Abstract
The genetic diversity of Cryptosporidium in reptiles was analyzed by PCR-restriction fragment length polymorphism and sequence analysis of the small subunit rRNA gene. A total of 123 samples were analyzed, of which 48 snake samples, 24 lizard samples, and 3 tortoise samples were positive for Cryptosporidium. Nine different types of Cryptosporidium were found, including Cryptosporidium serpentis, Cryptosporidium desert monitor genotype, Cryptosporidium muris, Cryptosporidium parvum bovine and mouse genotypes, one C. serpentis-like parasite in a lizard, two new Cryptosporidium spp. in snakes, and one new Cryptosporidium sp. in tortoises. C. serpentis and the desert monitor genotype were the most common parasites and were found in both snakes and lizards, whereas the C. muris and C. parvum parasites detected were probably the result of ingestion of infected rodents. Sequence and biologic characterizations indicated that the desert monitor genotype was Cryptosporidium saurophilum. Two host-adapted C. serpentis genotypes were found in snakes and lizards.
Cryptosporidium infections are common in reptiles and have been reported in at least 57 reptilian species (10). Unlike in other animals in which Cryptosporidium infection is usually self-limiting in immunocompetent individuals, cryptosporidiosis in reptiles is frequently chronic and sometimes lethal in snakes. Two Cryptosporidium spp. are recognized in reptiles (2, 15): Cryptosporidium serpentis in snakes and Cryptosporidium saurophilum in lizards, which differ from each other in morphology (oocysts of C. serpentis are bigger than those of C. saurophilum) and predilection sites (C. serpentis is a gastric parasite, whereas C. saurophilum is an intestinal parasite). Morphometric studies on isolates recovered from wild snakes and lizards have suggested the occurrence of at least five different morphotypes (12), indicating that it is likely other Cryptosporidium spp. may also exist in reptiles.
Until recently there have been few molecular characterizations of Cryptosporidium spp. from reptiles. Morgan et al. characterized 15 isolates of Cryptosporidium from snakes and lizards and found that the majority of animals were infected with C. serpentis, with the rest of the isolates belonging to oocysts of the Cryptosporidium parvum bovine genotype (two cases) and Cryptosporidium muris (one case), probably from ingested prey or feeder mice (9). Thus, it is difficult to differentiate parasitic Cryptosporidium oocysts from those merely passing through the gastrointestinal tract, and some of the previously observed morphotypes may represent oocysts of C. parvum and C. muris resulting from the ingestion of infected rodents (4). The extent of genetic diversity within C. serpentis organisms is also not clear, but C. serpentis infection in lizards is usually asymptomatic, whereas the infection in snakes frequently causes clinical diseases (1, 3). Minor genetic differences have been observed between isolates from snakes and those from lizards (16). A Cryptosporidium isolate from a desert monitor has recently been shown to be genetically distinct and was related to the intestinal Cryptosporidium group (17). It is unclear, however, whether oocysts from the desert monitor belong to C. saurophilum from lizards.
In this study, we analyzed 123 samples from snakes, lizards, and tortoises and characterized the small subunit (SSU) rRNA gene of Cryptosporidium-positive samples by PCR-restriction fragment length polymorphism (PCR-RFLP) and DNA sequencing. Results of the analysis suggest the existence of extensive genetic diversity and some host adaptations in Cryptosporidium isolates from reptiles.
MATERIALS AND METHODS
Samples.
A total of 123 diagnostic samples obtained from captive snakes, lizards, and tortoises from the United States, Switzerland, the Czech Republic, Ghana, and Australia were used in this study (Tables 1 through 3). These included 88 samples from snakes, 26 samples from lizards, and 9 samples from tortoises. Another 19 samples from cross-transmission studies (11 snake samples and 8 lizard samples) were also studied. With the exception of samples from the Saint Louis Zoo (78 snake samples, 7 lizard samples, and 3 tortoise samples), Louisville Zoo (6 tortoise samples), and transmission studies, all samples were previously diagnosed as Cryptosporidium positive by microscopy, and purified oocysts were used in molecular studies. Samples from the Saint Louis Zoo, Louisville Zoo, and National Zoological Park were mostly feces, with the exception of gastric washings from three snakes in the Saint Louis Zoo.
TABLE 1.
