Abstract
Lactobacillus reuteri ATCC 55730 is a probiotic (health-promoting) bacterium widely used as a dietary supplement. This study was designed to examine local colonization of the human gastrointestinal mucosa after dietary supplementation with L. reuteri ATCC 55730 and to determine subsequent immune responses at the colonized sites. In this open clinical investigation, 10 healthy volunteers and 9 volunteers with ileostomy underwent gastroscopy or ileoscopy and biopsy samples were taken from the stomach, duodenum, or ileum before and after supplementation with 4 × 108 CFU of live L. reuteri ATCC 55730 lactobacilli per day for 28 days. Biopsy specimen colonization was analyzed using fluorescence in situ hybridization with a molecular beacon probe, and immune cell populations were determined by immunostaining. Endogenous L. reuteri was detected in the stomach of 1 subject and the duodenum of 3 subjects (out of 10 subjects). After L. reuteri ATCC 55730 supplementation, the stomachs of 8 and the duodenums of all 10 subjects were colonized. Three ileostomy subjects (of six tested) had endogenous L. reuteri at baseline, while all six displayed colonization after L. reuteri supplementation. Gastric mucosal histiocyte numbers were reduced and duodenal B-lymphocyte numbers were increased by L. reuteri ATCC 55730 administration. Furthermore, L. reuteri administration induced a significantly higher amount of CD4-positive T-lymphocytes in the ileal epithelium. Dietary supplementation with the probiotic L. reuteri ATCC 55730 induces significant colonization of the stomach, duodenum, and ileum of healthy humans, and this is associated with significant alterations of the immune response in the gastrointestinal mucosa. These responses may be key components of a mechanism by which L. reuteri ATCC 55730 exerts its well-documented probiotic effects in humans.
Lactobacillus reuteri (13) is a heterofermentative bacterium that resides in the gastrointestinal tract of humans and animals (2, 11) and is considered to be one of the few true autochthonous (indigenous) Lactobacillus species in humans (14). L. reuteri ATCC 55730 (also designated SD2112) has been extensively studied and is widely used as a food additive to improve human gastrointestinal health. Oral administration delivers L. reuteri ATCC 55730 to the gastrointestinal tract, leading to shedding of live bacteria in the feces (16, 20). Clinical trials have shown that L. reuteri ATCC 55730 administration is safe (20, 21), significantly reduces the incidence and the severity of diarrhea of different origins (16, 17), reduces gastrointestinal illness and infections (G. Asli, A. Alsheikh, and Z. Weizman, Abstr. 36th Annu. Meet. Eur. Soc. Paediatr. Gastroenterol. Hepatol. Nutr., abstr. O041, 2003), and may be useful in the relief of constipation in elderly subjects (12).
Fecal colonization does not, however, provide information on the specific sites of colonization of L. reuteri ATCC 55730 along the gastrointestinal tract. Our aim was to document these specific sites and to examine possible alterations in the immune system associated with this colonization.
MATERIALS AND METHODS
Subjects.
The investigation had an open design and was performed in two parts: a gastroscopy session investigating the upper gastrointestinal tract and an ileoscopy session investigating the distal small bowel. Healthy adult subjects (>18 years of age) were recruited from the hospital staff at Roskilde County Hospital, Köge, Denmark, while ileostomy adult patients were recruited at the Gastroenterology Department. Subjects all had normal eating habits and gave written informed consent before entering the study. The exclusion criteria were (i) use of antibiotics 2 weeks before or during the study; (ii) the use of probiotics or probiotic foods 3 weeks before or during the study; (iii) ongoing treatment with nonsteroid anti-inflammatory drugs and/or proton pump inhibitors; (iv) regularly treated, severe organic disease; and (v) acute or chronic gastrointestinal illness in an active phase or with persistent mucosal abnormalities.
Study product.
Dietary supplementation with L. reuteri ATCC 55730 was the same for all subjects in the study. Each subject was asked to take two chewable tablets in the morning and two in the evening every day for 28 days. Each tablet contained 108 CFU of L. reuteri ATCC 55730 to give a total daily dose of 4 × 108 CFU of L. reuteri. Biopsies were performed at baseline (day 0) and again the day after L. reuteri supplementation was complete (day 28). The subjects thus acted as their own controls.
Biopsy procedures.
