Abstract
Functional genetic variations play important roles in shaping phenotypic differences among individuals through affecting gene expression, and thus, very likely to influence disease susceptibility, such as cancer susceptibility. One critical question in this era of post-genome wide association studies (GWAS) is how to assess the functional significance of the genetic variations identified from GWAS. In the current study, with lymphoblastoid cell lines (LCLs) from 74 non-related women with familial ovarian cancer and 47 unrelated controls matched on gender and race, we explored the associations between seven ovarian cancer risk variants identified from GWAS (rs3814113 on 9p22.2, rs2072590 on 2q31, rs2665390 on 3q25, rs10088218, rs1516982, rs10098821 on 8q24.21, and rs2363956 on 19p13) and whole genome mRNA expression profiles. We observed 95 significant trans-associations at a permutation level of 0.001. Compared to the other risk variants, rs10088218, rs1516982, and rs10098821 on 8q24.21 had the greatest number of significant associations (25, 16, and 38, respectively). Two possible cis-associations were observed between rs10098821 and c-Myc, and rs2072590 and HS.565379 (Permutated P = 0.0198 and 0.0399, respectively). Pathway enrichment analysis showed that several key biological pathways, such as cell cycle (P = 2.59×10−06), etc, were significantly overrepresented. Further characterization of significant associations between mRNAs and risk alleles might facilitate understanding the functions of GWAS discovered risk alleles in the genetic etiology of ovarian cancer.
Introduction
Recently, genome wide association studies (GWAS) have successfully identified a number of genetic variations which confer risk to human cancer [1]–[3]. However, most of the risk variants identified from GWAS reside in intergenic, intronic, and other non-coding regions of the genome [4]. Therefore, the observed associations have yet to be translated into a full understanding of the genes and genetic elements mediating disease susceptibility. How to study the functional significance of these GWAS hits poses a big challenge in this post-GWAS era. One of the options might be the investigation of the genetics of gene expression. Several landmark studies have unequivocally shown that many transcripts in the human genome are influenced by inherited variation [5]–[9]. Functional genetic variation, which leads to gene expression changes, may play a critical role in determining phenotypic differences among individuals, and thus, is very likely to influence disease susceptibility. As such, studying the associations between genetic variation and gene expression could potentially help prioritize fine-mapping efforts and provide a shortcut to disease biology.
Epithelial carcinoma of the ovary is one of the most common gynecologic malignancies in women [10]. Family history is the strongest risk factor for ovarian cancer. Compared to a 1.6% lifetime risk of developing ovarian cancer in the general population, women with one first-degree relative with ovarian cancer have a 5% risk. Familial clustering with an autosomal dominant pattern of inheritance (hereditary ovarian cancer) results from germ-line mutations in putative tumor suppressor genes (TSGs), such as the BRCA1/2 and MLH1/MSH2 genes [11]–[14]. However, known mutations in BRCA1/2 and mismatch repair (MMR) genes can only explain a small part of the familial aggregation of ovarian cancer (5–13%). This suggests that other genetic events may contribute to familial ovarian cancers. Several GWAS have been done in ovarian cancer and several risk variants have been identified, including rs3814113 on 9p22, rs2072590 on 2q31, rs2665390 on 3q25, rs10088218, rs1516982, rs10098821 on 8q24, and rs2363956 on 19p13 [1]–[3]. However, the functional significance of these risk variants is largely unknown. Thus, studying the associations between gene expression and ovarian cancer risk alleles identified from GWAS might help connect risk variants to their putative target genes/transcripts and biological pathways.
To study the associations between gene expression and ovarian cancer risk alleles, we obtained the whole genome mRNA expression profiles in 121 non-redundant lymphoblastoid cell lines (LCLs) derived from 74 non-related familial ovarian cancer patients who are non-carriers of known BRCA1/2 and MMR gene mutations, as well as 47 non-cancer unrelated family controls. We genotyped seven ovarian cancer risk variants discovered from GWAS in these 121 cell lines and studied their associations with gene expression variations. To our knowledge, this is the first genome-wide study to evaluate the associations between mRNA expression variations in LCLs of familial ovarian cancer cases and GWAS discovered ovarian cancer risk alleles [1]–[3].
Results
Lymphoblastoid cell lines were derived from the blood samples of 74 non-related women with familial ovarian cancer and 47 un-related cancer-free controls recruited for the GRFOCR (see Methods). Gene expression profiles were generated using the Illumina human HT-12 v3 Expression BeadChips. We filtered the processed data to include genes with expression above the background in at least 25% of the samples (n = 121). A total of 10,435 mRNA genes were retained for further analysis.