Distribution of Cryptosporidium spp. and genotypes in snakesa
| Snake
|
Source | Sample no. | Species and/or genotype identified by:
|
||
|---|---|---|---|---|---|
| Common name | Scientific name | RFLP | Sequence analysis | ||
| Amazon tree boa | Corallus hortulanus | Washington, D.C. | 64 | C. serpentis | C. serpentis type A |
| Ball python | Python regius | St. Louis Zoo | 812 | C. serpentis | C. serpentis type A |
| Ball python | Python regius | St. Louis Zoo | 1999 | C. parvum mouse genotype | ND |
| Black rat snake | Elaphe obsoleta obsoleta | St. Louis Zoo | 762 | C. parvum mouse genotype | ND |
| Black rat snake | Elaphe obsoleta obsoleta | St. Louis Zoo | 756 | C. serpentis | C. serpentis type A |
| Black rat snake | Elaphe obsoleta obsoleta | St. Louis Zoo | 757 | C. parvum mouse genotype | ND |
| Black rat snake | Elaphe obsoleta obsoleta | St. Louis Zoo | 758 | C. muris | C. muris |
| Black rat snake | Elaphe obsoleta obsoleta | St. Louis Zoo | 825 | C. parvum mouse genotype | ND |
| Boa constrictor | Boa constrictor ortoni | Maryland | 694 | C. serpentis | ND |
| Boa constrictor | Boa constrictor ortoni | St. Louis Zoo | 446 | C. serpentis | ND |
| Boa constrictor | Boa constrictor ortoni | St. Louis Zoo | 819 | C. serpentis | C. serpentis type A |
| Boa constrictor | Boa constrictor ortoni | St. Louis Zoo | 821 | C. serpentis | C. serpentis type A |
| Boa constrictor | Boa constrictor ortoni | St. Louis Zoo | 899 | C. serpentis | C. serpentis type A |
| Boa constrictor | Boa constrictor ortoni | St. Louis Zoo | 1158 | C. serpentis | C. serpentis type A |
| Boa constrictor | Boa constrictor ortoni | St. Louis Zoo | 1993 | C. muris | C. muris |
| Boa constrictor | Boa constrictor ortoni | St. Louis Zoo | 1996 | C. parvum mouse genotype | C. parvum mouse genotype |
| Boa constrictor | Boa constrictor ortoni | St. Louis Zoo | 2162 | Cryptosporidium sp. | Cryptosporidium sp. |
| Boelen's python | Morelia boeleni | St. Louis Zoo | 7632 | C. serpentis | ND |
| Boelen's python | Morelia boeleni | St. Louis Zoo | 7634 | C. serpentis | ND |
| Boelen's python | Morelia boeleni | St. Louis Zoo | 7636 | C. serpentis | ND |
| Bornmueller's viper | Vipera bornmuelleri | St. Louis Zoo | 816 | C. serpentis | C. serpentis type A |
| Bull snake | Pituophis melanoleucuc sayi | St. Louis Zoo | 815 | C. saurophilum | C. saurophilum type A |
| California king snake | Lampropeltis getulus californiae | St. Louis Zoo | 753 | C. serpentis and C. parvum mouse genotype | C. serpentis type A |
| California king snake | Lampropeltis getulus californiae | St. Louis Zoo | 754 | C. serpentis | C. serpentis type A |
| California king snake | Lampropeltis getulus californiae | St. Louis Zoo | 828 | C. serpentis | C. serpentis type A |
| Corn snake | Elaphe guttata guttata | Kansas | 18 | C. serpentis | C. serpentis type A |
| Corn snake | Elaphe guttata guttata | Maryland | 695 | C. serpentis | ND |
| Corn snake | Elaphe guttata guttata | St. Louis Zoo | 759 | C. serpentis and C. parvum mouse genotype | ND |
| Corn snake | Elaphe guttata guttata | St. Louis Zoo | 820 | C. serpentis | C. serpentis type A |
| Corn snake | Elaphe guttata guttata | St. Louis Zoo | 760 | C. serpentis | C. serpentis type A |
| Corn snake | Elaphe guttata guttata | St. Louis Zoo | 1444 | C. parvum mouse genotype | C. parvum mouse genotype |
| Corn snake | Elaphe guttata guttata | St. Louis Zoo | 1446 | C. parvum mouse genotype | ND |
| Corn snake | Elaphe guttata guttata | St. Louis Zoo | 1545 | C. serpentis | C. serpentis type A |
| Corn snake | Elaphe guttata guttata | St. Louis Zoo | 1779 | C. muris and C. parvum mouse genotype | C. muris and C. parvum mouse genotype |
| Corn snake | Elaphe guttata guttata | St. Louis Zoo | 1998 | C. parvum mouse genotype | ND |
| Corn snake | Elaphe guttata guttata | Switzerland | 1430 | C. serpentis | C. serpentis type A |
| Emerald tree boa | Corallus caninus | Maryland | 693 | C. serpentis | C. serpentis type A |
| Emerald tree boa | Corallus caninus | St. Louis Zoo | 1159 | C. parvum mouse genotype | ND |
| Fox snake | Elaphe vulpina gloydi | St. Louis Zoo | 755 | C. serpentis | C. serpentis type A |
| Fox snake | Elaphe vulpina gloydi | St. Louis Zoo | 818 | C. parvum mouse genotype | ND |
| Louisiana pine snakeb | Pituophis ruthveni | St. Louis Zoo | 691 | C. saurophilum | C. saurophilum type A |
| Louisiana pine snake | Pituophis ruthveni | St. Louis Zoo | 692 | C. saurophilum | C. saurophilum type A |
| Milk snake | Lampropeltis triangulum | St. Louis Zoo | 827 | C. parvum mouse genotype | ND |
| Mountain viper | Vipera wagneri | St. Louis Zoo | 822 | C. serpentis | C. serpentis type A |
| Mountain viper | Vipera wagneri | St. Louis Zoo | 2168 | C. serpentis | C. serpentis type A |
| Mountain viper | Vipera wagneri | St. Louis Zoo | 7010 | C. serpentis | C. serpentis type A |
| Prairie king snake | Lamproletis calligaster | St. Louis Zoo | 761 | C. parvum mouse genotype | ND |
| Taipan | Oxyuranus scutellatus | Australia | 1429 | C. serpentis | ND |
C. saurophilum, Cryptosporidium desert monitor genotype; ND, not sequenced.
Gastric washing from animal 692.
TABLE 3.
