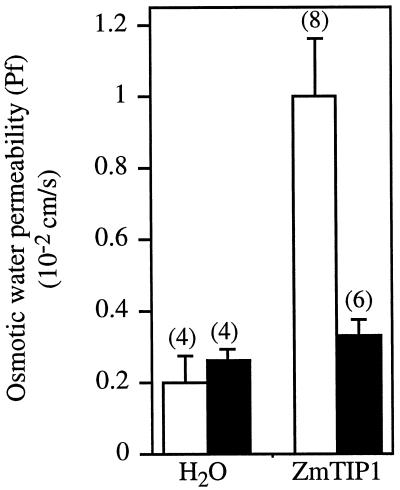
In vitro-transcribed cRNA encoding ZmTIP1 was injected into X. laevis oocytes and osmotically driven water transport into the oocytes was investigated 3 d after injection. Oocytes were exposed to hypoosmotic conditions by diluting the culture medium, and the changes in cell volume were recorded. In these conditions water-injected oocytes swelled slowly. In contrast, oocytes injected with ZmTIP1 cRNA rapidly increased their volume, indicating the presence of a facilitated water-transport pathway. The osmotic water-permeability coefficient (Pf) of the oocyte membrane increased 4- to 5-fold over the control value (Fig. 3).
Figure 3.
Osmotic water permeability (Pf) values of individual ZmTIP1 cRNA-injected oocytes derived from volume change measurements made over two independent preparations of oocytes. White bars, Control (no mercuric chloride); black bars, assay performed in the presence of 3 mm mercuric chloride with a 10-min preincubation. Data are expressed as the mean ± se, with the number of replicates indicated next to each bar in parentheses.
Mercuric chloride is a characteristic inhibitor of many water-channel proteins (Preston et al., 1992; Maurel et al., 1993). The mercury-sensitive sites of Arabidopsis γ-TIP and δ-TIP have been identified as Cys-118 and Cys-116, respectively, at a conserved position in a presumed membrane-spanning domain (Daniels et al., 1996). This Cys residue is conserved among the TIPs, including ZmTIP1, with the exception of α-TIP from bean (see arrow in Fig. 1). Water transport through ZmTIP1 is inhibited 70% by 3 mm mercuric chloride (Fig. 3). These results support the interpretation that ZmTIP1 forms channels in oocyte membranes that facilitate water transport.
ZmTIP1 Cross-Reacts with Different Aquaporin Antisera
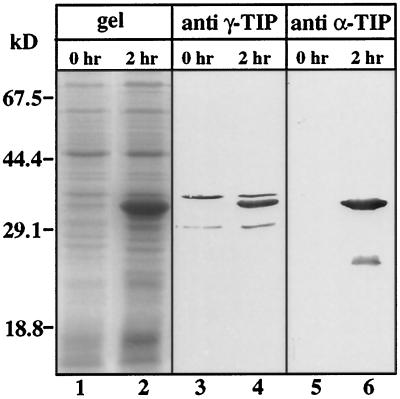
In the past our laboratory has raised antisera against the 30 carboxy-terminal amino acid residues of Arabidopsis γ-TIP (Höfte et al., 1992) and the whole α-TIP from bean (Johnson et al., 1989). To determine if these antisera cross-react with ZmTIP1, we fused the sequence encoding the carboxy-terminal 62 amino acid residues of ZmTIP1 to the GST gene in the pGEX-4T-1 plasmid vector, and the fusion protein was expressed in E. coli (see Methods). When the induced culture was allowed to express the fusion protein for 2 h, a strong band of 34 kD, corresponding to the GST-C-ZmTIP1 polypeptide, was produced as observed by SDS-PAGE analysis (Fig. 4, lane 2).
Figure 4.