For each sample, the seven variants identified from three ovarian cancer GWAS were genotyped using the StepOnePlus™ Real Time PCR system and Assays-on-Demand SNP Genotyping products (see Methods). We assessed the potential implications of these GWAS-discovered variants in ovarian cancer, by performing association analysis to analyze the correlations between mRNA expression variations and variant genotypes. Significant associations were identified by evaluating the relationships between variations of mRNA expression levels (with age and case-control status adjusted) and variant genotypes through 10,000 permutations. The number of significant associations at permutation level threshold of 0.05, 0.01 and 0.001 was summarized in Table 1 . The list of selected top-ranked significant associations (permutated P ≤ 0.001 and r2≥0.095) is shown in Table 2 . One of the most significant associations is observed between rs10098821 and IER3 gene (permutated P<0.0001). IER3, is a stress-inducible immediate early response gene, whose functions include cell proliferation and apoptosis regulation. It has been found that this gene is pro-apoptotic in the development of ovarian cancer [15]. rs10098821 explains about 13% of the variation in IER3’s expression level as measured by adjusted r2.
Table 1. Summary of significant association between GWAS discovered variant genotypes and mRNA gene expression phenotypes.
| rs2072590 | rs2665390 | rs10088218 | rs1516982 | rs10098821 | rs3814113 | rs2363956 | |
| P<0.05 | 585 | 378 | 821 | 618 | 959 | 274 | 394 |
| P<0.01 | 115 | 52 | 194 | 139 | 251 | 46 | 59 |
| P<0.001 | 6 | 5 | 25 | 16 | 38 | 2 | 3 |
Table 2. List of Top ranked significant associations between mRNA and variants (P<0.001).
| mRNAs | SNPs-ID | P –value* | r2 | mRNAs | SNPs-ID | P –value* | r2 |
| HS.571028 | rs2072590 | 0 | 0.16 | ARL1 | rs1516982 | 3.00E−04 | 0.10 |
| LRRC41 | rs2072590 | 0 | 0.16 | HS.340072 | rs1516982 | 3.00E−04 | 0.10 |
| APIP | rs2665390 | 0 | 0.13 | DOCK11 | rs2072590 | 4.00E−04 | 0.12 |
| ARL1 | rs10088218 | 0 | 0.13 | CISD2 | rs2665390 | 4.00E−04 | 0.10 |
| ENG | rs10088218 | 0 | 0.11 | KCNMB1 | rs10088218 | 4.00E−04 | 0.12 |
| FLJ21438 | rs10088218 | 0 | 0.10 | PIK3C2B | rs10088218 | 4.00E−04 | 0.10 |
| TMTC4 | rs10088218 | 0 | 0.12 | CD226 | rs10098821 | 4.00E−04 | 0.10 |
| GEMIN4 | rs10098821 | 0 | 0.13 | HIP1 | rs10098821 | 4.00E−04 | 0.10 |
| IER3 | rs10098821 | 0 | 0.13 | MCM7 | rs10098821 | 4.00E−04 | 0.10 |
| VGF | rs10098821 | 0 | 0.13 | RALGPS2 | rs10098821 | 4.00E−04 | 0.10 |
| VGF | rs10088218 | 1.00E−04 | 0.12 | ELMO1 | rs2363956 | 5.00E−04 | 0.12 |
| PLEKHA7 | rs10098821 | 1.00E−04 | 0.12 | EHMT1 | rs10098821 | 5.00E−04 | 0.10 |
| ZHX2 | rs10098821 | 1.00E−04 | 0.12 | IL32 | rs10098821 | 5.00E−04 | 0.10 |
| CCL4L1 | rs1516982 | 1.00E−04 | 0.14 | KCNMB1 | rs10098821 | 5.00E−04 | 0.11 |
| GEMIN4 | rs1516982 | 1.00E−04 | 0.10 | RIC8A | rs10098821 | 5.00E−04 | 0.11 |
| VGF | rs1516982 | 1.00E−04 | 0.13 | RNF44 | rs10098821 | 5.00E−04 | 0.10 |
| PI4K2A | rs2363956 | 2.00E−04 | 0.13 | ODF1 | rs1516982 | 5.00E−04 | 0.10 |