Cross-transmission of Cryptosporidium spp. between lizards and snakes
| Source of infection | Recipient host (tissues sampled) | Scientific name | Sample no. | Cryptosporidium parasite(s) identifieda |
|---|---|---|---|---|
| Inoculated with C. saurophilum of snake origin (sample 815) | Corn snake S1 (stomach) | Elaphe guttata guttata | 1336 | Negative |
| Corn snake S1 (intestine) | Elaphe guttata guttata | 1337 | Negative | |
| Corn snake S2 (stomach) | Elaphe guttata guttata | 1338 | Negative | |
| Corn snake S2 (intestine) | Elaphe guttata guttata | 1339 | Negative | |
| Leopard gecko, G1 (stomach) | Eublepharis macularius | 1340 | Negative | |
| Leopard gecko, G1 (intestine) | Eublepharis macularius | 1341 | C. saurophilum | |
| Leopard gecko G2 (stomach) | Eublepharis macularius | 1342 | C. saurophilum | |
| Leopard gecko G2 (intestine) | Eublepharis macularius | 1343 | C. saurophilum | |
| Inoculated with C. serpentis of Nile monitor (sample 1172a) | Burmese python | Python mollurus | 1172b | C. serpentis type B |
| Natural exposure via shared housing | Black rat snake | Elaphe obsolete | 940 | C. serpentis-Cryptosporidium sp.-C. saurophilum |
| Green python | Chondropython viridis | 941 | C. serpentis-Cryptosporidium sp.-C. saurophilum | |
| Milk snake | Lampropeltis triangulum | 939 | C. serpentis-Cryptosporidium sp.-C. saurophilum | |
| New Guinea viper boa | Candoia asper | 938 | C. serpentis-Cryptosporidium sp.-C. saurophilum | |
| Pine snake | Pituophis melanoleucus | 936 | C. serpentis-Cryptosporidium sp.-C. saurophilum | |
| Pine snake | Pituophis melanoleucus | 937 | C. serpentis-Cryptosporidium sp.-C. saurophilum | |
| Bearded dragon | Pogona vitticep | 944 | C. serpentis-C. saurophilum | |
| Gargoyle gecko | Rhodocodactylus auriculatus | 945 | C. serpentis-C. saurophilum | |
| Mountain chameleon | Chamaeleo montium | 942 | C. serpentis-C. saurophilum | |
| Mountain chameleon | Chamaeleo montium | 943 | C. serpentis-C. saurophilum |
C. saurophilum, Cryptosporidium desert monitor genotype.
After initial diagnosis of cryptosporidiosis in snakes in the Saint Louis Zoo at the end of 1998, a cryptosporidiosis control program was initiated, which involved the diagnosis and differentiation of Cryptosporidium infection by PCR and euthanasia of C. serpentis- or C. saurophilum-infected snakes. To monitor the effectiveness of this cryptosporidiosis control measure, 8 to 17 snakes at the zoo were examined for Cryptosporidium infection periodically for 1 year from May 1999 to April 2000, with continuous euthanasia of infected animals (Table 4). To identify the source of isolates belonging to the C. parvum mouse genotype and C. muris in snakes, fecal samples were also collected from 11 of the feeder mice used in the Saint Louis Zoo and examined for Cryptosporidium species and genotypes.
TABLE 4.
Effectiveness of a diagnosis-euthanasia strategy on the occurrence of Cryptosporidium infection in snakes at the Saint Louis Zooa
| Sample date | No. of samples | Total no. of positivesb | No. of isolates positive for the indicated parasite
|
|
|---|---|---|---|---|
| C. serpentis | C. saurophilum | |||
| May 1999 | 10 | 5 | 5 | |
| June 1999 | 17 | 8 | 7 | 1 |
| September 1999 | 8 | 0 | ||
| November 1999 | 10 | 0 | ||
| December 1999 | 10 | 1 | 1 | |
| March 2000 | 8 | 0 | ||
| April 2000 | 9 | 0 | ||
Euthanasia of infected animals started in March 1999 after the initial identification of cryptosporidiosis in snakes and lizards at the zoo between December 1998 and January 1999.
C. muris and the C. parvum mouse genotype are excluded from the number of positives.
Cross-transmission studies.
To evaluate the infectivity of C. saurophilum to snakes and lizards, two corn snakes and leopard geckoes were inoculated with 10,000 oocysts originating from a bull snake (sample 815). Forty-five days after the experimental infection, animals were euthanized, and the stomach and intestine and their contents were examined for Cryptosporidium oocysts by PCR-RFLP and DNA sequence analysis of the SSU rRNA gene. The infectivity for snakes of C. serpentis isolated from lizards was assessed by experimental infection of a captive-born Burmese python (Python mollurus) with oocysts isolated from a wild imported juvenile Nile monitor (Varanus niloticus) from Togo. Cryptosporidium oocysts in the python's feces were genotyped by DNA sequencing of the SSU rRNA gene 88 days after the inoculation. The cross-transmission of Cryptosporidium spp. between snakes and lizards was further evaluated by the differentiation of Cryptosporidium spp. and genotypes in a group of six snakes and four lizards that were housed in the same room by the SSU rRNA PCR-RFLP analysis (Table 3).
Morphometric measurements.
Oocysts of C. serpentis from a desert monitor (sample 806) and of C. saurophilum from a bull snake (sample 815) were measured under a differential interference contrast microscope at a magnification of ×1,000. Twenty oocysts were measured for C. saurophilum organisms, and 37 oocysts were measured for C. serpentis organisms. Mean length and width and the shape index were calculated along with the 95% confidence limits (CL) for each species.
DNA extraction.
Purified oocysts or fecal samples containing oocysts were used in DNA extraction. DNA was extracted from stool samples by alkaline digestion and phenol-chloroform extraction, followed by DNA purification with a commercial kit. Briefly, 33.3 μl of 1 M KOH and 9.3 μl of 1 M dithiothreitol were added to a 1.5-ml microcentrifuge tube containing 100 μl of stool or oocyst suspension. After incubation at 65°C for 15 min, the solution was neutralized with 4.3 μl of 25% hydrochloric acid and buffered with 80 μl of 2 M Tris-HCl (pH 8.3). The DNA was extracted with 250 μl of phenol-chloroform-isoamyl alcohol (Invitrogen, Carlsbad, Calif.) after thorough mixing and centrifugation in an Eppendorf (Hamburg, Germany) microcentrifuge at 5,000 × g for 5 min. The supernatant was transferred to a 2.0-ml Eppendorf tube containing 1.0 ml of ASL buffer from the QIAamp DNA Stool Mini Kit (QIAGEN, Valencia, Calif.). The DNA was further purified following the manufacturer-suggested procedures. DNA was stored at −70°C before it was used in molecular analysis.