In the gastroscopy group, 10 healthy volunteers were studied. A gastro-duodenoscopy was performed following an overnight fast, and biopsy samples were taken from the gastric corpus and antrum and from the third part of the duodenum. Samples of gastric juice were also taken. In the ileoscopy group, nine volunteers with ileostomy following colectomy for ulcerative colitis (three subjects) or Crohn's disease (six subjects) were studied. Two subjects with Crohn's disease had resection of 50 and 150 cm of small intestine. The remaining small bowel was without signs of inflammation in all subjects. Ileoscopies were performed and biopsy specimens were taken within the distal 20 cm of the small bowel.
The same physician took all the biopsy specimens, and each had an average approximate weight of 33 mg. Specimens were either prepared for histological examination or transported in cold phosphate-buffered saline (PBS) to BioGaia Laboratories (Lund, Sweden) for analysis of Lactobacillus counts (principally as described below for fecal samples). The time from sampling to analysis was less than 36 h. For histological analysis, biopsy specimens were formalin fixed and embedded in paraffin. Subsequently, 4-μm-thick sections were cut and stained histochemically (with hematoxylin-eosin, van Gieson, periodic acid-Schiff-Alcian, and periodic acid-Schiff-diastase stain) and immunohistochemically. The primary antibodies used were CD20 (B lymphocytes); CD3, CD4, and CD8 (T lymphocytes); CD68 (histiocytes); Helicobacter pylori; and Ki-67 (a proliferation marker) and were obtained from DakoCytomation (Glostrup, Denmark). Immunohistochemical staining was performed on a DAKO TechMate 500 immunostainer to obtain uniform staining.
A pathologist evaluated the biopsy specimens. Using histochemical staining, tissue damage was graded on a scale of 0 to 10 according to Madsen et al. (8), with grades representing the numerical sum of four criteria: ulceration of the mucosa, epithelial hyperplasia, and the amounts of mononuclear cells and neutrophil granulocytes in the lamina propria. On the basis of the immunohistochemical staining, the numbers of CD20-, CD3-, CD4-, CD8-, and CD68-positive cells in lamina propria were evaluated semiquantitatively. One section of each biopsy sample was used for each specific stain. Each slide was evaluated and scored in the following way: 1 = no cells detected, 2 = single cells detected, 3 = dispersed cells or aggregations of cells detected, 4 = several adjoining groups of cells detected. The whole biopsy section was evaluated, and cells in as many fields as possible were counted. The mean scores were then calculated for each cell type. In order to confirm the observations, the biopsy slides were reevaluated by the same pathologist after being blinded by an independent person and the overall concordance between the two analyses was calculated.
FISH probe and hybridization procedures.
A 16S rRNA-targeted molecular beacon probe for the detection of L. reuteri using fluorescence in situ hybridization (FISH) was developed. The probe was 100% matched to the 16S rRNA sequence available for L. reuteri. Specificity of the probe was carefully evaluated in a previous study (N. Carbajal and J. Abad, submitted for publication) by analysis of hybridization of the L. reuteri-beacon probe with Lactobacillus oris, Lactobacillus fermentum, and Lactobacillus rhamnosus—all of which have 16S rRNA sequences closely related to that of L. reuteri. No binding of the probe was observed, and thus a nonspecific reaction with other bacterial species can be excluded. The probe is species but not strain specific, and thus although it identifies L. reuteri under the conditions used, it cannot confirm that the strain on the biopsy samples is the L. reuteri ATCC 55730 given to the subjects.
Biopsy sections were deparaffinated by baking at 60°C for 1 h followed by rehydration with successive treatments with ethanol (100, 95, 80, 70, and 50%) and finally molecular-biology-grade water. The cells on the sections were lysed by incubation at 37°C for 1 h in a solution containing 25 mM Tris-HCl (pH 7.5), 10 mM EDTA, 600 mM sucrose, lysozyme (2 mg/ml), and mutanolysin (0.2 μl/50-μl mixture) in a humid chamber. After rinsing in PBS for 5 min, the tissue sections were treated with 50 μl of proteinase K in PBS (100 μg/ml) for 20 min at 37°C in a humid chamber. Proteinase K was then inactivated with 0.2% glycine (15 s), and the slides were washed in PBS for 5 min before being dehydrated in ethanol. Fifty microliters of denaturing-hybridization buffer was added [60% formamide, 14% dextran sulfate (60% solution), 7% SSC (pH 7.4) (1× SSC is 1.2 M NaCl plus 0.27 M sodium citrate), probe (2.5 ng/μl; 0.05 mg/ml), and poly(A) (0.5 μg/μl)] at a temperature of 75°C for 90 s. Hybridization was performed at 50°C for 2 to 5 h. The slides were then washed three times in prewarmed (37°C) 50% formaldehyde-4× SSC at 37°C for 5 min, and this was followed by two washes with 1× SSC for 5 min. Slides were examined with a confocal laser scanning system (Leica TCS SP) attached to a Leica DM IRBE microscope and a 100× PL APO 1.4-0.7NA oil immersion objective. Oregon Green-labeled samples were excited using an argon laser at 488 nm with the emission window set to 499 to 570 nm. Images were processed using Leica TCS software and Adobe Photoshop 5.5. The entire biopsy section was scanned at 40× power to detect fluorescence.