Coomassie blue-stained gel and immunoblot of extracts of E. coli expressing GST-C-ZmTIP1. Total E. coli protein extract before (lanes 1, 3, and 5) and after (lanes 2, 4, and 6) 2 h of induction of GST-C-ZmTIP1 expression by 2 mm isopropyl β-d-thiogalactopyranoside was fractionated by SDS-PAGE. Polypeptides were visualized with Coomassie blue (lanes 1 and 2) or transferred to nitrocellulose and immunostained using Arabidopsis γ-TIP antiserum (lanes 3 and 4) or bean α-TIP (lanes 5 and 6). The positions of molecular mass standards are indicated.
Immunoblot analysis of the same bacterial extract using the Arabidopsis γ-TIP antiserum showed that this serum detects the GST-C-ZmTIP1 polypeptide and cross-reacts with two E. coli proteins (Fig. 4, lanes 3 and 4). In the same way, the bean α-TIP antiserum cross-reacted with GST-C-ZmTIP (Fig. 4, lane 6). The reactivity of these two sera with ZmTIP1 came from the ZmTIP1 polypeptide (and not from the GST) because neither γ-TIP or α-TIP antisera elicited an immunostaining reaction with expressed GST protein (data not shown). Sequence identity in this carboxy-terminal region of ZmTIP1 with Arabidopsis γ-TIP and bean α-TIP was 73% and 55%, respectively (Fig. 1). These data indicate that antigenic epitopes recognized by γ-TIP and α-TIP antisera are conserved in TIPs from monocots and dicots.
A ZmTIP1 Cross-Reacting Serum Labels the Tonoplast
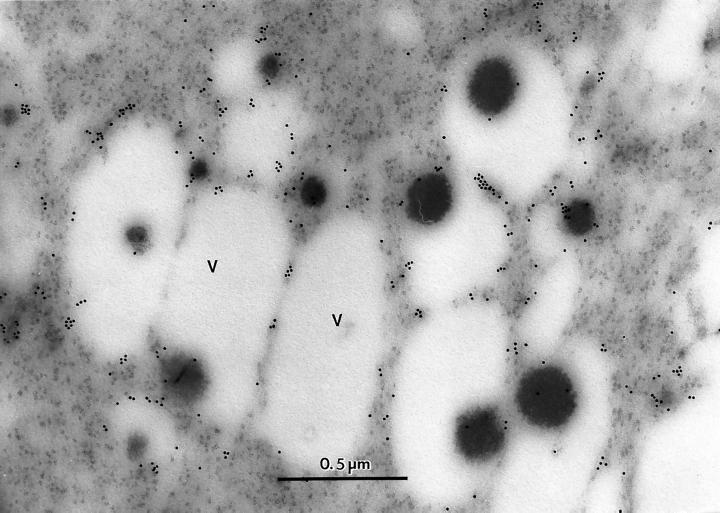
The cross-reactivity of ZmTIP1 with α-TIP antiserum allowed us to examine the subcellular localization of ZmTIPs by immunocytochemistry. We chose maize embryos for this localization because ZmTIP1 is highly expressed there (see below). Meristematic cells of embryos are characterized by the presence of numerous small vacuoles, which subsequently fuse and enlarge. Figure 5 shows abundant colloidal gold labeling of the interface between the cytoplasm and the vacuole where the tonoplast is located. This method of fixation, which minimizes the destruction of protein epitopes, does not allow for the visualization of the tonoplast. There was no specific labeling of the vacuolar content (Fig. 5) or of the plasma membrane (data not shown). These results indicate that ZmTIPs are localized in the tonoplast of maize vacuoles. We do not know if the labeling was caused by the presence of ZmTIP1 alone or by the presence of other aquaporins or MIPs that cross-react with this serum
Figure 5.
Immunocytochemical localization of ZmTIP1 in maize embryos. The gold particles are primarily at the interface of the cytoplasm and the vacuole (V). The vacuoles apparently contain aggregated protein. Magnification is ×60,000.