Species differentiation and genotyping.
Cryptosporidium spp. and C. parvum genotypes present were diagnosed by a PCR-RFLP technique (13, 16, 17). In this method, a segment (∼833 bp) of the Cryptosporidium SSU rRNA gene was amplified by nested PCR. Species and genotype diagnosis was made by restriction digestion of the secondary PCR product with SspI (New England BioLabs, Beverly, Mass.) and VspI (Promega, Madison, Wis.). Each sample was examined at least twice by independent PCR-RFLP analyses. To confirm the diagnosis of new Cryptosporidium spp. and to identify genetic heterogeneity within C. serpentis and C. saurophilum, secondary PCR products were sequenced in both directions on an ABI Prism 3100 analyzer (Applied Biosystems, Foster City, Calif.) by using forward and reverse primers, after PCR products had been purified with the Wizard PCR Prep Kit (Promega). Nucleotide sequences obtained from this study were aligned against each other by using the ClustalX (11) program and manual adjustment. A neighbor-joining tree was constructed from the aligned sequences as previously described by using the Treecon program, and genetic distances were calculated with the Kimura 2-parameter model (17).
Because of the presence of mixed Cryptosporidium species in samples from snakes and lizards that were housed together in cross-transmission studies, PCR products from one of the snakes (sample 938) and one of the lizards (sample 944) were cloned into a pGEM-T vector (Promega). Eight (for sample 944) or 15 (for sample 938) clones were sequenced for each PCR product to confirm the diagnosis.
Nucleotide sequence accession numbers.
The nucleotide sequences of the partial SSU rRNA gene have been deposited in the GenBank database under accession numbers AF093499, AF093501, AF112573, AY120913 through AY120915, and AY268581 through AY268584.
RESULTS
Cryptosporidium spp. in reptiles.
SSU rRNA PCR confirmed that all snake and lizard samples from Maryland (three snake samples), Washington, D.C. (one snake sample and four lizard samples), Kansas (one snake sample), Ghana (one lizard sample), the Czech Republic (five lizard samples), Switzerland (one snake sample and nine lizard samples), and Australia (one snake sample and one lizard sample) were positive for Cryptosporidium. The samples were previously diagnosed as Cryptosporidium positive by microscopy. In contrast, 3 of the 6 tortoise samples from the Louisville Zoo, 36 of 81 snake samples, 4 of 7 lizard samples, and 0 of 3 tortoise samples from the Saint Louis Zoo were positive for Cryptosporidium by PCR. These animals had not been previously screened for Cryptosporidium oocysts by microscopy.
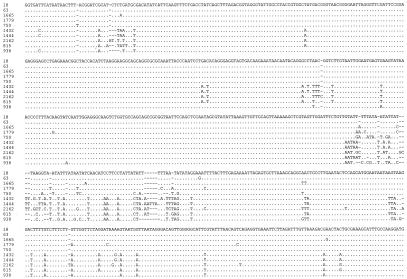
PCR-RFLP analysis of SSU rRNA PCR revealed banding patterns distinctive for C. serpentis and the Cryptosporidium desert monitor genotype (Tables 1 and 2). Cryptosporidium serpentis was found in 28 of 48 positive snake samples and 11 of 25 positive lizard samples, and the Cryptosporidium desert monitor genotype was identified in 3 of 48 positive snake samples and 9 of 24 positive lizard samples. Sequence analyses confirmed the results of RFLP analyses but also revealed genetic diversities within C. serpentis and the Cryptosporidium desert monitor genotype. Most desert monitor genotype isolates had identical SSU rRNA sequences, but isolates 1343 and 1786 had one single nucleotide polymorphism (SNP). Two genotypes (A and B) were seen in C. serpentis isolates, which differed from each other by one SNP (Fig. 1). One isolate from a lizard (sample 1665) had the C. serpentis RFLP banding pattern, was related to the two C. serpentis genotypes, but had significant differences in nucleotide sequence (Fig. 1).
TABLE 2.