| HS.572064 | rs2072590 | 2.00E−04 | 0.13 | TRPC1 | rs2363956 | 6.00E−04 | 0.12 |
| CCL4L1 | rs10088218 | 2.00E−04 | 0.11 | MFGE8 | rs10098821 | 6.00E−04 | 0.10 |
| BSDC1 | rs10098821 | 2.00E−04 | 0.11 | APOBEC3H | rs2072590 | 7.00E−04 | 0.10 |
| GART | rs10098821 | 2.00E−04 | 0.11 | EDEM1 | rs2072590 | 7.00E−04 | 0.11 |
| HS.340072 | rs10098821 | 2.00E−04 | 0.12 | HS.579631 | rs2665390 | 7.00E−04 | 0.10 |
| MAPKAP1 | rs10098821 | 2.00E−04 | 0.11 | NASP | rs10098821 | 7.00E−04 | 0.10 |
| RGL1 | rs1516982 | 2.00E−04 | 0.10 | MT1G | rs3814113 | 7.00E−04 | 0.15 |
| OPTN | rs10088218 | 3.00E−04 | 0.10 | DAP3 | rs2665390 | 8.00E−04 | 0.10 |
| ZHX2 | rs10088218 | 3.00E−04 | 0.10 | FANCE | rs1516982 | 8.00E−04 | 0.10 |
| CAV1 | rs10098821 | 3.00E−04 | 0.11 | GSTP1 | rs1516982 | 8.00E−04 | 0.13 |
| RGL1 | rs10098821 | 3.00E−04 | 0.11 | HS.340072 | rs10088218 | 9.00E−04 | 0.10 |
| STC2 | rs10098821 | 3.00E−04 | 0.10 | KRT17 | rs10098821 | 9.00E−04 | 0.10 |
| TBXAS1 | rs10098821 | 3.00E−04 | 0.10 | ATL2 | rs3814113 | 9.00E−04 | 0.11 |
Permutated P value.
Interestingly, the three variants from the 8q24 locus, namely rs10098821, rs10088218 and rs1516982, had the largest significant associations among all seven variants. At the 0.05 permutation threshold, the number of significant associations with these three variants was 959, 821 and 618. The number was 251, 194 and 139 at the more stringent permutation threshold of 0.01, and 38, 25 and 16 at the threshold of 0.001. These three variants share a number of significant mRNA gene expression associations. At the 0.05 permutation threshold, three hundred and twelve mRNAs, which account for 33% of the mRNA correlated with rs10098821, 38% of mRNA correlated with rs10088218, and 50% of mRNA correlated with rs1516982, are correlated with all three variants (Figure S1). For example, levels of FANCE (Fanconi anemia, complementation group E) expression is significantly associated with rs1516982 (permutated P = 8.0×10−4, adjusted r2 = 10.3%), rs10098821 (permutated P = 0.0037, adjusted r2 = 7.0%) and rs10088218 (permutated P = 0.0312, adjusted r2 = 3.4%), but none of the other four SNPs (Figure S2).
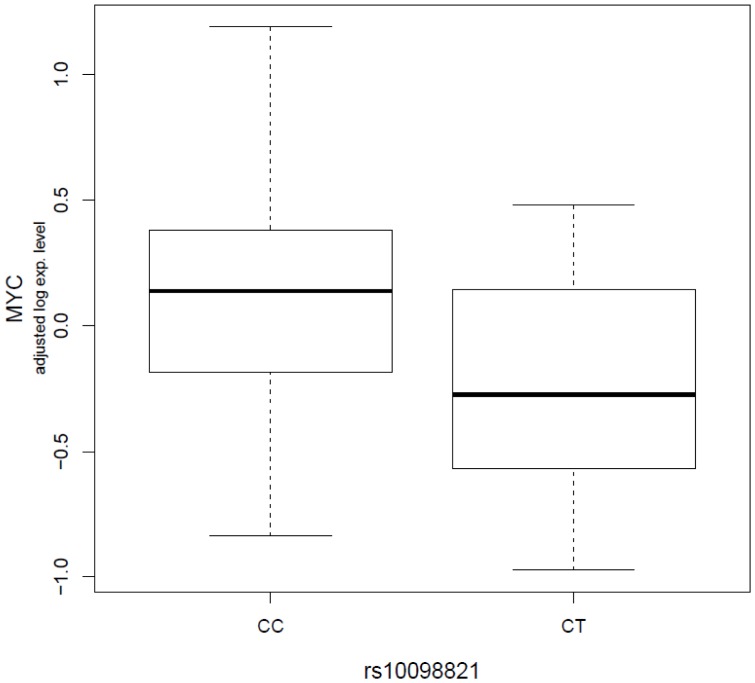
We observed two possible cis-associations in which the variant genomic location is within 1 Mb around the probe targeting gene. One cis-association is between rs10098821 and c-Myc gene, which is 806 kb away from the variant (permutated P = 0.0198, Figure 1 ), and the other is between rs2072590 and HS.565379, which is 697 kb away from the variant (permutated P = 0.0399, not shown). rs10098821 explained approximately 4.0% of the variation in c-Myc expression as measured by adjusted r2. Individuals with T variant alleles have statistically significantly lower expression of c-Myc compared to ones without T variant alleles. rs2072590 explained about 4.4% of the variation in HS.565379 expression. HS.565379 has been found to show tissue-specific expression in uterus and uterine tumor based on EST-based gene expression profiling [16].