Expression of ZmTIP1 in Different Tissues during Development
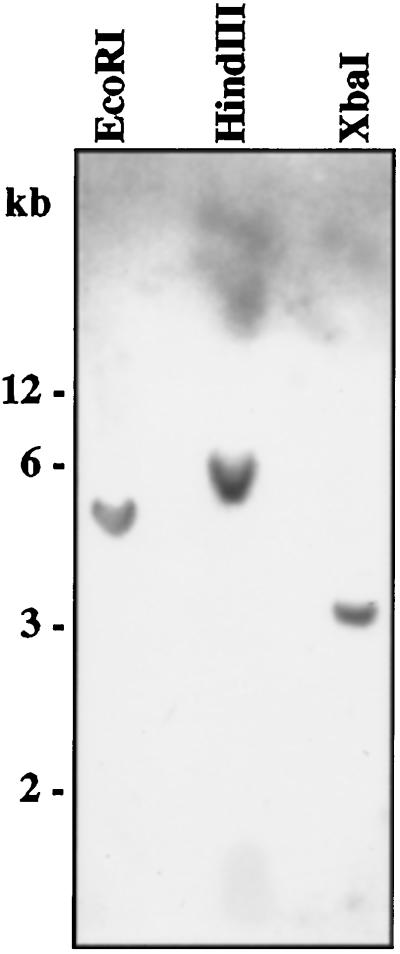
To analyze the expression pattern of ZmTIP1, a 203-bp DNA fragment from the 3′-untranslated region of ZmTIP1 cDNA was used as a probe. The specificity of the probe was tested by Southern hybridization (Fig. 6). In this experiment, only restriction enzymes that do not cut the 3′-untranslated sequence (EcoRI, HindIII, and XbaI) were used to digest genomic DNA samples. Hybridization at high-stringency conditions (0.1× SSC, 0.1% SDS, and 60°C) revealed only one band for each of the restriction digests. This result suggests that the probe is likely to be ZmTIP1 gene specific.
Figure 6.
Genomic Southern analysis. Total maize genomic DNA (15 μg per lane) was digested with EcoRI, HindIII, and BamHI and hybridized with labeled 3′-untranslated region of ZmTIP1 cDNA. The positions of the Mr markers are indicated.
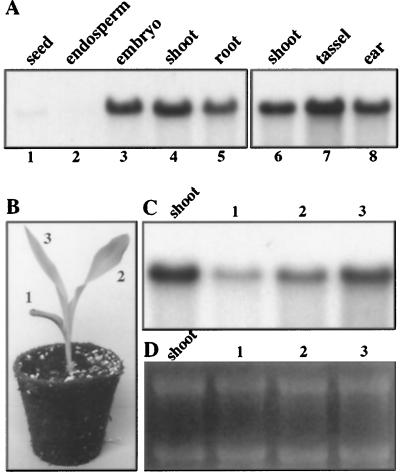
To characterize the pattern of expression of ZmTIP1, gel-blot analysis of total RNA from different maize tissues was performed. ZmTIP1 transcripts with a size of 1.15 kb were observed in all of the expanding tissues studied, from the embryo to the flower organs (Fig. 7A, lanes 3–8). The transcripts were absent from the endosperm, which represents more than 90% of the total seed extract (Fig. 7A, lane 2). To determine if the high ZmTIP1 expression seen in the expanding tissues persists in older organs, we performed an RNA-blot hybridization with total RNA obtained from leaves of light-grown seedlings at the three-leaf stage of development (Fig. 7, B–D). In these seedlings the first plumular leaf (leaf no. 1) was fully expanded, leaf no. 2 was close to the end of its growth, and the youngest leaf (leaf no. 3) was still growing rapidly (Fig. 7B). The ZmTIP1 transcript level was highest in the youngest expanding leaf (Fig. 7C), indicating a possible role of ZmTIP1 in leaf expansion. ZmTIP1 transcript abundance in the different leaves was lower in these green leaves compared with the level in the developing etiolated shoot.
Figure 7.