Distribution of Cryptosporidium spp. and genotypes in lizards and tortoises
| Animal
|
Source | Sample no. | Species and/or genotype identified bya:
|
||
|---|---|---|---|---|---|
| Common name | Scientific name | RFLP | Sequence analysis | ||
| Gecko | Gekkoninae sp. | Switzerland | 1433 | C. parvum bovine genotype | C. parvum bovine genotype |
| Gecko | Gekkoninae sp. | Switzerland | 1507 | C. saurophilum and C. parvum bovine genotype | C. saurophilum |
| Gecko | Gekkoninae sp. | Switzerland | 1508 | C. parvum bovine genotype | C. parvum bovine genotype |
| Leopard gecko | Eublepharis macularius | Czech Republic | 1665 | C. serpentis | C. serpentis like |
| Leopard gecko | Eublepharis macularius | Czech Republic | 7381 | C. saurophilum | C. saurophilum |
| Green iguana | Iguana iguana | Switzerland | 1431 | C. saurophilum | C. saurophilum |
| Green iguana | Iguana iguana | Switzerland | 1432 | C. parvum bovine genotype | C. parvum bovine genotype |
| Green iguana | Iguana iguana | Switzerland | 1506 | C. saurophilum and C. parvum bovine genotype | ND |
| Desert monitor | Varanus griseus | Czech Republic | 1667 | C. serpentis | C. serpentis type B |
| Desert monitor | Varanus griseus | St. Louis Zoo | 340 | C. saurophilum | C. saurophilum |
| Desert monitor | Varanus griseus | St. Louis Zoo | 600 | C. sepentis | |
| Desert monitor | Varanus griseus griseus | St. Louis Zoo | 806 | C. serpentis | C. serpentis type A |
| Mangrove monitor | Varanus indicus | St. Louis Zoo | 808 | C. parvum mouse genotype | ND |
| Monitor | Varanus sp. | Switzerland | 1434 | C. saurophilum | C. saurophilum |
| Monitor | Varanus sp. | Switzerland | 1504 | C. saurophilum and C. parvum bovine genotype | C. saurophilum |
| Monitor | Varanus sp. | Switzerland | 1505 | C. saurophilum | C. saurophilum |
| Nile monitor | Varanus niloticus | Czech Republic | 1172a | C. serpentis | C. serpentis type B |
| Savannah monitor | Varanus exanthematicus | Czech Republic | 844 | C. serpentis | C. serpentis type B |
| Savannah monitor | Varanus exanthematicus | Washington, D.C. | 40 | C. serpentis | C. serpentis type B |
| Savannah monitor | Varanus exanthematicus | Washington, D.C. | 41 | C. serpentis | ND |
| Savannah monitor | Varanus exanthematicus | Washington, D.C. | 63 | C. serpentis | C. serpentis type B |
| Savannah monitor | Varanus exanthematicus | Washington, D.C. | 521 | C. serpentis lizard type | C. serpentis type B |
| Plated lizard | Gerrhosaurus sp. | St. Louis Zoo | 1786 | C. saurophilum | C. saurophilum |
| Skink | Mabuya perrotetii | Ghana | 956 | C. serpentis | C. serpentis type B |
| Star tortoise | Geochelone elegans | Louisville Zoo | 747 | Cryptosporidium tortoise genotype | Cryptosporidium tortoise genotype |
| Star tortoise | Geochelone elegans | Louisville Zoo | 750 | Cryptosporidium tortoise genotype | Cryptosporidium tortoise genotype |
| Star tortoise | Geochelone elegans | Louisville Zoo | 751 | Cryptosporidium tortoise genotype | Cryptosporidium tortoise genotype |
C. saurophilum, Cryptosporidium desert monitor genotype; ND, not sequenced.
FIG. 1.
Sequence diversity in the SSU rRNA gene among Cryptosporidium in reptiles. Dots denote sequence identity to isolate 18, dashes represent nucleotide deletions. Isolate designations: 18, C. serpentis genotype A; 63, C. serpentis genotype B; 1665, a C. serpentis-like Cryptosporidium in lizard 1665; 1779, C. muris; 750, Cryptosporidium tortoise genotype; 1432, C. parvum bovine genotype; 1444, C. parvum mouse genotype; 2162, another new Cryptosporidium genotype in snake 2162; 815, C. saurophilum; 938, a new Cryptosporidium genotype in snake 938.
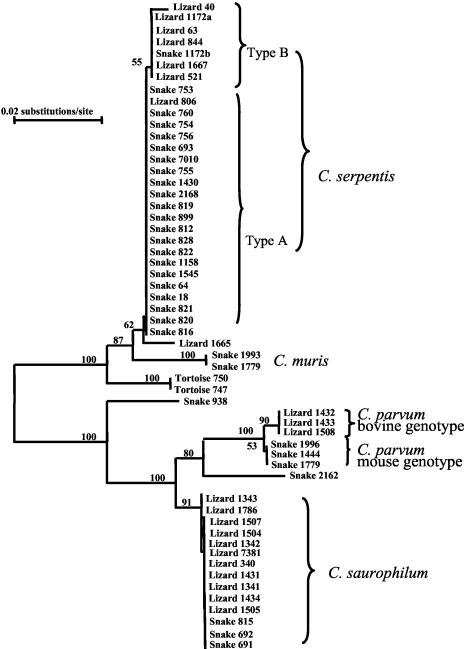
Several other Cryptosporidium spp. were also identified in reptiles. RFLP analysis showed banding patterns of the C. parvum mouse genotype in 12 snakes and 1 lizard, of the C. parvum bovine genotype in 6 lizards, and of C. muris in 3 snakes. DNA sequence analysis confirmed the identifications, as the sequences obtained were identical to previously reported sequences (16, 17). Two other new Cryptosporidium spp. were isolated in reptiles, as shown by both RFLP and sequence analyses. In RFLP analyses, one isolate from a snake (sample 2162) had an SspI band of over 800 bp but had a VspI band similar to the size of the band of the desert monitor genotype (between 600 to 700 bp). In contrast, three tortoise isolates had a slightly smaller SspI band size (near 800 bp) but had a VspI band similar to the band of C. serpentis (over 700 bp). DNA sequencing revealed that the sequences represented two new Cryptosporidium spp. A neighbor-joining analysis indicated that the tortoise genotype was related to C. serpentis and C. muris, whereas the new snake genotype was related to C. parvum (Fig. 2).
FIG. 2.
Genetic relationship between Cryptosporidium spp. in reptiles inferred by a neighbor-joining analysis of the partial SSU rRNA gene sequences by using the Kimura two-parameter model and the Treecon program. Numbers on branches are percentage bootstrap values of 1,000 replicates. Only values above 50% are shown.
Because C. muris and the C. parvum mouse genotype were seen in high frequency in snakes and lizards from the Saint Louis Zoo, whose diet contained mice, fecal samples were taken from 11 feeder mice and analyzed for Cryptosporidium. Three such samples were positive for Cryptosporidium by PCR analysis of the SSU rRNA gene. RFLP and sequence analyses showed the presence of the C. parvum mouse genotype in two mice and a mixed infection of C. muris and the C. parvum mouse genotype in one mouse.
The identification of the desert monitor genotype as C. saurophilum.