Figure 1. Significant cis-associations between rs10098821 genotype and c-Myc expression phenotypes.
The boxplot shows the relationship between log2 residuals of c-Myc expression levels (adjusted for age and case-control status) and genotype of the rs10098821. rs10098821 explained approximately 4.0% of the variation in c-Myc expression as measured by adjusted r2.
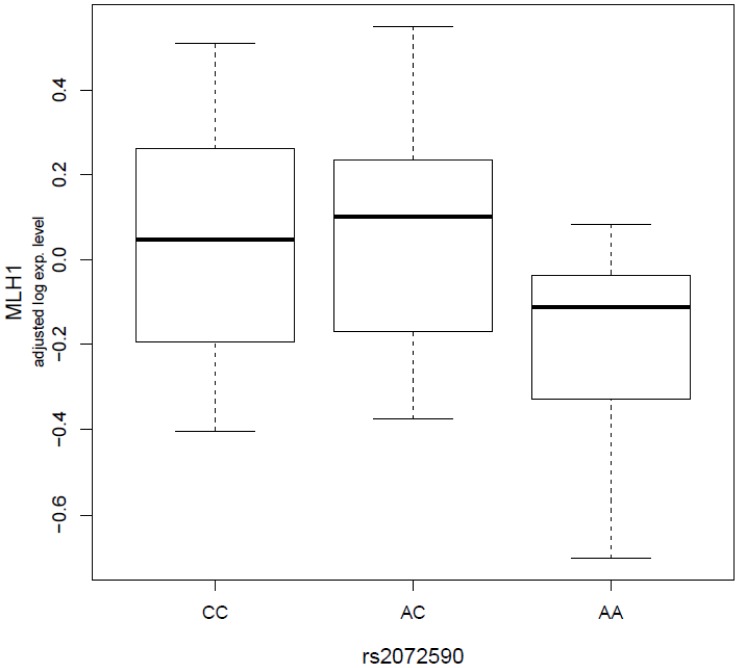
Then, we investigated whether there are any significant associations between these seven variants and known ovarian cancer risk genes, including BRCA1/2, MMR genes, p53, etc. We didn’t observe any significant association between these variants and the BRCA1/2 genes. However, we found several significant associations between the variants and the MMR genes and the p53 gene (Figure S3). For example, we found the expression level of the MLH1 gene is significantly associated with rs2072590, a variant on the 2q31 loci (permutated P = 0.0049, Figure 2 ). rs2072590 explained about 8.3% of the variation in MLHL1’s expression level. The expression of the p53 gene is significantly associated with rs2665390 (permutated P = 0.018, adjusted r2 = 0.036), rs1516982 (permutated P = 0.028, adjusted r2 = 0.035), and rs10088218 (permutated P = 0.049, adjusted r2 = 0.025). Additionally, the expression of the MSH5 gene is significantly associated with rs2363956 (permutated P = 0.0056, adjusted r2 = 0.075).
Figure 2. Significant associations between rs2072590 genotype and MLH1 expression phenotypes (permutated P = 0.0049, adjusted r2 = 8.3%).
The boxplot shows the relationship between log2 residuals of MLH1 expression levels (adjusted for age and case-control status) and genotype of the rs2072590.
Finally, to biologically characterize those mRNA genes significantly associated with GWAS discovered ovarian cancer risk alleles investigated here, we performed Gene Ontology (GO) enrichment analysis using the NCBI DAVID tool [17] As shown in Table 3 , the list of significantly enriched GO biological processes include “cell cycle” (P = 2.59×10−6), “regulation of apoptosis” (P = 4.37×10−5), and “programmed cell death” (P = 6.93×10−5). At the molecular function level, the significantly enriched GO terms include “nucleotide binding” (P = 7.43×10−9), “ATP binding” (P = 3.94×10−7), “transcription factor binding” (P = 1.19×10−5) and “DNA helicase activity” (P = 3.41×10−4).