Fecal analysis.
Stool samples were collected at baseline (day 0) and at the end of the study (day 28). Fecal samples from the ileostomy subjects were taken from the stoma output. Optional stool samples were provided by some of the subjects 14 days (day 42; n = 11) and 28 days (day 56; n = 8) after cessation of L. reuteri intake. Samples (no less than 5 g) were collected into a sterile container and placed immediately in a refrigerator. Within 24 h, 20 ml (1:5, wt/vol) of 0.1% peptone water was added, the sample was homogenized, and aliquots were dispensed into cryo-vials and frozen at −70°C before being shipped (frozen and on dry ice) to the BioGaia Laboratories for analysis of L. reuteri content. Thawed samples were diluted and plated on modified Rogosa Sharp plus 2% sodium acetate (MRS-3) agar containing vancomycin (50 mg/liter). Plates were incubated anaerobically at 37°C for 48 h, after which colonies were confirmed as L. reuteri using a BioGaia proprietary method (4a).
PCR.
DNA from L. reuteri ATCC 55730 in the tablets and certain randomly selected fecal isolates from the study (see Results) were analyzed by PCR using Bacterial Barcodes repPRO DNA Fingerprinting kit (BBCI, Houston, Tex.), and the fingerprints were analyzed using Bionumerics software.
Blood analysis.
Blood samples were taken at baseline and day 28, and the following were analyzed using standard clinical laboratory methods: hemoglobin, hematocrit, thrombocytes, leukocytes, C-reactive protein, potassium, sodium, creatinine, blood urea, plasma glucose, cholesterol, high-density lipoprotein, low-density lipoprotein, very-low-density lipoprotein, triglycerides, total bilirubin, urate, alanine aminotransferase, alkaline phosphatase, and lactate.
Symptoms.
At the baseline evaluation and on day 28, all subjects were asked to score subjective symptoms that they experienced during the previous 7 days. There were eight questions concerning physical discomfort, ability to work, abdominal pain, bowel distension, flatus, and stool consistency. Each subject answered all questions with a score ranking from 1 (no problem or symptom) to 7 (constant problems or symptoms).
Statistical analysis.
The Wilcoxon signed-rank test was used to compare symptoms, blood values, and histological differences between baseline and day 28, and a P of <0.05 was considered significant.
Ethical approval.
The protocol was approved by the Scientific Ethical Committee for Bornholms, Frederiksborg, Roskilde, Storstrøms, and West Sjaellands counties, Denmark, prior to start and was performed in accordance with the Declaration of Helsinki.
RESULTS
All subjects completed the study, and compliance was high (Table 1). While none of the subjects in the gastroscopy group had detectable L. reuteri in the feces at baseline, 28 days of L. reuteri tablet intake led to fecal shedding of live bacteria in all 10 subjects (Table 1). Fecal L. reuteri persisted at reduced but detectable levels 4 weeks after the end of intake. In the nine ileoscopy subjects, L. reuteri could be detected in the ileostomy output of two subjects at baseline and six subjects (including these two) after L. reuteri ATCC 55730 intake (Table 1). In the other three subjects fecal L. reuteri was not detectable. Again, in those that displayed fecal shedding, L. reuteri persisted 4 weeks after the end of intake (Table 1). DNA fingerprint analysis was performed on fecal L. reuteri isolates from three subjects on day 28, and all these isolates were found to have 98% similarity to the L. reuteri ATCC 55730 strain incorporated in the tablets. Although L. reuteri could be detected in a few biopsy samples using plating, there was no consistent detection of L. reuteri at 28 days, despite the presence of the bacteria in the feces of the subjects (Table 2). L. reuteri could not be detected in any of the samples of ventricle gastric juice.
TABLE 1.