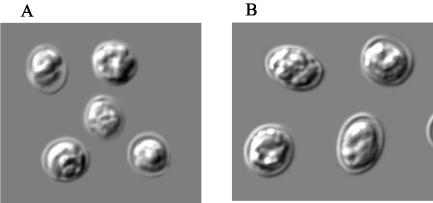
Previous characterization of the Cryptosporidium desert monitor genotype showed that the parasite is closely related to intestinal Cryptosporidium spp. Because the only known intestinal Cryptosporidium parasite in reptiles is C. saurophilum, morphometric measurements were done on the desert monitor genotype, and the data obtained were compared with those from C. serpentis and those previously reported for C. saurophilum. Oocysts of the desert monitor genotype were visibly smaller than those of C. serpentis (Fig. 3), with a mean length of 4.94 μm, a mean width of 4.49 μm, and a shape index (the length/width ratio) of 1.14. In comparison, oocysts of C. serpentis were 5.94 μm in length and 5.11 μm in width and had a shape index of 1.17 (Table 5). Thus, the morphometric measurements of the desert monitor genotype were similar to those previously reported for C. saurophilum (mean, 5.0 × 4.7 μm; range, 4.4 to 5.6 × 4.2 to 5.2 μm) (6).
FIG. 3.
Morphology of C. saurophilum (A) and C. serpentis (B) as seen under a differential interference contrast microscope (magnification, ×1,000).
TABLE 5.
Morphometric measurements (micrometers) of Cryptosporidium desert monitor genotype (C. saurophilum) in comparison with those of C. serpentisa
| Parameter | Desert monitor genotype (n = 20)
|
C. serpentis (n = 37)
|
||
|---|---|---|---|---|
| Mean | 95% CL | Mean | 95% CL | |
| Length | 4.94 | 4.81-5.07 | 5.94 | 5.82-6.06 |
| Width | 4.49 | 4.35-4.63 | 5.11 | 5.03-5.19 |
| Shape index | 1.14 | 1.11-1.17 | 1.17 | 1.14-1.20 |
n, number of oocysts.
Cross-transmission of Cryptosporidium between snakes and lizards.
To assess the ability for cross-transmission of Cryptosporidium spp. between snakes and lizards, an isolate of the Cryptosporidium desert monitor genotype originating from a snake was used to inoculate two corn snakes and two leopard geckoes that were free of Cryptosporidium infection by microscopy. Both geckoes started to shed oocysts 21 days after inoculation, but both snakes remained negative through the observation period. All four inoculated animals were euthanized 45 days after the inoculation, and tissue sections were taken from the stomach and intestine for histology and PCR-RFLP analysis. Cryptosporidium was not found in any of the tissue sections from the snakes. In contrast, Cryptosporidium in developmental stages was seen in hematoxylin and eosin-stained gastric tissues. PCR analysis of DNA from gastric and intestinal fragments confirmed that both snakes were negative for Cryptosporidium. Positive PCR amplifications, however, were obtained with DNA from the intestine of one gecko and from both the stomach and intestine of the other gecko. RFLP analysis of the PCR products with SspI and VspI indicated that the parasites present belonged to the Cryptosporidium desert monitor genotype (Table 3). The python inoculated with C. serpentis oocysts from a Nile monitor started to shed Cryptosporidium oocysts 88 days after inoculation. PCR-RFLP analysis of oocysts isolated from the snake confirmed that it belonged to C. serpentis (Table 3). Sequence analysis produced an SSU rRNA sequence identical to that of the isolate from the monitor, C. serpentis type B (Fig. 2).
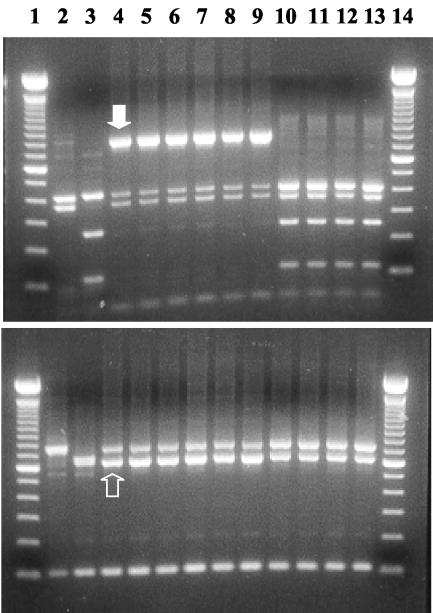
The cross-transmission of Cryptosporidium spp. between snakes and lizards was further assessed by the differentiation of Cryptosporidium spp. in a group of six snakes and four lizards that were housed in the same room. All snakes and lizards were positive for Cryptosporidium. PCR-RFLP analysis indicated that all animals were infected with multiple Cryptosporidium spp.; the six snakes were all infected with C. serpentis, a new Cryptosporidium genotype, and low levels of C. saurophilum, whereas the four lizards were all infected with C. serpentis and C. saurophilum (Fig. 4 and Table 3). Analysis of the cloned PCR products confirmed the diagnosis of C. serpentis and the new Cryptosporidium genotype in snakes and C. serpentis and C. saurophilum in lizards. Among the 15 clones of a PCR product from the snake isolate 938, 4 clones were identified as C. serpentis, and 11 clones were identified as the new Cryptosporidium genotype. Likewise, four of the eight clones of a PCR product from lizard isolate 944 belonged to C. saurophilum, and the remaining four clones belonged to C. serpentis. The new Cryptosporidium genotype was genetically related to the intestinal Cryptosporidium spp. and had a large SspI band (about 800 bp), similar to the tortoise genotype (Fig. 4, filled arrow), but had a VspI upper band (just over 600 bp) similar to that of C. saurophilum (Fig. 4, open arrow).
FIG. 4.