Table 3. Enriched GO Biological Processes for the genes with significant associations with GWAS discovered variants.
| GO Term | Count | Size | P-Value | FDR |
| GO:0006396∼RNA processing | 120 | 547 | 1.50×10−8 | 6.37×10−5 |
| GO:0043933∼macromolecular complex subunit organization | 144 | 710 | 9.22×10−8 | 1.96×10−4 |
| GO:0065003∼macromolecular complex assembly | 130 | 665 | 3.19×10−6 | 0.003382 |
| GO:0007049∼cell cycle | 148 | 776 | 2.59×10−6 | 0.003662 |
| GO:0016071∼mRNA metabolic process | 81 | 370 | 4.50×10−6 | 0.003819 |
| GO:0006259∼DNA metabolic process | 102 | 506 | 1.11×10−5 | 0.007811 |
| GO:0022613∼ribonucleoprotein complex biogenesis | 46 | 180 | 1.40×10−5 | 0.008466 |
| GO:0006397∼mRNA processing | 70 | 321 | 2.49×10−5 | 0.013134 |
| GO:0046907∼intracellular transport | 124 | 657 | 3.07×10−5 | 0.014409 |
| GO:0043067∼regulation of programmed cell death | 147 | 812 | 4.75×10−5 | 0.018195 |
| GO:0042981∼regulation of apoptosis | 146 | 804 | 4.37×10−5 | 0.018393 |
| GO:0010941∼regulation of cell death | 147 | 815 | 5.80×10−5 | 0.020328 |
| GO:0012501∼programmed cell death | 115 | 611 | 6.93×10−5 | 0.022407 |
| GO:0006915∼apoptosis | 113 | 602 | 8.96×10−5 | 0.023497 |
| GO:0008219∼cell death | 131 | 719 | 9.58×10−5 | 0.023645 |
| GO:0006986∼response to unfolded protein | 23 | 71 | 7.96×10−5 | 0.023869 |
| GO:0016265∼death | 132 | 724 | 8.87×10−5 | 0.024805 |
| GO:0034470∼ncRNA processing | 44 | 187 | 1.73×10−4 | 0.039963 |
| GO:0070271∼protein complex biogenesis | 96 | 505 | 2.05×10−4 | 0.044756 |
| GO:0006461∼protein complex assembly | 96 | 505 | 2.05×10−4 | 0.044756 |
| GO:0034660∼ncRNA metabolic process | 51 | 230 | 2.34×10−4 | 0.048516 |
| GO:0051789∼response to protein stimulus | 29 | 107 | 2.48×10−4 | 0.048976 |
Discussion
The genetic etiology of familial ovarian cancer is still a mystery. Known mutations in BRCA1/2 and MMR genes can only explain a small part of the familial aggregation of ovarian cancer. The results from recent GWAS studies have identified several common genetic variants conferring risk for ovarian cancer [1]–[3]. However, most of these variants are not in protein-encoding regions, so the functional significance of these variants is largely unknown. The current study presents an attempt to dissect the genetic susceptibility of familial ovarian cancer, as well as elucidate the potential functional significance of the identified risk variants from GWAS. Specifically, we investigated the associations between seven significant variants identified from ovarian cancer GWAS and global mRNA expression.
As expected, we have observed a larger number of distant (trans-) than local (cis-) associations. Among the two identified cis-associations, the association between rs10098821 at 8q24 and c-Myc is particularly interesting. Common variants at 8q24 have previously been shown to confer susceptibility to multiple cancer phenotypes, including prostate, colorectal, breast and bladder cancers [18]–[23], and previous functional studies have suggested that common variants in this region may be associated with transcriptional regulation of c-Myc [24]–[25]. Most risk associations at 8q24 are located 5′ of c-Myc, but the three most significant SNPs for ovarian cancer lie in an apparent gene desert which is >700 kb 3′ of c-Myc, suggesting either that c-Myc might not be the target susceptibility gene for ovarian cancer or that variants in this region are also capable of distant regulation of c-Myc. In a previous study [2], Goode et al compared c-Myc expression in 48 normal ovarian epithelial cell lines between individuals without rs10098821 variant alleles and ones with at least one rs10098821variant alleles. Using GAPDH as the reference mRNA, they found that the ones without rs10098821 variant alleles had higher c-Myc expression than ones with at least one rs10098821 variant alleles (Median of relative expression: 0.97 vs 0.62). However, the difference didn’t reach statistical significance (P = 0.43). Similar to their findings, we have observed that individuals without rs10098821 variant alleles had significantly higher levels of c-Myc expression compared to ones with at least one rs10098821 variant alleles (permutated P = 0.0198). As we have indicated above, rs10098821 is 3′ of MYC and lies about 0.8 Mb away. How this SNP might affect MYC expression is still unclear.