Recovery of L. reuteri in the feces of subjects before and after L. reuteri ATCC 55730 supplementationa
| Session | % Compliance (SD) | CFU of L. reuteri/g of fecal material (SD) at day:
|
|||
|---|---|---|---|---|---|
| 0 | 28 | 42 | 56 | ||
| Gastroscopy | 87 (7) (n = 10) | NDb (n = 10) | 4.0 × 104 (1.1 × 105) (n = 10) | 1.0 × 103 (5.3 × 102) (n = 7) | 1.2 × 103 (6.0 × 102) (n = 5) |
| Ileoscopy | 97 (8) (n = 9) | ND (n = 7); 1.0 × 103 (5.3 × 102) (n = 2)c | ND (n = 3); 8.0 × 103 (1.6 × 104) (n = 6)c | 1.0 × 103 (6.1 × 102) (n = 4) | 6.7 × 102 (4.2 × 102) (n = 3) |
Fecal or ileostomy output samples were taken from subjects at baseline (day 0) and after the daily administration of L. reuteri ATCC 55730 at 4 × 108 CFU/day for 28 days (day 28). Sample collection was optional on days 42 and 56 (i.e., 28 days after dosing of L. reuteri was stopped), and in all these samples L. reuteri was detected. Compliance was calculated from the reported intake of the subjects and is expressed as the average intake of tablets of each person as a percentage of the total per protocol dose over 28 days. Results are expressed as CFU per gram (wet weight) of fecal material, with standard deviations and number of observations in parentheses.
ND, not detected (<100 CFU/g).
Mean of samples where L. reuteri was detected.
TABLE 2.
FISH detection of L. reuteri in gastric and intestinal biopsy specimensa
| Subject group | Biopsy specimen |
L. reuteri detected by:
|
|||||
|---|---|---|---|---|---|---|---|
| FISH
|
Biopsy plating
|
Fecal plating
|
|||||
| Day 0 | Day 28 | Day 0 | Day 28 | Day 0 | Day 28 | ||
| Gastroscopy | Corpus | 1 of 10 | 7 of 10 | 2 of 10 | 1 of 10 | 0 of 10 | 10 of 10 |
| Antrum | 1 of 10 | 8 of 10 | 3 of 10 | 1 of 10 | |||
| Duodenum | 3 of 10 | 10 of 10 | 3 of 10 | 0 of 10 | |||
| Ileoscopy | Ileum | 3 of 6 | 6 of 6 | 4 of 9 | 1 of 9 | 2 of 9 | 6 of 9 |
Biopsy and fecal or ileostomy output samples were taken from subjects at baseline (day 0) and after the daily administration of L. reuteri ATCC 55730 at 4 × 108 CFU/day for 28 days (day 28). L. reuteri was detected in biopsy specimens by either FISH or plating using classical methods. Results are expressed as the number of subjects in which L. reuteri was detected out of the total number of subjects analyzed. Examples of FISH detection are shown in Fig 1.
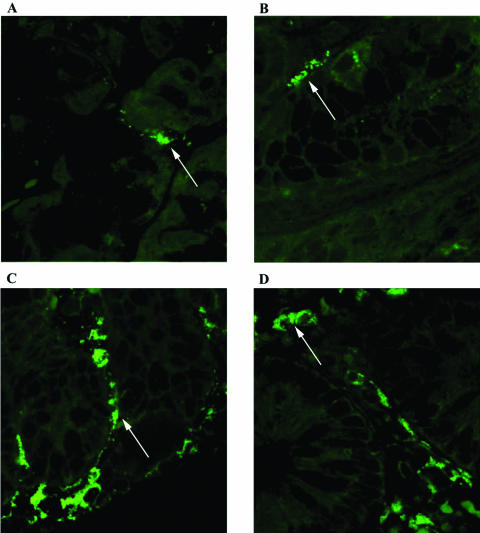
FISH analysis clearly demonstrated the presence of L. reuteri on the gastric, duodenal, and ileal epithelia. A low incidence of endogenous L. reuteri in the gastric corpus and antrum at baseline was dramatically elevated by L. reuteri ATCC 55730 consumption (Table 2). Similar results were seen in the duodenum (Table 2). In the ileoscopy group, only six subjects were analyzed using the FISH probes due to the lack of available biopsy material. The incidence of endogenous L. reuteri at baseline was somewhat higher (50%), increasing to 100% after L. reuteri ATCC 55730 administration (Table 2). Examples of typical findings are presented in Fig. 1.
FIG. 1.
FISH detection of L. reuteri on biopsy specimens from the human gastrointestinal tract. Biopsy specimens were taken from subjects after the daily administration of L. reuteri ATCC 55730 at 4 × 108 CFU/day for 28 days. L. reuteri was detected using a specific L. reuteri FISH molecular beacon probe. The bright green colonies of L. reuteri can be clearly seen (examples shown by arrows). Biopsy specimens were taken from the gastric corpus (A), gastric antrum (B), duodenum (C), and ileum (D).