Gel-blot analysis of ZmTIP1 mRNA in different vegetative and reproductive organs. Total RNA (20 μg) was extracted from the indicated organs (A, C, and D) and from 10-d-old maize plantlet leaves (1–3 in B–D), and separated by gel electrophoresis in the presence of ethidium bromide (D). After transfer the blots were hybridized with ZmTIP1 probe (A and C). Total shoot RNA was used as a control to compare the signal intensity in the different blots.
To more precisely determine the localization of ZmTIP1 expression in developing organs, the patterns of ZmTIP1 mRNA localization were determined by in situ hybridization of digoxigenin-labeled RNA probes using longitudinal sections through various organs (Fig. 8). The intensity of the red color indicates the abundance of mRNA. The controls probed with sense cRNA were white (Fig. 8, C, E, and I). In the primary root of a maize seedling the highest expression of ZmTIP1 was detected in the apical meristem and the cell-elongation zone (Fig. 8A). Cells close to the vascular bundles stained more intensely than the cortical cells. No transcripts were detected in the root cap or the quiescent center. At more distal regions from the root tip, ZmTIP1 expression decreased dramatically, and seemed to be restricted to the epidermis and a zone surrounding the vascular cylinder. More distally, strong signals were found in the new lateral root primordia at the periphery of the vascular cylinder (Fig. 8B). A weak expression was still detectable around the vascular bundle.
Figure 8.
Localization of ZmTIP1 mRNA by in situ hybridization. The controls, hybridized with sense RNA, are shown in C, E, and I; all other panels were hybridized with antisense RNA. A, Longitudinal section of 3-d-old root tip; B, longitudinal section in the zone of lateral root initiation; C, control, same section as shown in A; D and E, median sections of 3-d-old plumule; F, median section of an immature tassel; G, close-up of F; H and I, median section of an immature ear. Ca, Root cap; DZ, division zone; EZ, elongation zone; Gl, glume; Le, lemma; LF, lower floret; Lo, lodicule; LP, leaf primordium; S, stamen; VB, vascular bundle; TZ, transition zone; UF, upper floret. White and black arrows in B indicate RNA transcript signal in the lateral root meristems and the vascular bundle, respectively.
Figure 8D shows ZmTIP1 transcripts in the shoot apical region of a seedling plumule, where ZmTIP1 was most strongly expressed in leaf primordia and expanding leaves. No transcripts were detected in the coleoptile at this stage of seedling development (data not shown). In immature male and female inflorescences, ZmTIP1 expression was mainly localized in the developing spikelets (Fig. 8, F–H), but transcripts were also present around the vascular bundles. In the tassel spikelet, expression was highest in the stamen and lodicule primordia and in the adjoining vessel bundles, but signal was also present in the developing glume and lemma surrounding the florets (Fig. 8G). In the ear spikelet, ZmTIP1 expression was seen mainly in the upper and lower floret primordia (Fig. 8H).
DISCUSSION
The discovery of water-channel proteins in the membranes of plant cells allows the formulation of new mechanisms that may be used by plants to control water transport and osmotic adjustment (Maurel, 1997). The presence of highly conserved motifs in plant aquaporins permitted us to identify and clone by RT-PCR and RACE a tonoplast aquaporin cDNA from maize, ZmTIP1, which is closely related to the Arabidopsis γ-TIP aquaporin (76% amino acid identity). γ-TIP is an integral TIP expressed in the vegetative body of Arabidopsis (Höfte et al., 1992) that can form water channels in X. laevis oocyte membranes (Maurel et al., 1993). In the same way, ZmTIP1 increased the water membrane permeability of X. laevis oocytes. Both aquaporins are sensitive to mercuric chloride. On a dendogram, the ZmTIP1 amino acid sequence clusters with TIP homologs from two monocots, rice (Liu et al., 1994) and barley (Schünmann and Ougham, 1996), and together these three form a larger group with the cauliflower BobTIP26 and Arabidopsis γ-TIP, two dicot aquaporins. This group diverges from other identified tonoplast aquaporins such as the Arabidopsis δ-TIP (Daniels et al., 1996) and bean α-TIP (Johnson et al., 1990), which have different expression patterns. The clustering of γ-TIP homologs in two groups according to the plant classes suggests that a common but already specialized γ-TIP ancestor diverged during the evolution of the monocots and dicots.