Using these identified significant associations in the pathway analysis, we have found that the genes significantly associated with GWAS discovered ovarian cancer risk alleles are enriched in several key biological pathways, such as cell cycle, cellular response to stress/damage, energy metabolism, transcriptional factor binding, etc. Interestingly, most known familial ovarian cancer genes (i.e., BRCA1/2 and MMR) are key players in these key pathways. For example, it has been demonstrated that BRCA1 is the key regulator in sensing DNA stress/damage and subsequently promoting cell cycle arrest [26]. Although our association analysis cannot pinpoint the exact functions of these GWAS discovered variants, it provides a list of potential biological pathways for which one could focus on in future analysis.
There are several limitations to this study. First, many mRNAs are expressed in a tissue-restricted manner. The results from LCLs in this study are likely to represent a small subset of mRNA expression variations. Also, our ability to study the genetics of mRNA expression is limited by the fact that we only investigated seven variants in the analysis, although these seven variants have been associated with ovarian cancer risk in recent GWASs. Second, the effects on transcript abundance may be subtle and therefore below the sensitivity threshold of the microarray platform, and the sample size in our study is relatively small. Third, there is a concern about what the results actually mean when measuring expression in non-tumor tissue at a single point in time. The ultimate goal of our study is to identify the inherited genetic determinants of mRNA expression in normal tissues rather than somatic alterations of mRNA gene expression in tumor tissues. Studies have been shown that at least part of the mRNA gene expression is genetically determined. Therefore, even at a single time-point in non-tumor tissue, what we have observed from this study still provides useful information about how mRNA expression is genetically regulated. Forth, certain effects may only be revealed in certain contexts, such as perturbation of a particular pathway, and may occur through changes in gene transcripts mediated by alterations in microRNAs or non-coding RNAs rather than through direct effects on genes. In these cases, alternative assays will be required to implicate these genes. Finally, the significant associations are not further functionally characterized since all of the top associations are trans-associations. So far, there is still lack of established experimental methods to assess trans-regulation between SNPs and gene expression.
To the best of our knowledge, this study provides the first assessment of the expression level variation of mature human mRNAs in LCLs from familial ovarian cancer patients and healthy unrelated controls. Further studies are needed to identify the genetic causes and biological consequences related to the identified significant associations. Significant associations identified in this study may potentially facilitate better understanding of the genetic etiology of familial ovarian cancer.
Materials and Methods
Study Population
This study has been approved by the Institutional Research Board (IRB) of Roswell Park Cancer Institute. Written informed consents have been obtained from all study subjects. Data and samples from women with ovarian cancer and their relatives who were cancer-free were obtained from the Gilda Radner Familial Ovarian Cancer Registry (GRFOCR). Seventy-four non-related women with familial ovarian cancer were included in this study as the cases. They were identified from families with inherited ovarian cancer in which at least two first or second degree relatives had epithelial ovarian cancer diagnosed at any age. All of the women were non-carriers of BRCA1/2 or MLH1/MSH2 mutations. Over time, different methods have been used to determine the mutation status of BRCA1/2 in GRFOCR samples. For samples collected before 2002, mutation status was determined by screening all exons and intron/exon splice junctions of BRCA 1/2 by a combination of SSCP and HD analysis. Additionally, exon 11 of BRCA1 was assayed by the protein truncation test for stop codon generating mutations. If alterations were found, the altered fragment was sequenced. Since 2002, sequencing of exons and splice junctions was used. In the last 5 years, all samples (old and new) not showing a mutation were assayed for BRCA1 large-scale rearrangements. The cancer-free controls of GRFOCR were family relatives of the cases, including mothers, sisters, nieces, etc. However, in this study, we chose to use unrelated controls. Unrelated controls are women who are not relatives of any cases used in this study. Forty-seven unrelated controls were included. The cases and controls were matched on gender and race. All of the cases and controls were white women. The median age at cancer diagnosis for the 74 cases was 47 (ranging from 21 to 85), while the median age for the 47 controls at enrollment in GRFOCR was 58 (ranging from 26 to 89). All study subjects donated blood samples when they were enrolled in the GRFOCR. LCLs were established by EBV transformation using the isolated lymphocytes from the blood samples. The study was approved by the institutional IRB board.
Lymphoblastoid Cell Lines (LCLs) Culture and RNA Extraction
LCLs were maintained in RPMI 1640 (GIBCO BRL) media supplemented with 15% fetal calf serum and antibiotics at 37°C, 5% CO2 atmospheric condition and 95% humidity. Total cellular RNAs were isolated from LCLs using TRIzol reagent according to the protocols provided by the manufacturer (Invitrogen Corp., Carlsbad, CA, USA). Purified RNAs were further processed to remove any contaminating DNA (DNA-free kit, Ambion, Inc., Austin, TX, USA). The quality and quantity of the RNA was evaluated by 260/280 ratio using NanoDrop spectrophotometry (NanoDrop ND-1000 Technologies Inc.) and Agilent 2100 Bioanalyzer (Agilent Technologies).