The histology score was zero in all biopsy sections (Table 3) with the exception of one subject who had abnormal histology in the stomach throughout the study due to H. pylori infection (not discussed in this report). Further, all biopsy specimens had normal numbers of Ki-67-positive cells (a proliferation marker in the epithelium of the mucosa; data not shown). Thus, there was no evidence of any histological damage to the gastric or intestinal mucosal membrane after the intake of L. reuteri ATCC 55730 for 28 days.
TABLE 3.
Histological evaluation of gastric and intestinal biopsy specimensa
| Biopsy specimen | Day 0
|
Day 28
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Histology score | Mean score (SD) for:
|
Histology score | Mean score (SD) for:
|
|||||||||
| CD20 | CD3 | CD4 | CD8 | CD68 | CD20 | CD3 | CD4 | CD8 | CD68 | |||
| Corpus | 0c | 1.2 (0.4) | 2.1 (0.3) | 2.0 (0) | 2.1 (0.3) | 1.9 (0.3) | 0c | 1.1 (0.3) | 2.0 (0) | 2.0 (0) | 2.0 (0) | 1.4 (0.7)b |
| Antrum | 0c | 1.2 (0.6) | 2.1 (0.3) | 2.0 (0) | 2.1 (0.6) | 2.1 (0.3) | 0c | 1.4 (0.7) | 2.0 (0) | 2.1 (0.3) | 2.1 (0.3) | 1.7 (0.7)b |
| Duodenum | 0 | 1.4 (0.5) | 2.5 (0.7) | 2.7 (0.5) | 2.6 (0.5) | 2.6 (0.5) | 0 | 1.8 (0.4)b | 2.6 (0.7) | 3.0 (0) | 2.5 (0.5) | 2.6 (0.5) |
| Ileum | 0 | 2.1 (0.6) | 3.2 (0.4) | 3.2 (0.4) | 2.8 (0.4) | 2.8 (0.4) | 0 | 2.0 (0) | 2.8 (0.4) | 4.0 (0)b | 3.0 (0) | 2.8 (0.4) |
Biopsy specimens were taken from subjects at baseline (day 0) and after the daily administration of L. reuteri ATCC 55730 at 4 × 108 CFU/day for 28 days (day 28). Histological damage scoring was determined according to the method of Madsen et al. (8), and semiquantitative analysis of B lymphocytes (CD20 positive), T lymphocytes (CD3, CD4, or CD8 positive), and histiocytes (CD68 positive) was performed after specific staining of the sectioned biopsy samples. One biopsy specimen from each subject (n = 10 for corpus, antrum, and duodenum; n = 9 for ileum) was scored as follows: 1 = no cells detected, 2 = single cells detected, 3 = dispersed cells or aggregations of cells detected, and 4 = several adjoining groups of cells detected. Results are expressed as the means and standard deviations of the scores for each cell type and group.
P < 0.05 (Wilcoxon signed-rank test) compared to same cell type on day 0.
One subject had a score of 2 related to an H. pylori infection.
Most gastric biopsy samples displayed no detectable B lymphocytes (CD20-positive cells) and overall, there was no significant difference in the occurrence of B-lymphocytes in the stomach due to the administration of L. reuteri ATCC 55730 (Table 3). The occurrence of B lymphocytes in the duodenal biopsy specimens was significantly increased by L. reuteri ATCC 55730 administration (Table 3), but there were no significant changes in the occurrence of T lymphocytes (CD3-, CD4-, and CD8-positive cells) in the stomach or duodenum biopsy specimens during the study (Table 3). In both the corpus and the antrum epithelia, CD68-positive cells (histiocytes) were significantly reduced by L. reuteri ATCC 55730 administration, an effect not seen in the duodenum (Table 3). Concordance between the two investigations (first the not-blinded analysis and second the blinded analysis of histological samples) for the histiocyte observations in the corpus and antrum was only 40 and 65%, respectively, and for the duodenal B (CD20) cells it was only 60%. Thus, there is some uncertainty in these findings. Concordance in the CD3, CD4, and CD8 analyses was, however, much higher (82 to 89%).