ZmTIP1 Is Highly Expressed in Dividing Cells
Detailed analysis of ZmTIP1 transcript localization by in situ hybridization showed a high expression in zones of cell division and elongation of the roots, leaves, and reproductive organs. The high level of expression observed in conducting tissues is discussed in the accompanying paper (Barrieu et al., 1998). Dividing cells contain numerous small vacuoles in different stages of development. Stereological measurements with meristematic cells of Vicia faba showed that the combined volume of the spherical vacuoles represents 27% of the cell volume and that the combined surface area of these vacuoles is as large as that of the plasma membrane area (Steer, 1981). Meristematic cells must generate equal amounts of tonoplast and plasma membrane between rounds of cell division, a process that requires the synthesis of new membrane components. Vacuole biogenesis proceeds through the formation of provacuoles that fuse in an autophagic process (for review, see Marty, 1997).
Because TIPs are abundant in the tonoplast, one might expect a high level of TIP transcripts in dividing cells. The presence of TIPs in meristematic cells has been previously shown in root and shoot of beet (Marty-Mazars et al., 1995), in barley and pea root tips (Paris et al., 1996), and in cauliflower florets (Barrieu et al., 1998). Transcripts of the root-specific TobRB7 were also detected in meristematic cells but it is not known if this gene encodes a tonoplast or plasma membrane aquaporin (Yamamoto et al., 1991). The expression of ZmTIP1 reported here for dividing cells differs substantially from our previous finding with the close Arabidopsis homolog γ-TIP (Ludevid et al., 1992). Analysis of γ-TIP promoter-GUS gene fusion-transformed plant and whole-mount in situ hybridizations carried out with seedlings showed expression in vascular bundles and other tissues, but not in meristems. With respect to the GUS fusions it is likely that regulatory elements were missing from the promoter-GUS fusion construct.
ZmTIP1 Is Highly Expressed in Expanding Cells
In addition to the zone of cell division, root tips have a zone of cell elongation, and between these two there is a transition zone or distal elongation zone, in which the cells expand isodiametrically, growing in width as much as in length (Fig. 7A) (Ishikawa and Evans, 1995; Baluska et al., 1996). In this transition zone cells have to develop the necessary synthetic machinery for the biogenesis of new tonoplast and plasma membranes, cell wall components, new enzymatic complexes, and cytoplasmic structures that support the rapid growth in the elongation zone (Baluska et al., 1996). Cells of the distal elongation zone respond to a variety of signals, such as auxin (Ishikawa and Evans, 1993), water stress (Sharp et al., 1988) and gravistimulation (Ishikawa et al., 1991), differently from the cells in the main elongation zone. For instance, the elongation of cells in this zone is unaffected by reducing the water potential, whereas the rate of elongation in the main elongation zone is inhibited (Sharp et al., 1988). As observed by in situ hybridization experiments, the expression of the tonoplast aquaporin ZmTIP1 is high in these cells. It would be interesting to analyze the pattern of expression of ZmTIP1 and/or other aquaporin genes in this zone in response to external signals.
Rapid elongation of plant cells is based on an extensive uptake of solutes coupled with the uptake of water, resulting in the formation of a prominent vacuolar compartment. This mechanism maintains the turgor pressure that drives cell expansion. Rapid cell expansion may require a high hydraulic permeability of the tonoplast to support water entry into the vacuole. An important role for TIPs during this process was initially suggested by the observation that Arabidopsis γ-TIP is highly expressed in the zone of cell elongation in roots, hypocotyls, and leaves (Ludevid et al., 1992). Arabidopsis γ-TIP expression was shown to be up-regulated after application of GA3 in ga1, a GA-deficient dwarf mutant (Phillips and Huttly, 1994). Also, HvTIP1 transcripts were increased in the slender mutant of barley, characterized by a faster elongation rate of the leaves (Schünmann and Ougham, 1996). Maize ZmTIP1 expression is also high in young, developing leaves and zones of cell elongation in roots, as observed by RNA gel-blot analysis and in situ hybridization.