Genotyping Analysis for Ovarian Cancer Risk Alleles
Seven SNPs, which are identified from 3 ovarian cancer GWAS, were included in the genotyping analysis. They are rs3814113 on 9p22.2, rs2072590 on 2q31, rs2665390 on 3q25 in the intron of TCDD-inducible poly(ADP-ribose) polymerase (TIPARP) gene, rs2363956 on 19p13 in the ankyrin repeat and LEM domain containing 1 (ANKLE1) gene, and rs10088218, rs1516982, and rs10098821 on 8q24.21. rs2363956 is a nonsynomous SNP which leads to a Leu to Trp amino acid change. Genotyping analysis was carried out using StepOnePlus™ Real Time PCR system and Assays-on-Demand SNP Genotyping products for fluorogenic polymerase chain reaction allelic discrimination (Applied Biosystems). Each PCR reaction plate included negative controls, positive controls, and unknown samples. The minor allele frequencies for each SNP in the cases and unrelated controls were 0.346/0.298 (P = 0.65) for rs3814113, 0.3/0.368 (P = 0.84) for rs2072590, 0.081/0.060 (P = 0.60) for rs2665390, 0.149/0.107 (P = 0.13) for rs10088218, 0.167/0.119 (P = 0.06) for rs1516982, 0.127/0.071 (P = 0.07) for rs10098821, and 0.432/0.488 (P = 0.20) for rs2363956. The genotyping data have been deposited in NCBI’s Gene Expression Omnibus (GEO) with accession number GSE37582.
Gene Expression Microarray
Two hundred nanograms of total RNA from each sample were labeled and hybridized on Illumina human HT-12 v3 Expression BeadChips according to the manufacturer’s recommendations (Illumina Whole-Genome Gene Expression Guide). The expression profiles have been deposited in NCBI’s Gene Expression Omnibus (GEO) with accession number GSE37582.
Statistical Analysis
The raw intensity of the Illumina human HT-12 v3expression array was scanned and extracted using BeadScan, with the data corrected by background subtraction in the GenomeStudio module. The lumi package in the R-based Bioconductor Package was used to normalize the log2 transformed intensity data by using the Quantile normalization algorithm. For data quality control, we excluded the probes with detection P value>0.05 (the P values were generated in BeadStudio software) in at least 25% (n = 121) of the samples. A total of 10,435 mRNA genes passed the quality control step and were used for downstream analysis. The association of SNP genotype with residuals of expression level adjusted for age and case-control status was calculated using linear regression model as described before (27). Ten thousand permutations of the expression phenotypes relative to SNP genotypes were performed (28–29). To derive P-values adjusted for multiple testing, we determined the percentage of times out of 10,000 permutations that the observed P-value was exceeded in the permuted data analysis.
Supporting Information
Venn diagram showing the overlaps of mRNA genes significantly (permutated P<0.05) associated with the three GWAS discovered SNPs from 8q24 locus.
(TIF)
Significant associations between the genotypes of the three variants on 8q24 locus and FANCE expression phenotypes. The boxplot shows the relationship between log2 residuals of FANCE expression levels (adjusted for age and case-control status) and genotype of the rs1516982 (top, permutated P = 8.0×10−4, adjusted r2 = 10.3%), rs10098821 (middle, permutated P = 0.0037, adjusted r2 = 7.0%) and rs10088218 (bottom, permutated P = 0.0312, adjusted r2 = 3.4%).
(TIF)
Significant associations between the variants genotypes and p53 expression phenotypes. The boxplot shows the relationship between log2 residuals of p53 expression levels (adjusted for age and case-control status) and genotype of the rs2665390 (top, permutated P = 0.0181, adjusted r2 = 3.6%), rs1516982 (middle, permutated P = 0.0279, adjusted r2 = 3.5%) and rs10088218 (bottom, permutated P = 0.0494, adjusted r2 = 2.5%).