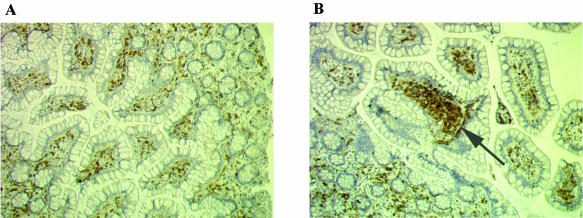
The levels of B lymphocytes, CD3- and CD8-positive T lymphocytes, and histiocytes in the ileal epithelium remained unchanged during the study, but there was a significantly higher number of CD4-positive T-lymphocytes in the ileum epithelium after supplementation with L. reuteri ATCC 55730 (Table 3). An example typical of these findings is shown in Fig. 2. Concordance between the two investigations of the ileum sections was generally high for all cell types examined, and for the CD4-positive cells concordance was 78%, indicating good reliability in these data.
FIG. 2.
CD4-positive cells (T lymphocytes) in ileal mucosa before and after L. reuteri ATCC 55730 administration. Biopsy specimens were taken from subjects at baseline and after the daily administration of L. reuteri ATCC 55730 at 4 × 108 CFU/day for 28 days, and the histological sections were stained for CD4+ cells. (A) Before L. reuteri ATCC 55730 administration (“dispersed cells or aggregations of cells”); (B) After L. reuteri ATCC 55730 administration (“several adjoining groups of cells” [arrow]).
Blood parameters were normal and no systematic changes were observed following supplementation with L. reuteri ATCC 55730 (results not shown). There were no significant differences in symptom scores due to L. reuteri supplementation except for a significantly higher flatus score in the gastroscopy group (results not shown).
DISCUSSION
Administration of L. reuteri ATCC 55730 at a dose of 4 × 108 CFU/day was well tolerated by both healthy individuals and subjects with an end ileostomy. Although the gastroscopy group did report an increase in flatus, these findings might be explained by a sudden greater awareness of symptoms in this group induced by the questionnaire. The lack of symptom reporting in the ileoscopy subjects, who are accustomed to registering their gastrointestinal symptoms, supports this idea. It should be emphasized that it is difficult to evaluate symptoms in an open study design.
A significant increase in live L. reuteri in feces after intake is a traditional indication of colonization (17, 20, 21), and this was also seen in our study. Surprisingly, we could detect L. reuteri in the feces of only six of the nine subjects with ileostomy after supplementation with the probiotic. Normally these subjects have smaller amounts of fecal bacteria since transit time is short (with little time for microbial multiplication) while the distribution volume is large. Thus, L. reuteri may have been present in their feces but in an undetectable (diluted) amount. The fecal L. reuteri recovered in random samples after supplementation was found to be genetically very similar to L. reuteri ATCC 55730, providing strong evidence that this was the colonizing strain in our subjects.
Using plating techniques, endogenous L. reuteri was detected in the feces of only 2 of the 19 (10.5%) subjects at the start of the study, while using FISH detection on biopsy specimens, baseline colonization was seen in 6 of 16 (37.5%) of the subjects. Thus, not only is the fecal plating method a very poor method to detect endogenous L. reuteri levels in the human gastrointestinal tract, but it also gives no information on the site of colonization. Fecal colonization is thus probably best used only as marker of compliance.
Detection of live bacteria in human biopsy specimens is difficult due to the practical issues of obtaining biopsy specimens, performing immediate analyses, and the lack of sensitivity of the plating methods (15), and our experience confirms these difficulties. However, others (6) have successfully studied colonization by lactobacilli of jejunal and rectal biopsy samples in 12 volunteers given a mixture of 19 different strains of lactobacilli (including L. reuteri at 5 × 108 CFU/day) for 10 days. L. reuteri (strain 108) was found in both the rectal and jejunal biopsy specimens after supplementation.
Our data, obtained by FISH, provide the first clear and direct evidence of colonization of the human stomach, duodenum, and ileum by L. reuteri. The appearance of L. reuteri lactobacilli at these sites after exogenous delivery of L. reuteri ATCC 55730 supports the idea that they come from the supplement. The demonstration that L. reuteri colonizes the stomach and duodenum combined with recent data that L. reuteri ATCC 55730 is a potent inhibitor of H. pylori growth (C. Johnson, H. Jonsson, and S. Roos, Abstr. 16th Int. Workshop Gastrointest. Pathol. Helicobacter, abstr. 16.33, p. 473, 2003) encourages further investigation of this bacteria on H. pylori infections at these sites in humans.