The high water permeability of the plant tonoplast was recently demonstrated directly with tonoplast vesicles of cultured tobacco cells (Maurel et al., 1997) and wheat roots (Niemietz and Tyerman, 1997). Tonoplast vesicles have channels that transport water with a low energy of activation and that are inhibited by mercuric chloride, whereas plasma membrane vesicles either do not have such channels or the channels are inactive. Together these experiments support the interpretation that TIPs permit the rapid influx of water into the vacuole of elongating cells. The much lower permeability of plasma membrane vesicles observed in the same studies (Maurel et al., 1997; Niemietz and Tyerman, 1997) may indicate that cells regulate the influx of water at the plasma membrane.
ACKNOWLEDGMENTS
We thank Dr. R.J. Schmidt and his collaborators for helpful discussions, and members of Dr. M.F. Yanofsky's laboratory for help with the in situ hybridization and use of their equipment.
Abbreviations:
- GST
glutathione S transferase
- MIP
major intrinsic protein
- PIP
plasma membrane intrinsic protein
- RACE
rapid amplification of cDNA ends
- RT-PCR
reverse transcription-coupled PCR
- TIP
tonoplast intrinsic protein
Footnotes
This work was supported by a cell biology grant from the National Science Foundation. F.C. was supported by a European Molecular Biology Organization fellowship.
LITERATURE CITED
- Baluska F, Volkmann D, Barlow PW. Specialized zones of development in roots. View from the cellular level. Plant Physiol. 1996;112:3–4. doi: 10.1104/pp.112.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrieu F, Chaumont F, Chrispeels M. High expression of the tonoplast aquaporin ZmTIP1 in epidermal and conducting tissues of maize. Plant Physiol. 1998;117:1153–1163. doi: 10.1104/pp.117.4.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrieu F, Thomas D, Marty-Mazars D, Charbonnier M, Marty F. Tonoplast intrinsic proteins from cauliflower (Brassica oleracea L. var. botrytis): immunological analysis, cDNA cloning and evidence for expression in meristematic tissues. Planta. 1998;204:335–344. doi: 10.1007/s004250050264. [DOI] [PubMed] [Google Scholar]
- Cone KC, Burr FA, Burr B. Molecular analysis of the maize anthocyanin regulatory locus C1. Proc Natl Acad Sci USA. 1986;83:9631–9635. doi: 10.1073/pnas.83.24.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MJ, Chaumont F, Mirkov TE, Chrispeels MJ. Characterization of a new vacuolar membrane aquaporin sensitive to mercury at a unique site. Plant Cell. 1996;8:587–599. doi: 10.1105/tpc.8.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MJ, Mirkov TE, Schroeder JI, Chrispeels MJ. The plasma membrane of Arabidopsis thaliana contains a mercury-insensitive aquaporin that is a homolog of the tonoplast water channel protein TIP. Plant Physiol. 1994;106:1325–1333. doi: 10.1104/pp.106.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evola SV, Burr FA, Burr B. The suitability of restriction fragment length polymorphisms as genetic markers in maize. Theor Appl Genet. 1986;71:765–771. doi: 10.1007/BF00276416. [DOI] [PubMed] [Google Scholar]
- Höfte H, Hubbard L, Reizer J, Ludevid D, Herman EM, Chrispeels MJ. Vegetative and seed-specific isoforms of a putative solute transporter in the tonoplast of Arabidopsis thaliana. Plant Physiol. 1992;99:561–570. doi: 10.1104/pp.99.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Evans ML. The role of the distal elongation zone in the response of maize roots to auxin and gravity. Plant Physiol. 1993;102:1203–1210. doi: 10.1104/pp.102.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Evans ML. Specialized zones of development in roots. Plant Physiol. 1995;109:725–727. doi: 10.1104/pp.109.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Hasenstein KH, Evans ML. Computer based video digitizer analysis of surface extension in maize roots: kinetics of growth rate changes during gravitropism. Planta. 1991;183:381–390. doi: 10.1007/BF00197737. [DOI] [PubMed] [Google Scholar]