(TIF)
Funding Statement
This work was supported by the National Institutes of Health (5R01CA136483 to HZ and P30 CA016056 to Roswell Park Cancer Institute); Ralph Wilson Medical Foundation (to HZ); Department of Defense Ovarian Cancer Program (OC073116 to HZ), and Roswell Park Alliance Foundation (to HZ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, et al. (2010) Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet 42: 880–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goode EL, Chenevix-Trench G, Song H, Ramus SJ, Notaridou M, et al. (2010) A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet 42: 874–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Song H, Ramus SJ, Tyrer J, Bolton KL, Gentry-Maharaj A, et al. (2009) A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet 41: 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Freedman ML, Monteiro AN, Gayther SA, Coetzee GA, Risch A, et al. (2011) Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet 43: 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Monks SA, Leonardson A, Zhu H, Cundiff P, Pietrusiak P, et al. (2004) Genetic inheritance of gene expression in human cell lines. Am J Hum Genet 75: 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, et al. (2004) Genetic analysis of genome-wide variation in human gene expression. Nature 430: 743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, et al. (2005) Genome-wide associations of gene expression variation in humans. PLoS Genet 1, e78. [DOI] [PMC free article] [PubMed]
- 8. Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, et al. (2003) Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302. [DOI] [PubMed] [Google Scholar]
- 9. Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, et al. (2003) Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302: 2141–2144. [DOI] [PubMed] [Google Scholar]
- 10. Yancik R (1993) Ovarian cancer. Age contrasts in incidence, histology, disease stage at diagnosis, and mortality. Cancer 71: 517–523. [DOI] [PubMed] [Google Scholar]
- 11. Narod SA, Ford D, Devilee P, Barkardottir RB, Lynch HT, et al. (1995) An evaluation of genetic heterogeneity in 145 breast-ovarian cancer families. Am J Hum Genet 156: 254–264. [PMC free article] [PubMed] [Google Scholar]
- 12. Ford D, Easton DF, Stratton M, Narod S, Goldgar D, et al. (1998) Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am J Hum Genet 62: 676–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Easton DF, Bishop DT, Ford D, Crockfor GF (1993) Genetic linkage analysis in familial breast and ovarian cancer: results from 214 families. Am J Hum Genet 52: 678–701. [PMC free article] [PubMed] [Google Scholar]
- 14. Lynch HT, Albano WA, Lynch JF, Lynch PM, Campbell A (1982) Surveillance and management of patients at high genetic risk for ovarian carcinoma. Obstet Gynecol 59: 589–596. [PubMed] [Google Scholar]
- 15. Han L, Geng L, Liu X, Shi H, He W, et al. (2011) Clinical significance of IEX-1 expression in ovarian carcinoma. Ultrastruct Pathol 35: 260–266. [DOI] [PubMed] [Google Scholar]
- 16. Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, et al. (2003) Database Resources of the National Center for Biotechnology. Nucl Acids Res 31: 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 18. Yeager M, Chatterjee N, Ciampa J, Jacobs KB, Gonzalez-Bosquet J, et al. (2009) Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet 41: 1055–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, et al. (2009) Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet 41: 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, et al. (2009) Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet 41: 1058–1060. [DOI] [PubMed] [Google Scholar]
- 21. Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, et al. (2008) Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet 40: 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, et al. (2008) Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst 100: 962–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kiemeney LA, Thorlacius S, Sulem P, Geller F, Aben KK, et al. (2008) Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet 40: 1307–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jia L, Landan G, Pomerantz M, Jaschek R, Herman P, et al. (2009) Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet 5: e1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, et al. (2009) The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet 41: 882–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu J, Lu LY, Yu X (2010) The role of BRCA1 in DNA damage response. Protein Cell 1: 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, et al. (2007) Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315: 848–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, et al. (2007) Population genomics of human gene expression. Nat Genet 39: 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, et al. (2010) Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464: 773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Venn diagram showing the overlaps of mRNA genes significantly (permutated P<0.05) associated with the three GWAS discovered SNPs from 8q24 locus.
(TIF)
Significant associations between the genotypes of the three variants on 8q24 locus and FANCE expression phenotypes. The boxplot shows the relationship between log2 residuals of FANCE expression levels (adjusted for age and case-control status) and genotype of the rs1516982 (top, permutated P = 8.0×10−4, adjusted r2 = 10.3%), rs10098821 (middle, permutated P = 0.0037, adjusted r2 = 7.0%) and rs10088218 (bottom, permutated P = 0.0312, adjusted r2 = 3.4%).
(TIF)
Significant associations between the variants genotypes and p53 expression phenotypes. The boxplot shows the relationship between log2 residuals of p53 expression levels (adjusted for age and case-control status) and genotype of the rs2665390 (top, permutated P = 0.0181, adjusted r2 = 3.6%), rs1516982 (middle, permutated P = 0.0279, adjusted r2 = 3.5%) and rs10088218 (bottom, permutated P = 0.0494, adjusted r2 = 2.5%).
(TIF)