Since we generally failed to recover live (culturable) bacteria from the biopsy specimens and since it is true that FISH can detect dead cells, we cannot be sure that the cells detected were living at the time of the biopsy. However, the sensitivity of the technique (using a molecular probe that anneals to bacterial RNA) is high when cells are growing (since the RNA comprises up to 80% of the total nucleic acid) and fades in dead cells. The L. reuteri cells on the biopsy specimens showed a high level of fluorescence, suggesting that they are growing, live cells. Further, there is a clear correlation between the appearance of L. reuteri on the biopsy specimens and shedding of live L. reuteri in the feces of the subjects after supplementation with the probiotic. Thus, there is no real reason to believe that the L. reuteri cells die and then “stick” to the mucosa in transit. Finally, higher magnifications of the FISH images reveal colony-like formations of the stained cells, suggesting growth at the time of biopsy. Thus, it is highly likely that the L. reuteri lactobacilli detected on the biopsy samples are snapshots of live and growing bacteria in situ on the mucosa.
New emerging evidence suggests that L. reuteri may be able to modulate the immune system in the gastrointestinal tract (4, 7, 8, 18, 19). Our data support such a hypothesis and provide a first firm indication of an in situ effect of L. reuteri on the immune system in the human gut. We observed an immune cell response in the mucosal biopsy specimens, with a significant increase in CD4-positive cells in the ileum after the intake of L. reuteri ATCC 55730. This observation is in good agreement with earlier observations in poultry (3), where an increased CD4/CD8 ratio in the ileum mucosa was found after intake of L. reuteri (poultry strain) in a model where colonization by Salmonella enterica serovar Typhimurium is markedly reduced by such supplementation (2, 3). Furthermore, Mao et al. (10) studied methotrexate-induced enterocolitis in rats and found that L. reuteri R2LC could increase both ileal and colonic secretory immunoglobulin A levels as well as CD4+- and CD8+-cell populations in the gut lamina propria and that these changes were associated with decreased intestinal permeability, increased mucosal mass, and recovery from enterocolitis (9). It is also worth noting that Ferreira et al. (5) have shown that activated T lymphocytes in the human small intestinal lamina propria are involved in enhancing proliferation of intestinal epithelial cells and that lactobacilli (1), including L. reuteri ATCC 55730 (2), have been shown to stimulate ileum mucosal growth in animals. L. reuteri is known to be an indigenous species in the human ileum (14), and thus stimulation of T-helper cells by this bacterium may be a central mechanism of symbiosis for improving the health of the host gut and a key mechanism of action for this probiotic.
In conclusion, this study shows colonization of the human gastrointestinal tract by L. reuteri ATCC 55730 delivered in a tablet formulation and consequent modulation of local immune cell populations. It seems likely that this response to exogenous L. reuteri may be involved in maintaining gastrointestinal well-being and defense against pathogens in an already-healthy recipient.
Acknowledgments
We acknowledge the receipt of a research grant from BioGaia AB, Lund, Sweden, in support of this work.
N. Valeur, P. Engel, and K. Ladefoged have no conflict of interest with regard to the use of BioGaia's proprietary strain of L. reuteri.
REFERENCES
- 1.Banasaz, M., E. Norin, R. Holma, and T. Mitvedt. 2002. Increased enterocyte production in gnotobiotic rats mono-associated with Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 68:3031-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casas, I. A., and W. J. Dobrogosz. 2000. Validation of the probiotic concept: Lactobacillus reuteri confers broad-spectrum protection against disease in humans and animals. Microb. Ecol. Health Dis. 12:247-285. [Google Scholar]
- 3.Casas I. A., F. W. Edens, and W. J. Dobrogosz. 1997. Lactobacillus reuteri: an effective probiotic for poultry, other animals and humans, p. 475-518. In S. Salminen and A. von Wright (ed.), Lactic acid bacteria. Marcel Dekker, New York, N.Y.
- 4.Christensen, H. R., H. Frokiaer, and J. J. Pestka. 2002. Lactobacilli differentially modulate expression of cytokines and maturation surface markers in murine dendritic cells. J. Immunol. 168:171-178. [DOI] [PubMed] [Google Scholar]
- 4a.Dobrogosz, W., and S. E. Lindgren. October 1994. U.S. patent 5,352,586.