- Johnson KD, Herman EM, Chrispeels MJ. An abundant, highly conserved tonoplast protein in seeds. Plant Physiol. 1989;91:1006–1013. doi: 10.1104/pp.91.3.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Höfte H, Chrispeels MJ. An intrinsic tonoplast protein of protein storage vacuoles in seeds is structurally related to a bacterial solute transporter (GlpF) Plant Cell. 1990;2:525–532. doi: 10.1105/tpc.2.6.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldenhoff R, Kolling A, Meyers J, Karmann U, Ruppel G, Richter G. The blue light-responsive ATHH2 gene of Arabidopsis thaliana is primarily expressed in expanding as well as in differentiating cells and encodes a putative channel protein of the plasmalemma. Plant J. 1995;7:87–95. doi: 10.1046/j.1365-313x.1995.07010087.x. [DOI] [PubMed] [Google Scholar]
- Kammerloher W, Fischer U, Piechottka GP, Schäffner AR. Water channels in the plant plasma membrane cloned by immunoselection from a mammalian expression system. Plant J. 1994;6:187–199. doi: 10.1046/j.1365-313x.1994.6020187.x. [DOI] [PubMed] [Google Scholar]
- Liu Q, Umeda M, Uchimiya H. Isolation and expression analysis of two rice genes encoding the major intrinsic protein. Plant Mol Biol. 1994;26:2003–2007. doi: 10.1007/BF00019511. [DOI] [PubMed] [Google Scholar]
- Ludevid D, Höfte H, Himelblau E, Chrispeels MJ. The expression pattern of the tonoplast intrinsic protein γ-TIP in Arabidopsis thaliana is correlated with cell enlargement. Plant Physiol. 1992;100:1633–1639. doi: 10.1104/pp.100.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrison JL, Leech RM. The subcellular and intra-organelle recognition of nuclear and chloroplast transcripts in developing leaf cells. Plant J. 1994;6:605–614. [Google Scholar]
- Marty F. The biogenesis of vacuoles: insights from microscopy. In: Leigh A, Sanders D, editors. The Plant Vacuoles, Vol 25. San Diego, CA: Academic Press; 1997. pp. 1–42. [Google Scholar]
- Marty-Mazars D, Clémencet MC, Dozolme P, Marty F. Antibodies to the tonoplast from the storage parenchyma cells of beetroot recognize a major intrinsic protein related to TIPs. Eur J Cell Biol. 1995;66:106–118. [PubMed] [Google Scholar]
- Maurel C. Aquaporins and water permeability of plant membranes. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:399–429. doi: 10.1146/annurev.arplant.48.1.399. [DOI] [PubMed] [Google Scholar]
- Maurel C, Reizer J, Schroeder JI, Chrispeels MJ. The vacuolar membrane protein γ-TIP creates water specific channels in Xenopus oocytes. EMBO J. 1993;12:2241–2247. doi: 10.1002/j.1460-2075.1993.tb05877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Tacnet F, Güclü J, Guern J, Ripoche P. Purified vesicles of tobacco cell vacuolar and plasma membranes exhibit dramatically different water permeability and water channel activity. Proc Natl Acad Sci USA. 1997;94:7103–7108. doi: 10.1073/pnas.94.13.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melroy DL, Herman EM. TIP, an integral membrane protein of the protein-storage vacuoles of the soybean cotyledon undergoes developmentally regulated membrane accumulation and removal. Planta. 1991;184:113–122. doi: 10.1007/BF00208244. [DOI] [PubMed] [Google Scholar]