Simultaneous presence of multiple Cryptosporidium spp. in a group of six snakes and four lizards housed together as revealed by PCR-RFLP analyses of the SSU rRNA gene. The upper panel shows the results of SspI digestion; the lower panel shows the results of VspI digestion. Lanes 1 and 14, 100-bp molecular markers; lane 2, positive control for C. serpentis; lane 3, positive control for C. saurophilum; lane 4, sample from pine snake 936; lane 5, sample from pine snake 937; lane 6, sample from a New Guinea viper boa (938); lane 7, sample from a milk snake (939); lane 8, sample from a black rat snake (940); lane 9, sample from a green python (941); lane 10, sample from mountain chameleon 942; lane 11, sample from mountain chameleon 943; lane 12, sample from a bearded dragon (944); lane 13, sample from a gargoyle gecko (945). Filled and open arrows are the SspI and VspI bands, respectively, for the new Cryptosporidium genotype in snakes. Three Cryptosporidium spp. (C. serpentis, a new Cryptosporidium genotype, and a trace of C. saurophilum) are seen in all six snakes (lanes 4 to 9), and two parasites (C. serpentis and C. saurophilum) are seen in all 4 lizards (lanes 10 to 13).
Effectiveness of the diagnosis-euthanasia control strategy.
At the Saint Louis Zoo, a diagnosis-euthanasia program was initiated in March 1999 after the identification between December 1998 and January 1999 of chronic cryptosporidiosis in snakes. To monitor the effectiveness of the control measures, samples were periodically taken from snakes for 1 year. Right after the initiation of the control measure, 5 of 10 and 8 of 17 snakes sampled were positive for C. serpentis or C. saurophilum in May and June 1999, respectively. Afterwards, only 1 of 45 snake samples taken at five different time periods was positive for C. serpentis (Table 4).
DISCUSSION
A total of nine Cryptosporidium spp. were found in captive snakes, lizards, and tortoises in this study. The most common parasites were C. serpentis and the Cryptosporidium desert monitor genotype identified before (16, 17). Both parasites were detected in snakes as well as lizards. Two other Cryptosporidium spp. previously reported in captive snakes, C. muris and the C. parvum mouse genotype, were also found in some snakes and one lizard. Another common Cryptosporidium parasite in mammals, the C. parvum bovine genotype, was also identified in six lizards from Switzerland. Four other Cryptosporidium spp. detected in this study, however, presented new Cryptosporidium spp.: a tortoise genotype identified in three tortoises, two new snake genotypes (one genotype identified in only one snake and the other genotype identified in six snakes), and another new Cryptosporidium genotype from a lizard (sample 1665), which was genetically distinct but was related to C. serpentis.
Molecular and biologic characterizations indicated that the Cryptosporidium desert monitor genotype was probably C. saurophilum. Phylogenetically, the desert monitor genotype belonged to the intestinal Cryptosporidium parasite group, indicating that it was most probably an intestinal Cryptosporidium parasite, which is in agreement with the initial description of C. saurophilum (6). Morphologically, oocysts of the desert monitor genotype were very similar to those of C. saurophilum in shape and size and were significantly smaller than oocysts of C. serpentis (6). Biologically, the desert monitor genotype preferentially infected lizards. Although the desert monitor genotype was found in a few snakes in this study, cross-transmission studies by oocyst inoculation or habitat sharing indicated that the infectivity to lizards was much higher than to snakes, which explains the failure of the establishment of the desert monitor genotype in two corn snakes inoculated with oocysts of this parasite. Infection with C. saurophilum may not be restricted to the intestine as previously suggested (6), because it was also found in gastric washings of several snakes infected with C. saurophilum and in the stomach tissue section of one experimentally infected lizard.
Oocysts of the C. parvum bovine and mouse genotypes and C. muris found in some of the snakes and lizards in this study probably do not represent true parasites of these animals. Instead, the oocysts were probably from rodents ingested by these carnivorous reptiles (4). This possibility was supported by the fact that none of the animals with these oocysts had clinical signs and by the presence of organisms belonging to C. muris and the C. parvum mouse genotype in some of the feeder mice which were fed to snakes and some lizards in the Saint Louis Zoo. C. muris and the C. parvum mouse genotype have previously been reported in captive snakes and lizards (9). Although the C. parvum bovine genotype has not been found in mice in the United States, it has been previously reported in mice in Australia (8). Thus, oocysts of the C. parvum bovine genotype seen in lizards in Switzerland could also be from ingested prey or feeder mice. Previously, it was shown that oocysts of the C. parvum bovine genotype were not infectious to snakes (4). Nevertheless, the possibility of organisms belonging to the C. parvum mouse and bovine genotypes and to C. muris infecting reptiles can only be totally ruled out by careful biologic and genetic studies.
Because the four new Cryptosporidium spp. found in this study have never been reported in other animals before, they probably were true parasites of these captive reptiles. The Cryptosporidium parasite in snake 1665 was clearly phylogenetically related to C. serpentis, even though significant differences between these two Cryptosporidium spp. (Fig. 1) were present. Likewise, the Cryptosporidium genotype found in three tortoises was related to C. serpentis and has also been found recently in a turtle in Portugal (L. Xiao and M. Alves, unpublished data). One of the two new Cryptosporidium spp. identified in snakes in this study was relatively common, because it was found in snake 938 and five other snakes in this study and was previously found in several storm water samples (genotype W11) in New York (13). The other snake genotype had only been found in one animal (snake 2162).
There were intraspecies genetic variations within C. serpentis and C. saurophilum. Two genotypes of C. serpentis were seen in the study, which differed from each other by one SNP. Likewise, most C. saurophilum isolates produced SSU rRNA sequences similar to the one for the desert monitor genotype reported previously (17). Two isolates, however, had one SNP. It is not clear whether the minor sequence difference in C. saurophilum was due to differences between copies of the SSU rRNA gene, as demonstrated in other Cryptosporidium spp. (7, 14). Even though C. serpentis was named for the Cryptosporidium parasite originally identified in snakes by Brownstein et al. (1) and C. saurophilum was named for a Cryptosporidium parasite in lizards (6), both parasites apparently have a host range broader than previously believed. Nevertheless, data from this study suggest the presence of host adaptation; most snakes (except for sample 1172, which was experimentally infected with an isolate from a lizard) had a C. serpentis genotype A sequence, whereas most lizards (except for sample 806) had a C. serpentis genotype B sequence.