- 5.Ferreira, R., L. E. Forsyth, and P. L. Richman. 1990. Changes in the rate of crypt epithelial cell proliferation and mucosal morphology induced by a T-cell-mediated response in human small intestine. Gastroenterology 98:1255-1263. [DOI] [PubMed] [Google Scholar]
- 6.Johansson, M. L., G. Molin, B. Jeppsson, S. Nobaek, S. Ahrné, and S. Bengmark. 1993. Administration of different Lactobacillus strains in fermented oatmeal soup: in vivo colonization of human intestinal mucosa and effect on the indigenous flora. Appl. Environ. Microbiol. 59:15-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maasen, C. B. M., C. van Holten-Neelen, F. Balk, M. J. H. den Bak-Glashouer, R. J. Leer, J. D. Laman, W. J. A. Boersma, and E. Claassen. 2000. Strain-dependent induction of cytokine profiles in the gut by orally administered Lactobacillus strains. Vaccine 18:2613-2623. [DOI] [PubMed] [Google Scholar]
- 8.Madsen, K. L., J. S. Doyle, L. D. Jewell, M. M. Tavernini, and R. N. Fedorak. 1999. Lactobacilli species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology 116:1107-1114. [DOI] [PubMed] [Google Scholar]
- 9.Mao, Y., S. Nobaek, B. Kasravi, D. Adawi, U. Stenram, G. Molin, and B. Jeppsson. 1996. The effects of Lactobacillus strains and oat fibre on methotrexate-induced enterocolitis in rats. Gastroenterology 111:334-344. [DOI] [PubMed] [Google Scholar]
- 10.Mao, Y., J. L. Yu, A. Lungh, G. Molin, and B. Jeppsson. 1996. Intestinal immune response to oral administration of Lactobacillus reuteri R2LC, Lactobacillus plantarum DSM 9843, pectin and oatbase on methotrexate-induced enterocolitis in rats. Microb. Ecol. Health Dis. 9:261-270. [Google Scholar]
- 11.Mitsuoka, T. 1992. The human gastrointestinal tract, p. 69-114. In B. J. B. Wood, (ed.), The lactic acid bacteria, vol. 1. The lactic acid bacteria in health and disease. Elsevier Science Publ. Ltd., Essex, England.
- 12.Ouwehand, A. C., H. Lagstrom, T. Suomalainen, and S. Salminen. 2002. Effect of probiotics on constipation, fecal azoreductase activity and fecal mucin content in the elderly. Ann. Nutr. Metab. 46:159-162. [DOI] [PubMed] [Google Scholar]
- 13.Reuter, G. 1965. Das Vorkommen von Laktobazillen in Lebensmitteln und ihr Verhalten im menschlichen Intestinaltrakt. Zentbl. Bakteriol. Parasitol. Infekt. Hyg. I Orig. 197S:468-487. [Google Scholar]
- 14.Reuter, G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2:43-53. [PubMed] [Google Scholar]
- 15.Rüssmann, H., V. A. J. Kempf, S. Koletzko, J. Heesemann, and I. B. Autenrieth. 2001. Comparison of fluorescent in situ hybridization and conventional culturing for detection of Helicobacter pylori in gastric biopsy specimenes. J. Clin. Microbiol. 39:30-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shornikova, A., I. A. Casas, H. Mykkanen, E. Salo, and T. Vesikari. 1997. Bacteriotherapy with Lactobacillus reuteri in rotavirus gastroenteritis. Pediatr. Infect. Dis. J. 6:1103-1107. [DOI] [PubMed] [Google Scholar]
- 17.Shornikova, A., E. Isolauri, I. A. Casas, H. Mykkanen, and T. Vesikari. 1997. Lactobacillus reuteri as a therapeutic agent in acute diarrhoea in young children. J. Pediatr. Gastroenterol Nutr. 24:399-404. [DOI] [PubMed] [Google Scholar]
- 18.Tejada-Simon, M. V., and J. J. Pestka. 1999. Proinflammatory cytokine and nitric oxide induction in murine macrophages by cell wall and cytoplasmic extracts of lactic acid bacteria. J. Food Prot. 62:1435-1444. [DOI] [PubMed] [Google Scholar]
- 19.Tejada-Simon, M. V., Z. Ustanol, and J. J. Pestka. 1999. Ex vivo effects of lactobacilli, streptococci and bifidobacteria ingestion on cytokine and nitric oxide production in a murine model. J. Food Protect. 62:162-169. [DOI] [PubMed] [Google Scholar]
- 20.Wolf, B. W., K. A. Garleb, D. G. Ataya, and I. A. Casas. 1995. Safety and tolerance of Lactobacillus reuteri in healthy adult subjects. Microb. Ecol. Health Dis. 8:41-50. [Google Scholar]
- 21.Wolf, B. W., K. B. Wheeler, D. G. Ataya, and K. A. Garleb. 1998. Safety and tolerance of Lactobacillus reuteri supplementation to a population infected with the human immunodeficiency virus. Food Chem. Toxicol. 36:1085-1094. [DOI] [PubMed] [Google Scholar]