Abstract
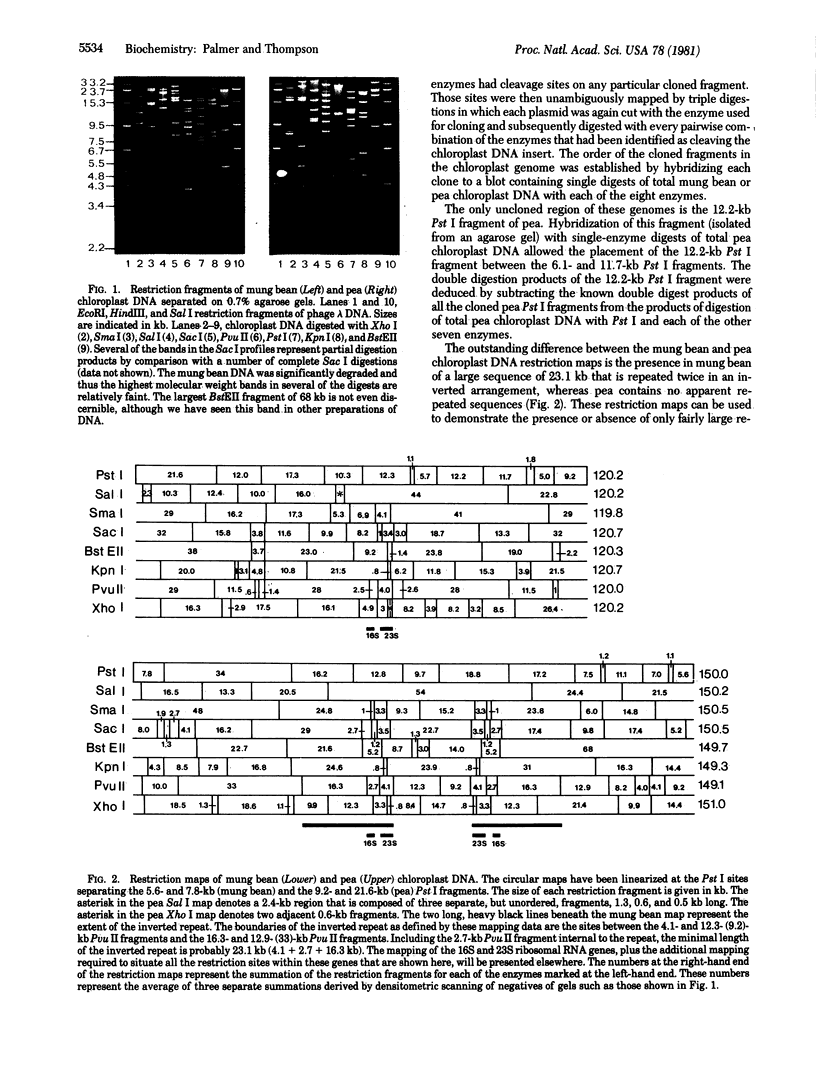
We have mapped all the cleavage sites for the restriction endonucleases BstEII, Kpn I, Pst I, Pvu II, Sac I, Sal I, Sma I, and Xho I on the circular chloroplast chromosomes from mung bean and pea. The mung bean chloroplast genome measures 150 kilobase pairs (kb) in length; it includes two identical sequences of 23 kb that contain the ribosomal genes and are arranged as an inverted repeat separated by single-copy regions of 21 and 83 kb. The pea chloroplast genome is only 120 kb in size, has only one set of ribosomal genes, and does not possess any detectable repeated sequences. The mung bean inverted repeat structure is common to all other nonleguminous higher plant chloroplast genomes studied, whereas the pea structure has been found only in the closely related legume Vicia faba. We conclude from these data that loss of one copy of the inverted repeat sequence has occurred only rarely during the evolution of the Angiosperms, and in the case of the legumes after the divergence of the mung bean line from the pea-Vicia line. We present hybridization data indicating that rearrangements that change the linear order of homologous sequences within the chloroplast genome have been quite frequent during the course of legume evolution.