Currently, there are no effective control strategies against cryptosporidiosis in reptiles. In a small-scale study, it was demonstrated that snakes with clinical and subclinical cryptosporidiosis could be effectively treated with hyperimmune bovine colostrum raised against C. parvum (5). A common control practice is to euthanize Cryptosporidium-infected snakes, which would prevent the spread of infection to other animals. This diagnosis-euthanasia strategy was apparently effective in the control of Cryptosporidium infection in snakes in the Saint Louis Zoo in this study. The effectiveness of the method was supported by the evident reduction of C. serpentis infection in snakes at the zoo. In addition to the premature death of infected animals, one problem with the control measure is the frequent presence of oocysts of C. muris and the C. parvum mouse genotype in snakes because of the use of feeder mice as part of the diet. Because it is difficult to differentiate oocysts of the pathogenic C. serpentis from those of nonpathogenic Cryptosporidium spp. that merely pass through the gastrointestinal tract, the diagnosis-euthanasia control strategy would lead to the killing of uninfected animals.
Acknowledgments
We thank Roy Burns of the Louisville Zoo and Richard J. Montali of the National Zoological Park for providing samples and Michael Cranfield of the Baltimore Zoo for facilitating the cross-transmission experiment.
This study was supported in part by grant no. 524/00/P015 of the Grant Agency of the Czech Republic
REFERENCES
- 1.Brownstein, D. G., J. D. Strandberg, R. J. Montali, M. Bush, and J. Fortner. 1977. Cryptosporidium in snakes with hypertrophic gastritis. Vet. Pathol. 14:606-617. [DOI] [PubMed] [Google Scholar]
- 2.Fayer, R., U. Morgan, and S. J. Upton. 2000. Epidemiology of Cryptosporidium: transmission, detection and identification. Int. J. Parasitol. 30:1305-1322. [DOI] [PubMed] [Google Scholar]
- 3.Fayer, R., C. Speer, and J. Dubey. 1997. The general biology of Cryptosporidium, p. 1-41. In R. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Boca Raton, Fla.
- 4.Graczyk, T. K., and M. R. Cranfield. 1998. Experimental transmission of Cryptosporidium oocyst isolates from mammals, birds and reptiles to captive snakes. Vet. Res. 29:187-195. [PubMed] [Google Scholar]
- 5.Graczyk, T. K., M. R. Cranfield, P. Helmer, R. Fayer, and E. F. Bostwick. 1998. Therapeutic efficacy of hyperimmune bovine colostrum treatment against clinical and subclinical Cryptosporidium serpentis infections in captive snakes. Vet. Parasitol. 74:123-132. [DOI] [PubMed] [Google Scholar]
- 6.Koudela, B., and D. Modry. 1998. New species of Cryptosporidium (Apicomplexa, Cryptosporidiidae) from lizards. Folia Parasitol. 45:93-100. [Google Scholar]
- 7.Le Blancq, S. M., N. V. Khramtsov, F. Zamani, S. J. Upton, and T. W. Wu. 1997. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol. Biochem. Parasitol. 90:463-478. [DOI] [PubMed] [Google Scholar]
- 8.Morgan, U. M., A. P. Sturdee, G. Singleton, M. S. Gomez, M. Gracenea, J. Torres, S. G. Hamilton, D. P. Woodside, and R. C. A. Thompson. 1999. The Cryptosporidium “mouse” genotype is conserved across geographic areas. J. Clin. Microbiol. 37:1302-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan, U. M., L. Xiao, R. Fayer, T. K. Graczyk, A. A. Lal, P. Deplazes, and R. C. Thompson. 1999. Phylogenetic analysis of Cryptosporidium isolates from captive reptiles using 18S rDNA sequence data and random amplified polymorphic DNA analysis. J. Parasitol. 85:525-530. [PubMed] [Google Scholar]
- 10.O'Donoghue, P. J. 1995. Cryptosporidium and cryptosporidiosis in man and animals. Int. J. Parasitol. 25:139-195. [DOI] [PubMed] [Google Scholar]
- 11.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Upton, S. J., C. T. McAllister, P. S. Freed, and S. M. Barnard. 1989. Cryptosporidium spp. in wild and captive reptiles. J. Wildl. Dis. 25:20-30. [DOI] [PubMed] [Google Scholar]
- 13.Xiao, L., K. Alderisio, J. Limor, M. Royer, and A. A. Lal. 2000. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl. Environ. Microbiol. 66:5492-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao, L., J. R. Limor, L. Li, U. Morgan, R. C. Thompson, and A. A. Lal. 1999. Presence of heterogeneous copies of the small subunit rRNA gene in Cryptosporidium parvum human and marsupial genotypes and Cryptosporidium felis. J. Eukaryot. Microbiol. 46:44S-45S. [PubMed] [Google Scholar]
- 15.Xiao, L., U. M. Morgan, R. Fayer, R. C. Thompson, and A. A. Lal. 2000. Cryptosporidium systematics and implications for public health. Parasitol. Today 16:287-292. [DOI] [PubMed] [Google Scholar]
- 16.Xiao, L. H., L. Escalante, C. F. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao, L. H., U. M. Morgan, J. Limor, A. Escalante, M. Arrowood, W. Shulaw, R. C. A. Thompson, R. Fayer, and A. A. Lal. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]