- Niemietz CM, Tyerman ST. Characterization of water channels in wheat root membrane vesicles. Plant Physiol. 1997;115:561–567. doi: 10.1104/pp.115.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman CH, Taylor CG, Conkling MA. Root-knot nematode-directed expression of a plant root-specific gene. Science. 1994;263:221–223. doi: 10.1126/science.263.5144.221. [DOI] [PubMed] [Google Scholar]
- Paris N, Stanley CM, Jones RL, Rogers JC. Plant cells contain two functionally distinct vacuolar compartments. Cell. 1996;85:563–572. doi: 10.1016/s0092-8674(00)81256-8. [DOI] [PubMed] [Google Scholar]
- Park JH, Saier MH., Jr Phylogenetic characterization of the MIP family of transmembrane channel proteins. J Membr Biol. 1996;153:171–180. doi: 10.1007/s002329900120. [DOI] [PubMed] [Google Scholar]
- Phillips AL, Huttly AK. Plant Mol Biol. 1994;24:603–615. doi: 10.1007/BF00023557. [DOI] [PubMed] [Google Scholar]
- Preston GM, Carroll TP, Guggino WB, Agre P. Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science. 1992;256:385–387. doi: 10.1126/science.256.5055.385. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmidt RJ, Burr FA, Burr B. Transposon tagging and molecular analysis of the maize regulatory locus opaque-2. Science. 1987;238:960–963. doi: 10.1126/science.2823388. [DOI] [PubMed] [Google Scholar]
- Schünmann PHD, Ougham HJ. Identification of three clones expressed in the leaf extension zone and with altered patterns of expression in the slender mutant of barley: a tonoplast intrinsic protein, a putative structural protein and photochlorophyllide oxidoreductase. Plant Mol Biol. 1996;31:529–537. doi: 10.1007/BF00042226. [DOI] [PubMed] [Google Scholar]
- Sharp RE, Silk WK, Hsiao TC. Growth of the maize primary root at low water potentials. I. Spatial distribution of expansive growth. Plant Physiol. 1988;87:50–57. doi: 10.1104/pp.87.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steer MW (1981) Understanding Cell Structure. Cambridge University Press, Cambridge, UK, 126 pp
- Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang S, Yin X, Guzzo-Arkuran C, Jones V, Davidson A. Loss of resolution of gel electrophoresis of RNA: a problem associated with the presence of formaldehyde gradients. BioTechniques. 1993;14:380–381. [PubMed] [Google Scholar]
- Weig A, Deswarte C, Chrispeels MJ. The major intrinsic protein family of Arabidopsis has 23 members that form three distinct groups with functional aquaporins in each group. Plant Physiol. 1997;114:1347–1357. doi: 10.1104/pp.114.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westgate ME, Boyer JS. Osmotic adjustment and the inhibition of leaf, root, stem and silk growth at low water potentials in maize. Planta. 1985;164:540–549. doi: 10.1007/BF00395973. [DOI] [PubMed] [Google Scholar]
- Yamada S, Katsuhara M, Kelly W, Michalowski C, Bohnert H. A family of transcripts encoding the water channel proteins: tissue-specific expression in the common ice plant. Plant Cell. 1995;7:1129–1142. doi: 10.1105/tpc.7.8.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YT, Taylor CG, Acedo GN, Cheng C-L, Conkling MA. Characterization of cis-acting sequences regulating root-specific gene expression in tobacco. Plant Cell. 1991;3:371–382. doi: 10.1105/tpc.3.4.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu GL, Steudle E. Water transport across maize roots. Simultaneous measurement of flows at the cell and root level by double pressure probe technique. Plant Physiol. 1991;95:305–315. doi: 10.1104/pp.95.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]