Keywords: inverted repeat, restriction endonuclease maps, ribosomal genes, sequence arrangement
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adelberg E. A., Bergquist P. The stabilization of episomal integration by genetic inversion: a general hypothesis. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2061–2065. doi: 10.1073/pnas.69.8.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedbrook J. R., Bogorad L. Endonuclease recognition sites mapped on Zea mays chloroplast DNA. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4309–4313. doi: 10.1073/pnas.73.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedbrook J. R., Kolodner R., Bogorad L. Zea mays chloroplast ribosomal RNA genes are part of a 22,000 base pair inverted repeat. Cell. 1977 Aug;11(4):739–749. doi: 10.1016/0092-8674(77)90288-4. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisaro D., Siegel A. Sequence Homology between Chloroplast DNAs from Several Higher Plants. Plant Physiol. 1980 Feb;65(2):234–237. doi: 10.1104/pp.65.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovenberg W. A., Kool A. J., Nijkamp H. J. Isolation, characterization and restriction endonuclease mapping of the Petunia hybrida chloroplast DNA. Nucleic Acids Res. 1981 Feb 11;9(3):503–517. doi: 10.1093/nar/9.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant D. M., Gillham N. W., Boynton J. E. Inheritance of chloroplast DNA in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1980 Oct;77(10):6067–6071. doi: 10.1073/pnas.77.10.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann R. G., Possingham J. V. Plastid DNA-the plastome. Results Probl Cell Differ. 1980;10:45–96. doi: 10.1007/978-3-540-38255-3_3. [DOI] [PubMed] [Google Scholar]
- Jurgenson J. E., Bourque D. P. Mapping of rRNA genes in an inverted repeat in Nicotiana tabacum chloroplast DNA. Nucleic Acids Res. 1980 Aug 25;8(16):3505–3516. doi: 10.1093/nar/8.16.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R., Tewari K. K. Inverted repeats in chloroplast DNA from higher plants. Proc Natl Acad Sci U S A. 1979 Jan;76(1):41–45. doi: 10.1073/pnas.76.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R., Tewari K. K. The molecular size and conformation of the chloroplast DNA from higher plants. Biochim Biophys Acta. 1975 Sep 1;402(3):372–390. doi: 10.1016/0005-2787(75)90273-7. [DOI] [PubMed] [Google Scholar]
- Lamppa G. K., Bendich A. J. Chloroplast DNA Sequence Homologies among Vascular Plants. Plant Physiol. 1979 Apr;63(4):660–668. doi: 10.1104/pp.63.4.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N. Dictyostelium 17S, 25S, and 5S rDNAs lie within a 38,000 base pair repeated unit. Cell. 1976 Nov;9(3):431–438. doi: 10.1016/0092-8674(76)90088-x. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. H., Kutter E., Nakanishi M. A restriction map of the bacteriophage T4 genome. Mol Gen Genet. 1980;179(2):421–435. doi: 10.1007/BF00425473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Sato S., Hutchinson C. A., 3rd, Harris J. I. A thermostable sequence-specific endonuclease from Thermus aquaticus. Proc Natl Acad Sci U S A. 1977 Feb;74(2):542–546. doi: 10.1073/pnas.74.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thomas J. R., Tewari K. K. Conservation of 70S ribosomal RNA genes in the chloroplast DNAs of higher plants. Proc Natl Acad Sci U S A. 1974 Aug;71(8):3147–3151. doi: 10.1073/pnas.71.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. R., Tewari K. K. Ribosomal-RNA genes in the chloroplast DNA of pea leaves. Biochim Biophys Acta. 1974 Aug 15;361(1):73–83. doi: 10.1016/0005-2787(74)90210-x. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensink P. C., Tabata S., Pachl C. The clustered and scrambled arrangement of moderately repetitive elements in Drosophila DNA. Cell. 1979 Dec;18(4):1231–1246. doi: 10.1016/0092-8674(79)90235-6. [DOI] [PubMed] [Google Scholar]
- van Ee J. H., Vos Y. J., Planta R. J. Physical map of chloroplast DNA of Spirodela oligorrhiza; analysis by the restriction endonucleases PstI, xhoI, SacI. Gene. 1980 Dec;12(3-4):191–200. doi: 10.1016/0378-1119(80)90101-8. [DOI] [PubMed] [Google Scholar]